Abstract
Auditing biomedical terminologies often results in the identification of inconsistencies and thus helps to improve their quality. In this paper, we present a method based on Semantic Web technologies for auditing biomedical terminologies and apply it to the NCI thesaurus. We stored the NCI thesaurus concepts and their properties in an RDF triple store. By querying this store, we assessed the consistency of both hierarchical and associative relations from the NCI thesaurus among themselves and with corresponding relations in the UMLS Semantic Network. We show that the consistency is better for associative relations than for hierarchical relations. Causes for inconsistency and benefits from using Semantic Web technologies for auditing purposes are discussed.
Introduction
Over the past years, many studies have audited biomedical terminologies. It has been shown that terminologies often present inconsistencies, in particular in the semantic categorization of concepts [1], but also among the hierarchical relations [2]. One study analyzes the quality of the NCI thesaurus from a formal point of view and investigated its compliance with ontological principles [3]. Like [1], we investigate the consistency between relations asserted in a terminology and corresponding relations in the UMLS Semantic Network, with application to the NCI thesaurus as in [3]. In contrast to these studies, we leverage Semantic Web technologies for auditing purposes.
The objective of the Semantic Web [4] is to facilitate data exchange and to represent information formally so that machines can process it automatically. Semantic Web technologies, such as metadata, languages, and Web services, are the means to this end. RDF1 (Resource Definition Framework) is a notation developed by the W3C (World Wide Web Consortium), the main international standards organization for the World Wide Web. RDF enables the representation of resources in the form of subject-predicate-object expressions, called triples in RDF parlance. RDF has been used widely for representing terminologies, as well as for knowledge sharing and integration [5]. OWL2 (Web Ontology Language) is a W3C standard for representing ontologies.
The objective of this work is to assess the consistency of both hierarchical and associative relations from the NCI thesaurus among themselves and with corresponding relations in the UMLS Semantic Network. We illustrate the benefit of using Semantic Web technologies for auditing purposes by showing that simple queries to an RDF semantic store help identify errors.
Resources
The National Cancer Institute thesaurus (NCIt) is a biomedical terminology that provides broad coverage of the cancer domain [6]. It is distributed as a component of the NCI Center for Bioinformatics ca-CORE. It provides vocabulary for various domains related to cancer research. Its rich network of semantic relations supports the integration of information. The NCIt contains nearly 60,000 concepts. In this study, we used the latest version of the NCIt (2007_05E) available in the most recent version of the UMLS. In practice, we used the OWL representation of the NCIt.
The Unified Medical Language System® (UMLS®) [7] includes two sources of semantic information: the Metathesaurus® and the Semantic Network. The UMLS Metathesaurus is assembled by integrating close to 150 sources vocabularies. It contains about 1.5 million concepts and more than 32 million relations among these concepts. There are some 7 million hierarchical relations in the Metathesaurus. The Semantic Network (SN) is a much smaller network of 135 semantic types organized in a tree structure. The semantic types have been aggregated into fifteen coarser semantic groups [8], which represent subdomains of biomedicine (e.g., Anatomy, Disorders). Each Metathesaurus concept is assigned at least one semantic type from the SN, independently of its hierarchical position in a source vocabulary. Version 2007AC of the UMLS is used in this study.
Methods
The method developed for auditing the NCIt can be summarized as follows. We first converted NCIt data into RDF triples. We complemented the triple store with information from the UMLS, especially semantic types (STs) and semantic groups (SGs). Then, we queried the triple store to check the consistency of hierarchical and associative relations among NCIt concepts.
Defining consistency
A relation (c1 rnci c2) involving a relationship rnci between two NCIt concepts c1 and c2 is deemed consistent if rnci is equivalent to or a sub-property of the relationship rsn asserted between the STs st1 and st2 categorizing c1 and c2, respectively. If these concepts are assigned more than one ST, the relation between c1 and c2 must be consistent with every relation asserted between the STs categorizing c1 and c2. Completeness and accuracy of the categorization is assumed.
Creating the triple store
For each NCIt concept, we extracted from the OWL file its code, its preferred name, its ST(s), and its parent(s). As mentioned earlier, RDF triples comprise a subject, a predicate, and an object. Here, the subject is the concept under investigation. The relations in which this concept participates provide the predicate (relationship) and the object (related concept). The following properties are used as predicates in our triple store: hasName, hasNCIST, and subClassOf.
Information from the UMLS was used to complement the triple store. For each concept, we first recovered the Concept Unique Identifier (CUI) associated with the NCIt code. With the CUI, we could obtain the UMLS ST(s) categorizing the concept. We integrated the CUI and the ST(s) through the predicates hasID and hasUMLSST, respectively. The associative relations existing among NCIt concepts were also recovered from the UMLS Metathesaurus. Although the asserted relations are also represented in the OWL file, the UMLS offers the advantage of representing both asserted and inferred relations. Finally, the UMLS STs and SGs, ST relationships, and relations (direct and inferred) existing among STs were integrated into the triple store.
The Mulgara™ semantic store3 is an Open Source, scalable RDF database. We chose it to store the triples created from the NCIt and UMLS.
Hierarchical relationships
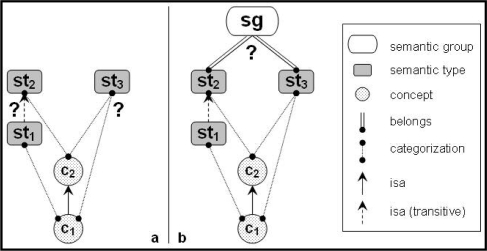
Hierarchical relations from the NCIt were examined both at the semantic type (ST) and the semantic group (SG) level. For a pair (c1, c2) of hierarchically related concepts, where c1 is the child of c2, we examined the STs of these concepts, st1 and st2, respectively, and checked whether st1 is the same as or a descendant of st2, direct or not. When the concepts are assigned multiple STs, the STs categorizing c1 must all be same as or descendants (direct or not) of at least one ST categorizing c2 (Figure 1.a). At the SG level, we simply checked whether the STs categorizing c1 and c2 belong to the same SG (Figure 1.b).
Figure 1.
Auditing hierarchical relationships a) at the semantic type level; b) at the semantic group level.
Interestingly, two categorizations are available for each NCIt concept, one coming from the NCIt, the other from the UMLS. We applied this method separately to each source of STs.
Associative relationships
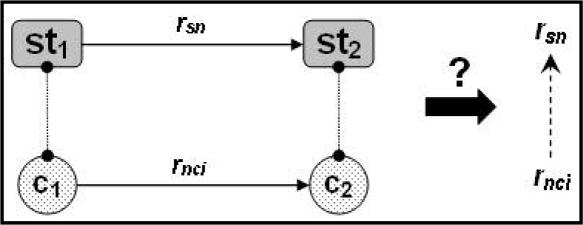
In order to evaluate the consistency of associative relations in the NCIt, we compared the relationship existing between two concepts with the relationship between their corresponding ST(s) (Figure 2). Such comparison requires a correspondence between NCIt relationships and ST relationships. As no authoritative mapping among relations is available, we established such a mapping for a subset of all NCIt relationships. In practice, we selected 19 relationships linking concepts of the “Disease” kind to other concepts. We primarily exploited the domain and range of each NCIt relationship, which we compared to STs. When a match could be found in the SN, we used the relationship(s) existing between the corresponding STs (or, if necessary, the inverse relationship). For each relationship, we then examined several pairs of concepts to assess our mapping. At the end of this process, each NCIt relationship was mapped to (i.e., equivalent to or a subproperty of) some ST relationship.
Figure 2.
Auditing associative relationships.
The consistency of each NCIt relation rnci between two concepts c1 and c2 was assessed as follows. The STs st1 and st2 categorize c1 and c2, respectively, and are linked by the relationship rsn. In this context, the relationship rnci is expected to be equivalent to or a sub-property of rsn for the NCIt relation to be consistent semantically with the ST relationship.
Inference rules and queries
One benefit of using a triple store, compared to a relational database, is the availability of support for subsumption reasoning. We created three inference rules to infer new knowledge (in practice, new triples) from the existing store. The first rule asserts the reflexivity and transitivity of the subPropertyOf relationship defined between NCIt associative relationships. More precisely, given an associative relationship ra, reflexivity implies that ra sub-PropertyOf ra is true and transitivity guarantees that if ra subPropertyOf rb and rb subPropertyOf rc, then ra subProper-tyOf rc. Another rule implements the reflexivity of sub-PropertyOf between ST relationships. The last rule implements the disjointness among SGs by creating an isDisjoint relationship between each SG and the 14 others. The auditing was implemented, not through ad hoc programs, but through simple queries against the triple store. Some of these queries take advantage of the triples generated by the inference rules.
Results
Triple store content
A total of 785,408 triples were loaded in the triple store (4 minutes). An example of some triples found in the triple store when querying for the concept Melanomatosis (C9499) is displayed in Figure 3 (Mulgara presentation).
Figure 3.
Triples obtained for the concept C9499.
The application of inference rules generated 368 additional triples. More precisely, 68 triples were created through the rule defining the transitivity and reflexivity of subPropertyOf for NCIt associative relationships. 90 triples were added through the rule implementing the reflexivity of subPropertyOf for ST relationships. In addition, 210 triples were generated by the rule asserting the disjunction of SGs. Overall, our triple store contains 785,776 triples.
Hierarchical relations
Out of the 58,843 NCIt concepts, all of them are categorized by at least one UMLS ST and 58,657 concepts (99.7%) are assigned at least one NCIt ST. As expected, the consistency is better at the SG level, which represents a higher level of abstraction. The categorization provided by the UMLS reveals more inconsistencies than that provided by the NCIt (30% vs. 20%) at the ST level. In contrast, the results are roughly similar at the SG level (11 vs. 12 %). Detailed results are given Table 1.
Table 1.
Number of inconsistent hierarchical relations at semantic type and semantic group levels. Results are given for UMLS and NCIt categorizations.
| Source | Level | Incompatible | Total | |
|---|---|---|---|---|
| UMLS | ST | 17,480 | 30% | 58,843 |
| SG | 6,656 | 11% | ||
| NCIt | ST | 11,624 | 20% | 58,657 |
| SG | 7,222 | 12% | ||
An example of inconsistent relation is the concept Salivary Gland Fistula which is a child of Gastrointestinal Fistula Adverse Event, categorized by Anatomical Abnormality and Finding, respectively. As the ST Anatomical Abnormality is neither a child, nor a descendant of Finding, the relation between the two concepts is inconsistent at the ST level. The relation is however consistent at the SG level since both STs belong to the SG Disorders.
Associative relations
Overall, the consistency of associative relations in the NCIt is good. Results are nearly perfect when the NCIt categorization is used, while the UMLS reveals few inconsistencies. Some of the results are given in Table 2. For each relationship, its frequency and the number of inconsistent relations in which it participates are displayed. The table also compares the results obtained with STs assigned in the NCIt and the UMLS.
Table 2.
Number of relations whose concepts are categorized by the UMLS and the number of inconsistent relations among all. The same results are given for NCIt categorization.
| NCIt relationship | SN relationship | # relations UMLS | Incompatible UMLS | # relations NCIt | Incompatible NCIt |
|---|---|---|---|---|---|
| gene_associated_with_disease | location_of | 1,017 | 0 | 981 | 0 |
| disease_has_abnormal_cell | has_location | 7,085 | 8 | 7,095 | 7 |
| disease_has_finding | has_manifestation | 5,960 | 39 | 5,959 | 2 |
| disease_has_normal_cell_origin | has_location | 5,913 | 0 | 5,821 | 0 |
| disease_has_associated_anatomic_site | has_location / produces | 6,208 | 301 | 6,184 | 0 |
| disease_has_primary_anatomic_site | has_location / produces | 4,831 | 250 | 4,552 | 0 |
| eo_disease_has_associated_eo_anatomy | has_location / produces | 1,313 | 0 | 1,324 | 0 |
| regimen_has_accepted_use_for_disease | treats / result_of | 229 | 0 | 171 | 0 |
| disease_is_stage | has_result | Ø | Ø | 1,043 | 0 |
| disease_is_grade | has_evaluation | Ø | Ø | Ø | Ø |
| eo_disease_has_property_or_attribute | has_property | Ø | Ø | Ø | Ø |
For eight relationships, the consistency was perfect whether the categorization came from the UMLS or the NCIt. For two NCIt relationships, no relation existed between st1 and st2, preventing the consistency from being assessed.
The relationship disease_is_stage provided results only with NCIt STs. This can be explained by the fact that the UMLS does not categorize the target concepts (disease stages) in the same way as the NCIt does. Indeed, the NCIt assigned these concepts the ST Clinical Attribute whereas the UMLS categorized them as Intellectual Product. The original concepts are of type Disease or Syndrome, which has a relationship with the NCIt ST but not with the UMLS ST.
For two relationships, very few inconsistencies were identified using the NCIt categorization. The analysis of the nine cases reveals that these inconsistencies resulted from wrong NCIt categorization. For instance, the concept Balloon Cell Nevus, categorized as Neoplastic Process, is linked to the concept Balloon Nevus Cell through the relationship dis-ease_has_abnormal_cell. Balloon Nevus Cell should have been assigned the ST Cell. Instead, the NCIt categorized it as a Neoplastic Process. A reflexive relationship exists for Neoplastic Process but this relationship is not an ancestor (direct or not) of has_location, the relationship to which disease_has_ abnormal_cell is mapped.
For the six remaining relationships, only the UMLS categorization exhibits inconsistencies. These errors were too many to check individually but are probably also the result of inappropriate categorization or missing relations in the SN. For example, the relation Chronic Idiopathic Myelofibrosis disease_has_associated_anatomic_site Hematopoietic and Lymphatic System is inconsistent since the source concept is categorized as Neoplastic Process, whereas the target concept is assigned the ST Body System, which is unrelated to Neoplastic Process.
Discussion
Categorization issues
As expected, the NCIt categorization exhibits better results than those obtained with the UMLS categorization. This can be explained by the fact that concepts and the NCIt STs come from the same source. Moreover, consistency in the NCIt may be enforced by the editing tools. Like the concept structure, categorization in the UMLS may differ from that asserted in the biomedical sources it integrates. In fact, the UMLS editors assign STs to concepts corresponding to synonymous terms from several sources that may provide slightly different contexts. Conversely, codes that are distinct in a given terminology may be aggregated into a unique UMLS concept, which can result in slightly different categorizations. For instance, the concepts Hard Palate (C12230) and Malignant Hard Palate Neoplasm (C3528) are integrated in the UMLS concept C0153375, whose ST is Neoplastic Process. In the NCIt, C3528 is assigned the ST Neoplastic Process, whereas C12230 is categorized by the ST Body Part, Organ, or Organ Component.
As shown with associative relations, concept categorization is not always accurate, which is a source of confusion and misinterpretation of the knowledge represented in the NCIt. Our approach could thus be useful for biomedical terminology developers in order to improve the quality of their data.
Relaxing consistency
In this study, we adopted a strict definition of inconsistency for auditing hierarchical relations. Many concepts are categorized by more than one ST. We considered that a relation between c1 and c2 was inconsistent if at least one of the STs of c1 was not the same as or a descendant (direct or not) of the ST(s) of c2. A more relaxed definition of inconsistency would assess consistency if at least one ST of c1 is compatible with at least one ST of c2. Following this definition, 46,963 relations are consistent according to the UMLS categorization at the ST level, decreasing the percentage of inconsistency by one third (from 30% to 20%).
Limitations
This work is incomplete as we audited only a subset of the associative relations in the NCIt. We focused on a specific category (“Disease”), in order to test our approach. This is a limitation of our study since we may not have encountered all possible issues with NCIt relationships. In future work, we plan to complete the mapping we created between NCIt and ST relationships.
This study revealed a small number of inconsistencies involving associative relations. However, the methodology followed to establish the mapping could be questioned. We mainly relied on the domain and range defined in the NCIt OWL file for each of its relationships and chose a ST relationship suitable for linking the corresponding kinds. We did so even if none of the existing relationships in the SN really corresponded. For instance, the relationship eo_disease_maps_to_human_disease has Disease as domain and range. We thus selected one of the relationships linking Disease or Syndrome, e.g., co-occurs_with, although it would have been better to simply make this relationship a subproperty of conceptually_related_to. This choice, however, would have been inconsistent with NCIt and UMLS categorizations. In [9], authors propose to adapt and complement the ST relationships with NCIt relationships. Our work could benefit from and complement this previous study.
Semantic Web technologies
One significant difference between our approach and that of previous studies [1,2] is that no ad hoc programming was required to carry out this work. Indeed, we only had to define simple queries against an RDF triple store in order to check the consistency of NCIt relations. For performance reasons, some queries had to be “grounded” (i.e., anchored with the particular concept under investigation) and run for each concept.
Since the NCIt was available in OWL, we could have enriched its representation with UMLS categorization and converted the UMLS SN into OWL. OWL reasoners could have been used to perform the kinds of queries we ran against the RDF store. Our rationale for using RDF instead of OWL is essentially that the performance of OWL reasoners is still limited, especially with large terminological structures such as the NCIt. While current reasoners could handle the 60,000 classes of the NCIt, this method would not be applicable to larger terminologies, e.g., SNOMED Clinical Terms (310,000 classes). Approaches based on RDF guarantee that virtually any biomedical terminology can be audited using this method, provided that it is available in (or amenable to transformation into) RDF and integrated in the UMLS.
In conclusion, we audited hierarchical and associative relations asserted between concepts in the NCIt according to the relations defined in the UMLS Semantic Network between the STs categorizing these concepts. We showed that the consistency is limited for hierarchical relations (20–30% inconsistency), but better for associative relations. The use of Semantic Web technologies largely facilitated the auditing, especially by eliminating the need for ad hoc programming.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM).
Footnotes
References
- 1.McCray AT, Bodenreider O. A conceptual framework for the biomedical domain. In: Green R, Bean CA, Myaeng SH, editors. The semantics of relationships: an interdisciplinary perspective. Dordrecht; Boston: Kluwer Academic Publishers; 2002. pp. 181–98. [Google Scholar]
- 2.Cimino JJ, Min H, Perl Y. Consistency across the hierarchies of the UMLS Semantic Network and Metathesaurus. Journal of Biomedical Informatics. 2003;36(6):450–61. doi: 10.1016/j.jbi.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI Thesaurus. Methods Inf Med. 2005;44(4):498–507. [PubMed] [Google Scholar]
- 4.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Scientific Am. 2001 May;:34–43. [Google Scholar]
- 5.Nardon FB, Moura LA. Knowledge sharing and information integration in healthcare using ontologies and deductive databases. Medinfo. 2004;11:62–6. [PubMed] [Google Scholar]
- 6.Hartel FW, De Coronado S, Dionne R, Fragoso G, Golbeck J. Modeling a description logic vocabulary for cancer research. Journal of Biomedical Informatics. 2005;38(2):114–29. doi: 10.1016/j.jbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. Journal of Biomedical Informatics. 2003;36(6):414–32. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Coronado S, Tuttle MS, Solbrig HR. Using the UMLS Semantic Network to Validate NCI Thesaurus Structure and Analyze its Alignment with the OBO Relations Ontology. Proc AMIA Symp. 2007:165–9. [PMC free article] [PubMed] [Google Scholar]