Abstract
MorphoSaurus, a concept-based document search engine, was incorporated into an EHR system in order to support search across the whole corpus of patient discharge letters and other clinically relevant documents. A user survey showed a general satisfaction with the system and revealed novel usages for information stored in discharge letters. The retrieval system was also used to identify relevant documents for a five-year retrospective survey of suspicious syphilis cases in the department. This retrieval scenario was used to assess the performance of MorphoSaurus against a manually created gold standard. A substring search for the German words “syphilis” and “lues” was used as baseline. The system yielded a precision p = 20.1% and a recall r = 100%. The values for the substring “syphilis” were p = 65.5% and r = 47.5%, for “lues” p = 15.4% and r = 87.7%. The results support the use of the proposed recall-oriented search across EHR documents to acquire valid and complete data for epidemiology studies in hospital populations.
INTRODUCTION
Traditionally, electronic health record (EHR) systems provide a patient or case centered view on data and documents. Although highly desired by clinicians, data aggregation to detect inter-patient dependencies is not a standard functionality of common EHR systems [1] in Germany. Such aggregations are rather reserved to hospital administrators for the purpose of business statistics, or clinical epidemiologists for assessing the distribution of diseases in an institution. These services, however, depend on coded data that represent only small parts of the patient’s information. Furthermore they often lack quality and completeness for medical questions.
The retrieval of cases that are relevant for retrospective studies or for the recruitment of patients for clinical trials therefore tends to be a time consuming and error-prone endeavor if some or all relevant criteria are only available in free text and not in a structured form. Thus, reliable and efficient methods to extract structured information from clinical documents using information extraction techniques may greatly enhance this process.
The usefulness of such tools, however, goes beyond the support of research. Integrated into a medical workplace, they can also help physicians retrieve past, related, or similar cases by free text search, a technique that has become an integral part of today's life due to the omnipresence of the Web. Finally the retrieval of “similar cases” may be of a great didactic and heuristic value for residents or medical students. Traditionally, in German hospitals, discharge letters are considered very valuable, as they summarize all aspects of a clinical case. They are not only produced at the discharge of in-patients but are also commonly produced to give a summary account of one or more ambulatory episodes. It is common practice to look first at these documents in order to get a general idea of a new patient’s past history.
Dealing with medical language, document retrieval and information extraction is challenged by several factors. Especially in Germanic languages, medical terms often exhibit complex forms of composition, derivation and inflection, as well as the continuous generation of new acronyms, abbreviations and proper names. Moreover, spelling and syntax rules are not always respected. Due to these peculiarities, current information retrieval approaches that are usually based on simple comparison of entire or automatically stemmed words are inappropriate because they produce results that are incomplete, inaccurate, or outside the desired scope [2,3,4,5], at least for morphologically rich languages such as German, a language that provides a remarkable wealth of synonyms, inflections and spelling variants (see Table 1 for an incomplete selection of terms for colon cancer.)
Table 1.
Term Variation in German
| Kolon-Ca | Dickdarmkarzinom |
| Colon-Ca | Dickdarm-CA |
| Kolonkarzinom | Karzinom des Dickdarms |
| Koloncarcinoms | Dickdarmkrebs |
| Coloncarcinomen | Kolonkrebs |
| Colonkarzinomen | Kolonkrebses |
| Npl des Dickdarms | Büsartiger Dickdarmtumor |
These language idiosyncrasies constitute a big obstacle for an effective document retrieval or information extraction system in an EHR context. In contradistinction to the retrieval of documents or text messages from a Web search, where – due to the overwhelming amount of data – a precision-oriented approach offers advantages over a recall oriented approach, the latter one is more acknowledged in a EHR setting where a “needle in a haystack” scenario is much more common, especially when documents with rare features are targeted.
In this article we will report on a pilot implementation of a full-text document retrieval system in a clinical environment. We will support the usefulness of this system by three kinds of evidence:
Assessment of the users’ satisfaction with the system measured by a survey [6];
Participative observation focusing on the integration of the search engine in their daily practice; and
A case study on a clinical-epidemiological research question.
MATERIAL AND METHODS
The core technology of the EHR document retrieval prototype is based on morphologic analysis and semantic indexing of both queries and documents. The MorphoSaurus system [7] uses subwords as lexical units. A subword is defined as a minimum lexical unit for a meaningful term in a domain. This premise implies that the meaning cannot be anymore split. Thus, we can consider the term “Kolonkarzinom” as a composition of two subwords, “kolon” and “karzinom” because the meaning results from the meaning of the two constituents. However, the delineation of subwords (in contrast to morphemes) is semantically determined and follows the principle of semantic compositionality. E.g., as the meaning of “hypophysis” cannot be derived from “hypo” + “physis”, “hypophys-” is included into the lexicon. In MorphoSaurus each subword entry is classified according to attributes such as language (English, German, French, Spanish, Portuguese, Swedish, and Italian) and type (stem, prefix, suffix, invariant). The semantic layer of MorphoSaurus is represented by subword equivalence classes, identified by so called MIDs (MorphoSaurus identifiers). Each lexical entry is associated with exactly one equivalence class. Equivalence classes group lexical variants, synonyms, and translations. The delimitation of semantic classes is a laborintensive task that requires considerable knowledge of the domain terminology.
The lexical resources of MorphoSaurus were mainly created by medical experts. Semi-automated techniques were applied to bootstrap the resources in several languages [8]. Currently, over 100,000 lexical entries exist. MorphoSaurus assures a high performance extraction of subwords and their mapping by using finite-state techniques for lexiconbased decomposition, derivation and deflection such as described in [9]. Furthermore, the MorphoSaurus system incorporates the following features:
Assignment of a sequence of MIDs to certain semantically composed lexemes, e.g. #urinalys → #urin #analys (due to the contraction of the word “unrinalysis”)
Mappings between MID sequences to handle synonymy / translation at a multi word level, e.g. #mucus #viscous #disorder → #cyst #fibro;
Context-based sense disambiguation [10]; and
Acronym detection and disambiguation [11].
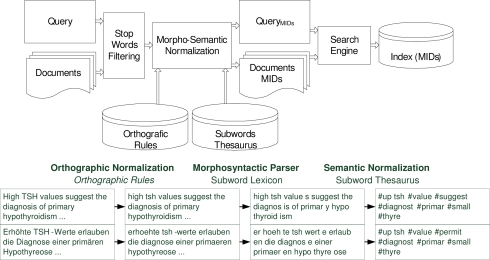
Figure 1 (top) depicts the insertion of the MorphoSaurus engine into a document retrieval scenario and the step-wise abstraction from free text to a sequence of semantic identifiers (bottom). Firstly, the input is normalized to 7-bit ASCCII, and then the words are split into subword fragments using a finite-state automaton driven by the lexicon. Finally, each meaning-bearing subword is substituted, according to the semantic layer of MorphoSaurus: (i) by one MID, (ii) by a sequence of MIDs, or (iii) by a group of MIDs representing different meanings.
Figure 1.
MorphoSaurus Processing Architecture (top), Morpho-Semantic Normalization pipeline (bottom):
In a pilot study, the MorphoSaurus search engine was used for supporting semantically enhanced document retrieval in the Department of Dermatology of the University Medical Center Freiburg. A simple and intuitive search interface (Figure 2) allows a full text search in the whole corpus of about 30,000 dermatological discharge letters, surgical reports and immuno-dermatologic findings. A database of images was also included into the scope.
Figure 2.
Search engine for Discharge Summaries
Up to now, about 35 physicians and medical students used the MorphoSaurus interface in their daily routine. As the system should show the clinical impact of biomedical data mining, an evaluation engine was implemented. Users were intermittently encouraged by an online user survey to assess the value of the system and its further potential for their work. Search terms that were applied by the users were recorded anonymously. Three different major topics (clinical, scientific, educational impact) were assessed, using a structured questionnaire with open free-text comments. In a participative observation, the last author (MM) was observing the usage on-site. He conducted informal interviews in order to elicit the users’ experiences and to collect their suggestions for a possible enhancement of this tool.
To demonstrate the potential of the system, a retrospective clinical-epidemiological study targeting cases of suspected or manifest syphilis has been conducted. Parameters of interest were prevalence, age distribution, population groups, signs, symptoms, HIV co-incidence and guideline compliance. The identification of a complete set of relevant cases was the main challenge in this study – traditional database-driven retrieval techniques scanning for distinctive diagnosis codes reveal a small amount of cases and are not very reliable. We used this retrieval task to assess the potential of the proposed retrieval technology comparing three different approaches.
- Gold standard:Search using alternative expressions and manual document checking of the documents retrieved. To this end, an experienced dermatologist identified the following substrings, at least one of which was supposed to occur in each syphilis-relevant document using substring match (not respecting word boundaries).
*lues* *fta abs* *luet* *schanker* *syphil* *pallidum* *fta-abs* Baseline:Substring search across the whole document corpus, using the German word stems “syphilis” and alternatively “lues”.
MorphoSaurus: Searching the documents by simply entering “syphilis” or “lues” into the SuitSearch interface. This corresponds to the expected behavior of common users. The underlying assumption is that synonymous terms are mapped to the same semantic ID.
The effectiveness of both baseline and MorphoSaurus queries was measured against the gold standard using standard information retrieval measures (Table 2).
Table 2.
Measures for document retrieval effectiveness
| Relevant documents (according to gold standard) | Non relevant documents (according to gold standard) | |
|---|---|---|
| Documents retrieved | A | B |
| Documents not retrieved | C | D |
| Precision | A / (A+B) | |
| Recall | A / (A + C) | |
| F-Score | 2 * (Precision * Recall) / (Precision + Recall) | |
RESULTS
User acceptance
The users’ acceptance of the interface was remarkably high and enthusiastic from the beginning: doctors were very happy to have a highly convenient access to ten thousands of documents with the possibility to find and browse independently of the individual patient record. This new route to clinical data has given rise to several retrospective surveys that measure the clinical outcome of certain therapies. Typical clinical use scenarios e.g. comprised searching for cases with similar diagnosis/treatments to a current case, selecting patients that got a certain treatment (like immunostimulatory drugs in an oncological setting) and aggregation of cases with symptom complexes in order to detect clinically relevant syndromes. For educational purposes, typical clinical cases were extracted and presented to medical students. Furthermore, some doctors created new use scenarios, such as searching good discharge letters as blueprints for new letters they had to create.
User survey
The survey revealed 21 user evaluation data sets (16 physicians, 3 students, 2 information specialists). 81% stated that the system could enhance their clinical performance. 90% thought that this kind of biomedical data mining and the integration of narrative text with dermatological images have a very positive impact on their scientific work. The impact on dermatologic education has been less well-accepted, only 52% the users saw a potential benefit. The reason was that they generally wished the integration of more clinical data, like radiology images and reports, laboratory results and other clinical findings.
Document filtering
The document filtering by substring match identified 226 documents out of a total corpus of about 30,000 documents. This corpus contained not only discharge letters but to a minor extent also findings reports and surgery reports. A careful manual check of this selection finally yielded 40 relevant documents that belonged to 31 patients. Using this gold standard we get the following results for four different retrieval scenarios.
The first one (“Substring match”) refers to the match of the documents against selected substrings or regular expressions – as specified above – but without a manual relevance check. This result would correspond to a scenario in which a careful compilation of all relevant terms or term fragments is done prior to the retrieval.
The query includes all documents but is very nonspecific (Table 3). The set of false positives also included one case in which the substring “*lues*” matched a patient name.
Table 3.
Search for *lues* OR *luet* OR *syphil* OR *fta-abs* OR *fta abs* OR *schanker* OR *pallidum*
| Optimized substring match | Relevant documents | Non relevant documents |
|---|---|---|
| Documents retrieved | 40 | 186 |
| Documents not retrieved | 0 | 29,774 |
| Precision | 17.7% | |
| Recall | 100.0 % | |
| F-Score | 30.0% | |
| Query preparation | Laborious | |
The second and the third scenario (“Token match”) refer to the match of the documents against one query token, corresponding to the common Web search engine scenario. It is interesting that there is a high dependence on the word chosen. Neither query retrievs all documents. Whereas the “syphilis” query is quite selective, the “lues” query is rather nonspecific with 19 times more false positive hits. In our example this is easily explained by the fact that the expression “Lues-Serologie” is common in many non relevant documents, as this lab test is rather common.1 Although the “syphilis” query has the highest F-Score, it cannot be recommended, since more than half of relevant documents are not found (Table 4).
Table 4.
Search with simple token match
| Token match “syphilis” | Relevant documents | Non relevant documents |
|---|---|---|
| Documents retrieved | 19 | 10 |
| Documents not retrieved | 21 | 29950 |
| Precision | 65.5 % | |
| Recall | 47.5 % | |
| F-Score | 55.1 % | |
| Query preparation | No | |
Finally, the MorphoSaurus enhanced query behaves slightly better than the substring match query.
Independently of the term searched for (“syphilis” or “lues”) as it also retrieves all relevant documents (Recall = 100%) and has a slightly higher precision. The big advantage here is that such a query can be formulated “on the fly” and does not require any laborious preparation (Table 5).
Table 5.
Search for the expressions “lues” and, alternatively, “syphilis”: Both searches yield exactly the same results
| MorphoSaurus match “lues” or “syphilis” | Relevant documents | Non relevant documents |
|---|---|---|
| Documents retrieved | 40 | 159 |
| Documents not retrieved | 0 | 29801 |
| Precision | 20.1% | |
| Recall | 100.0 % | |
| F-Score | 33.5% | |
| Query preparation | no | |
CONCLUSION
The introduction of a semantic search engine for hospital discharge letters in a clinical setting yielded two types of preliminary evaluation results. Firstly, the user observation and inquiry showed a very good acceptance among physicians and revealed that the new option of WWW-like querying discharge letters across patients may change the way physicians deal with clinical documents. Secondly, the assessment of the search engine against a manually created gold standard showed that it retrieved all relevant documents, however at the cost of a low precision. A much higher precision was found with one baseline query (“syphilis”), however at the cost of a poor recall. Although this query scenario had a higher F-value than the semantic search it was not useful for the task under scrutiny because one third of the documents were not found at all. This example shows the limited validity of one standardized performance measure (F-measure with equal weight) if it is not interpreted in the light of a specific retrieval task. The performance of the semantic search is similar to the technique that manually composes a set of appropriate search strings. The latter is, however, rather laborious, requires a perfect understanding of the terminology and the ability to deal with a complex retrieval syntax. Due to the variability of medical language (cf. Table 1) this task may be very time consuming. One can therefore not expect the use of this technique in the clinical routine.
The results support the use of the proposed recalloriented search across EHR documents to acquire valid and complete data for epidemiology studies in hospital populations. However, the improvement of precision is an important goal for the future.
| Token match “lues” | Relevant documents | Non relevant documents |
|---|---|---|
| Documents retrieved | 35 | 192 |
| Documents not retrieved | 5 | 28,768 |
| Precision | 15.4 % | |
| Recall | 87.5 % | |
| F-Score | 26.2 % | |
| Query preparation | No | |
Footnotes
We are currently expanding the search engine by a negation detection feature so that documents containing “Lues Serologie negativ” would be excluded
REFERENCES
- 1.Müller M, Markó K, Daumke P, Paetzold J, Roesner A, Klar R. Biomedical data mining in clinical routine. MEDINFO. 2007:340–344. [PubMed] [Google Scholar]
- 2.Airio E. Word normalization and decompounding in mono and bilingual IR. Information Retrieval. 2006;9(3):249–271. [Google Scholar]
- 3.Braschler M, Ripplinger B. How effective is stemming and decompounding for German text retrieval. Information Retrieval. 2004;27(3–4):291–316. [Google Scholar]
- 4.Daumke P, Markó K, Poprat M, Schulz S, Klar R. Biomedical information retrieval across languages. Medical Informatics & Internet in Medicine. 2007;32(2):131–147. doi: 10.1080/14639230701197587. [DOI] [PubMed] [Google Scholar]
- 5.Honeck M, Hahn U, Klar R, Schulz S. Text Retrieval Based on Medical Subwords. Stud Health Technol Inform. 2002;90:241–245. [PubMed] [Google Scholar]
- 6.Daumke P, Markó K, Paetzold K, Müller M. Biomedical Data Mining in a Hospital information System. Technology and Health Care. 2007;15(5):308. [PubMed] [Google Scholar]
- 7.Markó K, Schulz S, Hahn U.MorphoSaurus - Design and Evaluation of an Interlingua-based, Cross-language Document Retrieval Engine for the Medical Domain Meth Inf Med4/200544537–545. [PubMed] [Google Scholar]
- 8.Markó K, Schulz S, Medelyan O, Hahn U. Bootstrapping Dictionaries for Cross-Language Information Retrieval. Proceedings of the 28th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; Salvador, Brazil. 2005. pp. 528–535. [Google Scholar]
- 9.Markó K, Daumke P, Schulz S, Klar R, Hahn U. Large-Scale Evaluation of a Medical Cross-Language Information Retrieval System. MEDINFO. 2007:392–396. [PubMed] [Google Scholar]
- 10.Markó K, Schulz S, Hahn U. Unsupervised Multilingual Word Sense Disambiguation via an Interlingua. Proc. 20th National Conf. on Artificial Intelligence; 2005. pp. 1075–1080. [Google Scholar]
- 11.Markó K, Daumke P, Hahn U. Cross-Lingual Alignment of Biomedical Acronyms. MIE. 2006:857–862. [PubMed] [Google Scholar]
- 12.Scott RE. e-Records in health Preserving our future. Int J Med Inf. 2007;76(5–6):427–431. doi: 10.1016/j.ijmedinf.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Lobach DF, Detmer DE.Research Challenges for Electronic Health Records American Journal of Preventive Medicine 200732 (5, Supplement 1): S104–S11. [DOI] [PubMed] [Google Scholar]
- 14.Wen HC, Ho YS, Jian WS, Li HC, Hsu YHE. Scientific production of electronic health record research, 1991–2005. Computer Methods and Programs in Biomedicine. 2007;86(2):191–196. doi: 10.1016/j.cmpb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Oard DW, He D, Wang J. User-assisted query translation for interactive cross-language information retrieval. Information Processing & Management. 2007 [Google Scholar]
- 16.Gey FC, Kando N, Peters C. Cross-Language Information Retrieval: the way ahead. Information Processing & Management. 2005;41(3):415–431. [Google Scholar]
- 17.Kishida K. Technical issues of cross-language information retrieval: a review. Pergamon Press, Inc; 2005. pp. 433–455. [Google Scholar]
- 18.Franz M, McCarley JS, Ward RT. Ad hoc, cross-language and spoken document information retrieval at IBM TREC-8. Gaithersburg, MD: National Institute of Standards and Technology; 2000. [Google Scholar]
- 19.Markó K, Hahn U, Schulz S, Daumke P, Nohama P. Interlingual Indexing across Different Languages. 7th International Conference “Recherche d'Information Assistée par Ordinateur” (RIAO'04); 2004. pp. 82–99. [Google Scholar]
- 20.Markó K, Daumke P, Schulz S, Hahn U. Cross-language MeSH Indexing using Morpho-Semantic Normalization. AMIA Annu Symp Proc. 2003:425–429. [PMC free article] [PubMed] [Google Scholar]