Abstract
Limited resources and the sheer volume of concepts make auditing a large terminology, such as SNOMED CT, a daunting task. It is essential to devise techniques that can aid an auditor by automatically identifying concepts that deserve attention. A methodology for this purpose based on a previously introduced abstraction network (called the p-area taxonomy) for a SNOMED CT hierarchy is presented. The methodology algorithmically gathers concepts appearing in certain overlapping subsets, defined exclusively with respect to the p-area taxonomy, for review. The results of applying the methodology to SNOMED’s Specimen hierarchy are presented. These results are compared against a control sample composed of concepts residing in subsets without the overlaps. With the use of the double bootstrap, the concept group produced by our methodology is shown to yield a statistically significant higher proportion of error discoveries.
Introduction
SNOMED CT [1] has proven to be an important resource to the healthcare and biomedical community since its origination in 2002. It is widely used in health information systems whose quality can impact the healthcare industry in general and patients in particular. Therefore, assuring the quality of SNOMED’s content is essential, especially as it continues to expand [2,3]. Automated tools that can enhance the efficiency and efficacy of SNOMED auditing are invaluable. Specifically, tools that can scour SNOMED’s 376,000 concepts (July 2007 release) and suggest potential errors to an auditor are in high demand.
In this paper, we present a methodology for locating concepts that have a high likelihood of being erroneous. The work proceeds from the premise that “complex” concepts are a natural place to look for errors. Of course, one needs to quantify this notion of complexity. For example, one straightforward measure would be the number of lateral relationships exhibited by a concept; the more relationships, the more complex the concept. Or it could be based on the number of parents.
We go beyond such straightforward measures and define complexity in terms of a previously introduced abstraction network, called the partial-area taxonomy [3,4], for the SNOMED hierarchy. In this context, the complex concepts are those residing in overlapping portions of the high-level groupings that form the basis of the partial-area taxonomy. The concepts are characterized by lying at points in the IS-A hierarchy where multiple paths originating from several “significant” ancestors converge. (This notion of significance will be fleshed out below.) Our methodology, overall, serves to algorithmically identify the complex concepts as an aid to the auditor.
As a test, our methodology is applied to SNO-MED’s Specimen hierarchy. The collection of suggested complex concepts is reviewed by domain-expert auditors in an attempt to find errors. As a basis of comparison, they are also presented with a control collection of concepts gathered by other means. The outcomes of these audits are reported. They support our hypothesis that the complex concepts—as we have defined them—are more error-prone than concepts at large.
Background
Various auditing techniques have been developed and applied to SNOMED. Among these are techniques that proceed from foundational ontological and linguistic principles [5,6]. In general, algorithms making use of description-logic formalisms—on which SNOMED is based—have been utilized to discover terminological inconsistencies [7] and synonymy [8].
We have previously formulated an auditing methodology for a SNOMED hierarchy based on two programmatically derived abstraction networks: the area taxonomy and the partial-area taxonomy (“p-area taxonomy” for short) [3,4]. Both reflect the attribute relationship distribution in a SNOMED hierarchy at a high level, while the latter further serves to reveal groupings of concepts with common ancestry. (We will use “relationship” to refer to an attribute (“lateral”) relationship. The hierarchical relationship is “IS-A.”) The methodology presented in this paper is based on aspects of the “p-area taxonomy,” so in the following we give the details of these two networks.
Let us start with some definitions. The set of relationships of a given concept C will be written relshps(C). That is, if r∈ relshps(C), then C is in the domain of r. The area taxonomy is derived from a partition of the concepts in a SNOMED hierarchy based on their respective sets of relationships. Let {r1, r2,…, rn} be a set of relationships. The area defined with respect to this set of relationships is:
That is, the area is defined as the set of all concepts that have exactly the relationships r1, r2,…, rn—no more, no less. When there is no confusion, we will denote an area as its set {r1, r2,…, rn} since that is the defining characteristic. Note that areas are disjoint, and collectively they form a partition of a hierarchy’s set of concepts. Two example areas from the Specimen hierarchy are {substance}, containing 81 concepts, and {morphology, procedure}, containing two concepts. The number of areas does not explode combinatorially because most combinations of relationships yield areas that are empty. Such empty areas are ignored. Some concepts have no relationships at all; hence, it makes sense to include Area (∅) (denoted simply as ∅).
In the area taxonomy, each area is represented as a node. Like the underlying concept hierarchy from which it is derived, the area taxonomy’s nodes are laid out in a hierarchical (directed acyclic graph) configuration. The hierarchical relationship between areas is called child-of, and each occurrence is derived directly from the IS-As in the SNOMED hierarchy, as follows. A root of an area is a concept in that area whose parents all reside in other areas. Blood specimen from patient, for example, is a root of {identity, substance}. A child-of from an area, say, A to an area B indicates that some root of A has a parent in B. There is a child-of from {identity, substance} to {identity} expressing the fact that the former’s root Blood specimen from patient has a parent Specimen from patient in the latter.
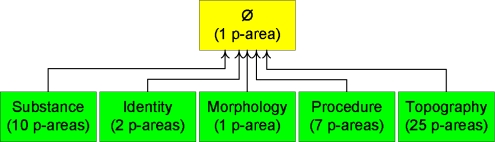
The area taxonomy of SNOMED’s Specimen hierarchy (July 2007 release) comprises 24 areas distributed over five levels. The top two levels can be seen in Figure 1. A box is an area. The relationships in the area’s name are listed in the box without braces. The arrows are the child-of ’s. The top level area is ∅. The five green boxes on Level 1 are areas having one relationship each. We also indicate in parentheses the number of partial-areas (“p-areas”)—to be defined shortly—that an area contains. For example, {substance} has ten p-areas; {identity} has two.
Figure 1.
Top two levels of the area taxonomy
An area can very well contain multiple roots. The roots are considered especially significant since each serves to generalize its entire group of descendants in the area, which can constitute a large swath of the area. (If the area has a single root, then it is the whole area.) A root therefore makes an excellent proxy for its descendants, and we use this fact as the basis for the partial-area (p-area) taxonomy. Let O be a root of an area A, and let desc(X, Y) denote the fact that concept X is a descendant of concept Y. The partial-area (p-area) defined with respect to O is:
That is, the p-area is a subset of the area consisting of the root O and all its descendants in the area. A p-area is denoted by its root O since, in this case, it is the defining characteristic. Note that the set of p-areas does not form a partition of the area. That is, a given concept can be a member of two or more p-areas. We will exploit this potential overlap among p-areas in our methodology below. Example p-areas in the area {substance} include Body substance specimen, Fluid Sample, and Plant Specimen, containing 47 concepts, 44 concepts, and 1 concept, respectively.
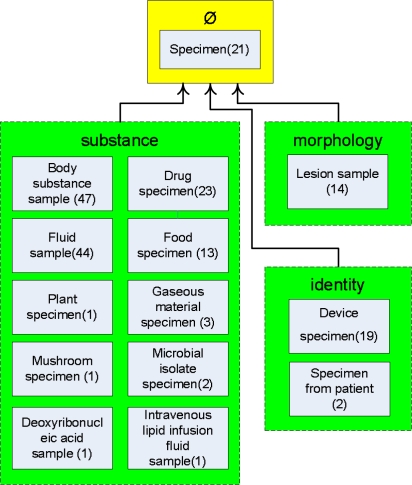
The p-area taxonomy refines the area taxonomy with the inclusion of p-area nodes (boxes) embedded inside their respective area nodes (now drawn as dashed boxes). Figure 2 shows a small portion of the p-area taxonomy of the Specimen hierarchy. Only three areas on Level 1 are displayed. In each p-area node, the number in parentheses is its number of concepts. For example, we see that area {identity} has two p-areas, Device specimen and Specimen from patient, containing 19 and two concepts, respectively. The complete p-area taxonomy of the Specimen hierarchy has a total of 361 p-areas.
Figure 2.
A portion of the p-area taxonomy
Methods
The root concepts of an area, each of which induces a p-area, are of considerable significance in the makeup of a SNOMED hierarchy. They are the first concepts in the hierarchy (starting from the top) to be defined with the area’s combination of relationships—whether those relationships are explicitly introduced or inherited. In this sense, they are cornerstones in the successive build-up of knowledge that is a hierarchy. Each respective p-area adds to this a hierarchical focus defined by a common ancestor, namely, the root. When a p-area is particularly small (e.g., one or two concepts), it denotes an uncommon convergence of relationship structure and hierarchical locality that very well may signal an error, as we have previously shown [3,4].
It is from the significance of the roots that we derive our notion of “complex concept” that underpins our new auditing methodology. The definition of p-area does not preclude two or more from having nonempty intersections. That is, two p-areas are not necessarily disjoint. A concept in an intersection of p-areas lies at a point in the hierarchy beneath multiple roots (of a single area) and elaborates the semantics of their combination. For this reason, concepts in p-area intersections are those that we deem to be complex for the purpose of auditing consideration. These are the concepts identified as deserving auditing priority. We will refer to such concepts as overlapping concepts.
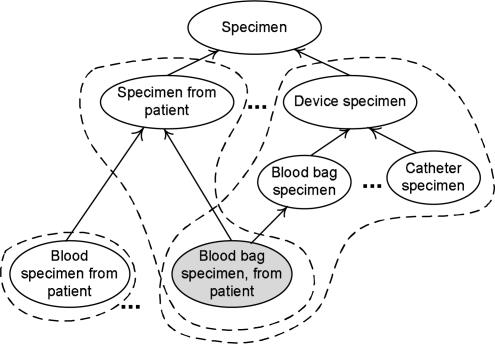
To further motivate the focus on overlapping concepts and see their inherent complexity, let us look at some examples. In the area {identity} (Figure 2), we find the two p-areas Device specimen and Specimen from patient. Figure 3 shows seven concepts, five of which are from these two p-areas. The p-areas are delineated by the dashed bubbles. The ancestor Specimen is in area ∅. The concept Blood bag specimen, from patient (highlighted in gray) is an overlapping concept sitting in both p-areas. It has two parents, Specimen from patient, the root of one p-area, and Blood bag specimen, a child of the root Device specimen of the other p-area. It inherits the relationship identity with accompanying targets from both its parents. Hence, it has two occurrences of identity, one directed to Patient and the other directed to Blood bag. Note that its sibling concept Blood specimen from patient belongs to the area {identity, substance} with a substance relationship directed to Blood. It constitutes a p-area of one concept in an area of six concepts. The overlapping concept Blood bag specimen, from patient is clearly more complex than its parent Blood bag specimen, which is non-overlapping, since the latter elaborates the semantics of one root, Device specimen, while the former elaborates the semantics of two, Device specimen and Specimen from patient.
Figure 3.
Overlapping concepts in area {identity}
The area {substance} (Figure 2) contains ten p-areas, including Body substance sample and Fluid sample. It also has quite a few overlapping concepts which can be gathered from the fact that the sum of the numbers of concepts in the ten p-areas (136) is much higher than the actual number of concepts in the area (81). An example is Body fluid sample, a child of Body substance sample as well as Fluid sample. Furthermore, all descendants of Body fluid sample residing in {substance} are overlapping concepts belonging to the p-areas Body substance sample and Fluid sample. All these specializations of Body fluid sample, e.g., Amniotic fluid specimen and Lymph sample, are more complex than concepts that are only fluid samples, e.g., Water specimen, or only body substance samples, e.g., Calculus specimen. The increased complexity is due to the dual specialization inherited from the roots of these two p-areas.
The amount of overlapping may increase as we traverse downward along the IS-A hierarchy. For example, one of the children of Body fluid sample, Blood specimen, has another parent Drug specimen, which is the root of its own p-area. In this case, Blood specimen is the specialization of three roots and thus resides in three separate p-areas. In {substance}, we find 15 concepts belonging to exactly two p-areas, and 20 concepts belonging to three p-areas. From this, we get its actual number of concepts: 136 − 1 · 15 − 2 · 20 = 81.
The additional complexity of overlapping concepts together with the theme of complex concepts having a higher likelihood of being in error leads us to the following hypothesis that we wish to investigate.
Hypothesis:
Overlapping concepts are more likely to have errors than concepts residing in p-areas without overlaps.
Following the paradigm of “group based” auditing [3], our methodology includes for review both the overlapping concepts as well as concepts in their immediate neighborhoods, consisting of parents, children, siblings, and targets of relationships. This may help to discover error propagations, which would be missed if the review were limited to the overlapping concepts alone. Examples of the kinds of errors we expect to find in an application of our methodology include incorrect IS-As and relationship targets.
To test our methodology and study the above hypothesis, we audit all the overlapping concepts of SNOMED’s Specimen hierarchy. As a basis for comparison, we also audit a control sample comprising concepts gleaned from p-areas having no intersections with other p-areas. Both kinds of concepts are audited with the same rigor by the same auditors.
To compare overlapping concepts with those in the control sample, we look at the proportion of erroneous concepts. We use the p-area as the unit of analysis, and we aggregate across levels (because of the small number of concepts at Level 2). We employ the double bootstrap [9] to calculate the statistical significance of the difference of the proportions.
Results
The Specimen hierarchy of SNOMED consists of 1,073 active concepts, of which 162 are overlapping. Most of these reside in Level l areas, i.e., those having one relationship. In fact, roughly one third (155 out of 468) of the Level 1 concepts are overlapping. And these are found primarily in {topography} and {substance}. The results of auditing the Level 1 overlapping concepts are given in Table 1. For each area, we list its total number of concepts C (Column 2), number of overlapping concepts V (Column 3), and number of erroneous concepts E c (Column 4). For example, {substance} has 81 concepts, 35 overlapping concepts, and 11 erroneous concepts.
Table 1.
Results of auditing areas at Level 1
| Area | C | V | Ec |
|---|---|---|---|
| substance | 81 | 35 | 11 |
| morphology | 14 | 0 | 0 |
| topography | 333 | 116 | 71 |
| procedure | 20 | 3 | 3 |
| identity | 20 | 1 | 0 |
| Total: | 468 | 155 | 85 |
Most overlapping concepts in area {topography} are found in intersections of p-areas involving Tissue specimen of 126 concepts. We have tabulated these results separately in Table 2. For example, the p-area Specimen from eye has 18 concepts. Its intersection with Tissue specimen has 12 of them. Eight of those are in error.
Table 2.
Results of auditing intersections involving p-area Tissue specimen
| Second P-area | C | V | Ec |
|---|---|---|---|
| Specimen from eye | 18 | 12 | 8 |
| Ear sample | 2 | 1 | 0 |
| Specimen from breast | 8 | 4 | 2 |
| Cardiovascular sample | 13 | 3 | 1 |
| Products of conception tissue sample | 12 | 1 | 1 |
| Genitourinary sample | 73 | 20 | 17 |
| Dermatological sample | 6 | 2 | 0 |
| Spec. from digestive system | 74 | 29 | 18 |
| Musculoskeletal sample | 35 | 22 | 15 |
| Respiratory sample | 41 | 6 | 5 |
| Endocrine sample | 12 | 3 | 0 |
| Specimen from central nervous system | 4 | 1 | 1 |
| Spec. from thymus gland | 2 | 1 | 0 |
| Specimen from trophoblast | 2 | 1 | 0 |
Overlapping concepts appear in the p-areas of areas with two relationships, but in far fewer numbers. In fact, there are only seven of them. Six are in {topography, procedure}, and the other is in {topography, morphology}.
The control sample was taken from p-areas that had no intersections with other p-areas and contained more than one concept. The reason for the second requirement is that, as we alluded to, p-areas of one concept are already known to be error-prone [2,4]. Thus, they do not make for a proper control sample.
Due to a lack of enough such p-areas with no intersections on Level 1, we use a control sample of 78 concepts, half the number of Level 1 overlapping concepts. From Level 2, we gathered seven concepts. Hence, there are 155 + 7 = 162 overlapping concepts, and the control sample has 78 + 7 = 85 concepts.
Table 3 gives the results of the auditing carried out on these two groups of concepts. Note that E (Column 3) denotes the total number of errors. This value differs from Ec, the number of erroneous concepts (Column 5), because a given concept can have more than one error. The average erroneous-concept rate among the overlapping concepts was 55%, and among the control sample it was 29% (Column 6). The difference was significant at the 0.05 level. Let us point out that erroneous concepts in the overlapping group had close to two errors on average (last column).
Table 3.
Auditing results for overlapping (“Over”) concepts vs. control (“Ctrl”) sample
| C | E | E/C | Ec | Ec/C | E/Ec | |
|---|---|---|---|---|---|---|
| Over | 162 | 158 | .98 | 89 | 55% | 1.8 |
| Ctrl | 85 | 31 | .36 | 25 | 29% | 1.2 |
Table 4 lists the number of different kinds of errors found for overlapping concepts. For example, 48 cases of missing children were discovered. Table 5 provides a sample of the errors. Note that some concepts are listed with two errors.
Table 4.
Kinds of errors and their counts
| Kind of Error | # |
|---|---|
| Ambiguous concept | 1 |
| Missing child | 48 |
| Missing parent | 30 |
| Missing relationship | 21 |
| Missing sibling | 4 |
| Incorrect child | 5 |
| Incorrect parent | 44 |
| Incorrect target of relationship | 5 |
| Total: | 158 |
Table 5.
Error samples
| Concept | P-areas | Error Type | Correction |
|---|---|---|---|
| Blood spec. | Body substance smp. / Fluid smp. / Drug spec. | Incorrect child | Remove child: Erythrocyte spec. |
| Body fluid smp. | Body substance smp. / Fluid smp. | Missing child | Add child: Tissue fluid smp. |
| Cartilage biopsy smp. | Tissue spec. / Musculoskeletal smp. | Missing relationship | Add rel: Procedure to Biopsy |
| Missing parent | Add parent: Biopsy smp. | ||
| Female genital tissue smp. | Tissue spec. / Genitourinary smp. | Missing child | Add child: Tissue spec. from ovary |
| Meconium spec. | Body substance smp. / Fluid smp. | Incorrect parent: Body fluid smp. | Correct parent: Fecal smp. |
| Synovial cytologic material | Tissue spec. / Musculoskeletal smp. | Incorrect parent: Musculoskeletal smp. | Correct parent: Synovial smp. |
| Missing parent | Add parent: Cytologic material |
Discussion
The auditing was performed by two of the authors (GE, JX) who are MDs with experience in medical terminologies. Their error report, obtained by a consensus from their individual findings, was reviewed by another author (KAS), the Chief Terminologist of IHTSDO. Only confirmed (by KAS) errors were reported here. Our interest was not in studying the auditing process per se, but in the distribution of the unquestionable errors resulting from it. These errors were corrected in SNOMED’s July ’08 release.
As we can see from Table 3, according to all reported measures, there is a significantly higher return for the auditing effort obtained for the overlapping concepts compared to concepts in p-areas without overlaps. Such higher return seems to justify concentrating auditing efforts on the more complex overlapping concepts. The results confirm the hypothesis we stated. More experiments with different and larger hierarchies of SNOMED and similar terminologies, e.g., NCIT [2], are needed to further confirm our finding.
This finding also confirms the auditing theme that complex concepts have relatively more errors. Another manifestation of this theme, in [4], was the group of concepts residing in “strict inheritance” p-areas. It is suggested that the design of taxonomies and the auditing of the complex concepts discussed here and in [4] should become integral parts of the design cycle for terminologies such as SNOMED and NCIT.
It is notable that the Specimen hierarchy underwent three auditing efforts by our team. The previous two were reported in [3,4]. Nevertheless, the present auditing still yielded a high return of errors. One explanation may be that concentrating on the overlapping concepts reveals truly new fertile ground, and those concepts did not get appropriate attention in our previous efforts directed at small p-areas. A second explanation is that the group-based approach, where the neighbors of overlapping concepts were reviewed, had a hand in the success. We note that an error may appear in a parent or a child of an overlapping concept, and this is considered an error for the overlapping concept.
Design patterns are currently being considered as an integral part of a machine-readable concept model for SNOMED. When expressed as tighter constraints in editing tools, they will be a way to prevent the introduction of new errors. Our auditing techniques are clearly applicable in finding errors that have occurred, and thereby suggesting constraints and patterns that are needed to help support content editors.
Conclusion
We proceeded from the assumption that “complex” concepts warrant particular attention in quality assurance activities pertaining to terminologies like SNOMED. We presented an auditing methodology in which we took such complex concepts to be those residing in special overlapping subsets of a SNOMED hierarchy defined with respect to an abstraction network called the p-area taxonomy. These so-called overlapping concepts in the Specimen hierarchy were identified programmatically and then put through a rigorous audit. Comparing these auditing results with results from a control set, we found a statistically significant higher error rate among the overlapping concepts. Thus, our auditing methodology based on overlapping concepts can be seen as an important addition to the existing suite of terminology auditing regimens.
Acknowledgments
This work was partially supported by the NLM under grant R-01-LM008912-01A1.
References
- 1.IHTSDO: SNOMED CTAvailable at http://www.ihtsdo.org/our-standards/snomed-ct Accessed December 31, 2007.
- 2.Min H, Perl Y, Chen Y, Halper M, Geller J, Wang Y. Auditing as part of the terminology design life cycle. JAMIA. 2006;13(6):676–690. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. JBI. 2007;40(5):561–581. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, Spackman KA. Analysis of error concentrations in SNOMED. In: Teich JM, Suermondt J, Hripcsak G, editors. Proc. AMIA Annual Symposium; Chicago, IL: 2007. 2007. pp. 314–318. [PMC free article] [PubMed] [Google Scholar]
- 5.Ceusters W, Smith B, Kumar A, Dhaen C. Ontology-based error detection in SNOMED-CT. In: Fieschi M, Coiera E, Li YC, editors. Proc. Med-info 2004; San Francisco, CA. 2004. pp. 482–486. [PubMed] [Google Scholar]
- 6.Bodenreider O, Smith B, Kumar A, Burgun A. Investigating subsumption in DL-based terminologies: a case study in SNOMED CT. In: Hahn U, Schulz S, Cornet R, editors. Proc. KR-MED 2004; Whistler, Canada. 2004. pp. 12–20. [Google Scholar]
- 7.Schlobach S, Huang Z, Cornet R, Van Harmelen F. Debugging incoherent terminologies. J. Autom. Reasoning. 2007;39:317–349. [Google Scholar]
- 8.Cornet R, Abu-Hanna A. Auditing description-logic-based medical terminological systems by detecting equivalent concept definitions. Int J. Med. Informatics. 2008;77(5):336–345. doi: 10.1016/j.ijmedinf.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Boca Raton, FL: CRC Press; 1993. [Google Scholar]