Abstract
The prevalence of electronic medical record (EMR) systems has made mass-screening for clinical trials viable through secondary uses of clinical data, which often exist in both structured and free text formats. The tradeoffs of using information in either data format for clinical trials screening are understudied. This paper compares the results of clinical trial eligibility queries over ICD9-encoded diagnoses and NLP-processed textual discharge summaries. The strengths and weaknesses of both data sources are summarized along the following dimensions: information completeness, expressiveness, code granularity, and accuracy of temporal information. We conclude that NLP-processed patient reports supplement important information for eligibility screening and should be used in combination with structured data.
Introduction
Eighty-six percent of all clinical trials are delayed in patient recruitment from 1 to 6 months and 13% are delayed by more than 6 months1. The broad deployment of electronic medical records (EMR) systems has potential to accelerate subject pre-screening for clinical trials research by enabling access to rich clinical data, which often exist in both structured and free-text formats. Although it is well-known that administrative data such as ICD9 codes have poor granularity and accuracy for identifying patients with certain diseases2, the utility of ICD9 codes for clinical trials pre-screening is understudied. In fact, they are popular data sources for EMR-based patient eligibility alert systems3. Only a few systems used textual clinical notes4. An understudied research topic is: What are the strengths and weaknesses of using structured or textual clinical data for clinical trials eligibility pre-screening? Only recently did one study compare ICD9 codes against natural language processed (NLP) patient notes for identifying clinical phenotypes5. That study concluded that ICD9 codes are inadequate for eligibility screening, but did not provide qualitative analyses of the tradeoffs of ICD9 codes and NLP-processed notes. In the rest of this paper, using a case study, we provide a detailed qualitative analysis of the strengths and weaknesses of both data sources for clinical trial pre-screening.
Research Design
1. The Clinical Trial Case
We selected an ongoing clinical trial in our institution, for which only 7 eligible subjects were identified through manual chart review in the year 2007. Eligible patients must meet one of the four following criteria (A or B or C or D):
A. Documented previous ischemic stroke (all of the following must be satisfied): (a) A focal ischemic neurological deficit persistent for more than 24 hours; (b) considered being of ischemic origin. Onset within previous 5 years but not within 8 weeks prior to enrollment; (c) patients with previous atrial fibrillation should be excluded; (d) A CT scan or MRI must have been performed to rule out hemorrhage and non ischemic neurological disease.
B. Symptomatic carotid artery disease with ≥ 50% stenosis;
C. Asymptomatic carotid stenosis ≥ 70%
D. History of carotid revascularization (surgical or catheter-based).
2. Data Source and Tools
The data source for this study was the Clinical Data Warehouse (CDW), which is the research database of New York Presbyterian Hospital (NYP) and Columbia University Medical Center (CUMC). The CDW contains a broad range of clinical information, such as diagnoses, procedures, discharge summaries, laboratory tests, and pharmacy data for the past 20 years. It is updated nightly to be synchronized with the real time EMR system used in NYP. Our study sample was selected from all the patient records of the year 2005, which contained 9,022,255 ICD9 diagnoses for 269,647 unique patients. Furthermore, 21,833 patients from this population had 29,016 electronic textual discharge summaries in the CDW.
We used the Medical Language Extraction and Encoding System (MedLEE)6 to process the narrative discharge summaries. MedLEE generates structured, computer-recognizable, XML-coded concepts using semantic classes for problem, finding, body location, certainty, degree, region, and so on. MedLEE also extracts a series of modifiers linked to concepts, such as certainty, status, location, quantity, and degree. Applicable concepts are further encoded as concepts recognizable by The Unified Medical Language System (UMLS) 7.
3. Two Eligibility Queries: ICD9 vs. MedLEE
We created an ICD9 query and a MedLEE query to search for eligible patients. In the MedLEE query, we included the following terms: “ischemic stroke”, “carotid endarterectomy / revascularization”, “cerebral vascular disease/accident”, “carotid narrowness /stenosis /occlusion”, and “thrombotic stroke”. Notes containing “atrial fibrillation / flutter”, “cerebral hemorrhage”, and “transient ischemic attack” were excluded. The ICD9 query included corresponding ICD9 codes for the above concepts and was reviewed by two internal medicine physicians. The actual UMLS and ICD9 codes used in the queries are available in an online supplement http://www.dbmi.columbia.edu/~chw7007/ICD.htm.
Results
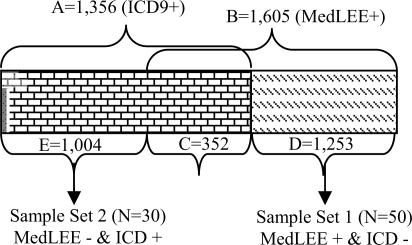
The ICD9 query returned 1,356 patients (set A). The MedLEE query returned 1,605 patients (set B). The intersection between set A and B contains 352 patients (set C) who were retrieved by both queries. Therefore, 1,253 patients (set D) were identified by the MedLEE query but missed by ICD9 query while 1,004 patients (set E) were identified by ICD9 query but missed by the MedLEE query. The relationships between set A through E are shown in Figure 1.
Figure 1.
Patients Retrieved by Both Queries:
Set A=E+C=1,356: retrieved by ICD9;
Set B=C+D=1,605: retrieved by MedLEE;
Set C= 352: retrieved by both ICD9 and MedLEE;
Set D=1,253: retrieved only by MedLEE;
Set E=1,004: retrieved only by ICD9.
To characterize the eligibility information in the two data sources and to analyze the causes of the above query results discrepancies, we randomly selected 50 patients from set D to form sample set 1 and 30 patients from set E to form sample set 2 for manual chart review. The results and corresponding analyses are summarized in Table 1 and Table 2 respectively, with details following each table.
Table 1.
Review of Sample Set 1: MedLEE Test Positive (+) & ICD9 Test Negative (–)
| MedLEE (+) ICD9 (–) | Causes of Discrepancies | Percentage |
|---|---|---|
| MedLEE True Positive =28 (56%) | MedLEE provided additional information about past medical histories. | 19 (38%) |
| MedLEE provided additional information from lab reports. | 2 (4%) | |
| MedLEE provided etiological information for diagnoses. | 7 (14%) | |
| MedLEE False Positive =22 (44%) | Inclusion and exclusion criteria were met in different notes. | 4 (8%) |
| There was incompleteness and inconsistency between notes and ICD9 codes. | 1 (2%) | |
| There were documentation errors in notes. | 17 (34%) |
Table 2.
Review of Sample Set 2: MedLEE Test Negative (–) & ICD9 Test Positive (+)
| MedLEE (–) ICD9 (+) | Causes of Discrepancies | Percentage |
|---|---|---|
| ICD9 True Positive =27 (90%) | Fragmented information scattered across multiple notes | 12 (40%) |
| Defects in MedLEE Query | 15 (50%) | |
| ICD9 False Positive =3 (10%) | Diagnoses satisfying exclusion criteria missed by ICD9 codes | 3 (10%) |
1. MedLEE Query’s True Positive Cases
(Eligible patients identified by MedLEE but not by ICD9)
1.1: MedLEE provided additional information about past medical histories:
Four patients had “carotid endarterectomy”, 2 patients had “carotid stenosis”, and 13 patients had “stroke” specified in the past medical history section that met criterion A, B/C or D respectively, for which these patients did not have corresponding ICD9 diagnoses.
1.2: MedLEE provided additional information from laboratory reports:
Two patients had “carotid stenosis” in their laboratory reports, such as “carotid artery stenosis L 60–79%” and “Doppler of her carotid which showed 99% right internal carotid artery stenosis…to follow-up as an outpatient for further evaluation of the carotid stenosis”. However, neither of them had corresponding ICD9 diagnoses.
1.3. MedLEE provided etiological information that was unavailable in ICD9 codes:
Seven patients had “stroke” specified in their past medical histories and met criterion A. Five of them had residual symptoms; four of them had hemiplegia with the body location information specified. None of the 7 patients had ICD9 diagnoses for “stroke”, but for “Hemiplegia Unspecified Side”. The ICD9 codes used in our query did not include “Hemiplegia” because it can be caused by multiple etiologies; adding it to the query would increase the false positive rate. Therefore, the lack of etiological information in ICD9 codes lowered the sensitivity of our ICD9 query. Meanwhile we can see that the ICD9 codes were not precisely used on these patients. In the notes, four of these patients had hemiplegia side specified, but the code used was “unspecified side”. Although the lack of specific details makes no big differences for billing purposes, it hampers the accuracy of trial eligibility queries.
2. MedLEE’s False Positive Cases
(Ineligible patients excluded by the ICD9 query but included by the MedLEE query)
2.1. Inclusion and exclusion criteria were satisfied in different notes:
Four patients had “stroke” (inclusion criteria) and “atrial fibrillation” (exclusion criteria) specified in different notes. Since we applied MedLEE on one note each time, we included these patients immediately after recognizing “stroke” in one of the notes, but failed to combine the results for the exclusion criteria for “atrial fibrillation” that were satisfied in other notes.
2.2. There was incomplete information and inconsistency between notes and ICD9 diagnoses:
One patient had “stroke” recorded in his discharge summary, but also had an ICD9 code for “Transient Cerebral Ischemia NOS”, which made this patient ineligible by criterion A. Without further information to prove that the ICD9 code was wrong, we counted the patient as false positive for MedLEE query.
2.3. There were documentation errors:
Seventeen patients were considered eligible by MedLEE due to documentation errors, such as complicated negations, unusual abbreviations, wrong grammars, and spelling errors in discharge summaries. Such documentation errors were not only hard for computer to interpret, but also difficult for human experts to interpret. An example is “?afib, CT scan r/o bleding/strokes”.
3. ICD9’s True Positive
(Eligible patients identified by the ICD9 query but missed by the MedLEE query)
3.1. Fragmented information was scattered across multiple notes of the same or different types:
Twelve patients had the ICD9 diagnoses for “Cerebral Vascular Disease”, but no pertinent information was specified in their discharge notes. The diagnosis may be elaborated in discharge notes dated other than year 2005 or in other data sources such as admission notes, progress notes, laboratory reports that were not part of our sample. We counted these patients as eligible or true positives for the ICD9 query.
3.2. There were defects in our MedLEE queries:
Our MedLEE query failed to retrieve 15 eligible patients because we did not specify appropriate attributes in the query. For example, for the note that contains “CT/MRI demonstrated with small acute infarct in R upper posterior left basal ganglia/corona radiate”, MedLEE has the capacity to extract the location information “basal ganglia” for term “infarction”; however, due to our oversight, we did not add the specific location constraints in our query and hence missed the patient. Such human errors can be avoided if the person who formulates the query can accurately translate the eligibility criteria into corresponding semantic classes and attributes for the MedLEE query and thoroughly considers all possible combinations of semantic classes and attributes.
4. ICD9 Query’s False Positive
(Ineligible patients included by the ICD9 query but appropriately excluded by the MedLEE query)
Three patients with inclusion ICD9 codes for “Cerebral Vascular Disease/Accident” also met exclusion criteria (cerebral hemorrhage or atrial fibrillation) specified only in the discharge summaries. Therefore, these patients were mistakenly considered positive by the ICD9 query. The MedLEE query successfully excluded the patients who met exclusion criterion A based on their notes.
Discussion
Our manual review sample sizes were small: 50 patients from set D and 30 patients from set E. A larger sample analysis would strengthen the validity of the findings. In spite of this, in our sample set 1, 56% of the patients were identified eligible using the NLP-processed notes. If we apply this ratio to all the patients who had diagnoses in notes but did not have corresponding ICD9 diagnoses (N=1,253), we may identify 702 more eligible patients than using the ICD9 query alone. The results indicated that NLP-processed notes provided richer information than ICD9 codes for clinical trial pre-screening.
Next, we further analyze how NLP-processed notes complement ICD9 diagnoses for clinical trials pre-screening and the strength and weakness of both data sources with regard to information completeness, expressiveness, concept encoding granularity, and the accuracy of temporal information.
1. Information Completeness
For clinical trials pre-screening, textual notes can supplement valuable observations and etiological knowledge related to ICD9 diagnoses. Discharge summaries contain multiple sections. The “past medical history” and “review of systems” sections document patients’ major past diseases or procedures with details for the onset time and severity of the diseases. The “Laboratory Data” section includes doctors’ interpretations and diagnoses for the results of major tests such as CT, MRI and angiogram. The “Hospital Course” section records patients’ disease progression, medications, procedures, and symptoms developed during the course. The “Admission /Discharge Diagnosis” section documents diagnoses related to a current admission of the patient. Therefore, applying NLP methods to such patient information in discharge summaries can potentially enable us to have comprehensive and thorough information about the patient’s disease onset and evolution over time, which is necessary for eligibility screening. However, we also noticed that by relying on discharge summaries only, we missed approximately 40% of eligible candidates from our sample set 2. In addition, single discharge notes may not include all the information we need. In our sample set 1, 8% of the patients were false positive because “atrial fibrillation” was documented in separate discharge summary notes, but we only ran the MedLEE query on one note each time.
Past medical history information about patients often are included in textual notes but the appropriate ICD9 codes could be missing in the EMR for multiple reasons. First, some diagnoses were made before the clinical data warehouse or our EMR system were installed and used. Second, we do not have past records for a new patient in the system. Third, some mobile patients use multiple hospitals and do not consistently visit our hospital so their medical records are discontinuous in our EMR system. In these cases, clinical notes processed by NLP tools can supplement information about patients’ past medical histories. In our sample set 1, 38% of the patients had “stroke”, “carotid stenosis” or “carotid endarterectomy” specified in their past medical history and were eligible for the clinical trial.
NLP-processed clinical notes can also supplement information about disease conditions without diagnoses. As administrative data, ICD9 codes are primarily used for billing purposes and are assigned at admission. If the diseases were in the past, non-progressive and/or unrelated to patients’ current admission, or asymptomatic, clinicians might not give diagnoses but briefly describe them in the past medical history, review of systems, or laboratory results. In our sample set 1, 4 patients had “carotid stenosis” specified in their notes, 2 of them had narrowness described in the laboratory report and 1 of them was instructed to follow up with an outpatient clinic. Since these patients were not admitted for stenosis, they did not have a corresponding ICD9 diagnosis. In such scenarios, querying the NLP processed notes enables us to identify patients who are potential clinical trial candidates but do not have complete diagnoses for all of their disease conditions.
2. Information Expressiveness
Laboratory results in the notes provide specific details that cannot be represented by using ICD9 codes. When trying to find patients with specific degrees of carotid stenosis (50% stenosis if symptomatic (Criteria 1B) or 70% stenosis if asymptomatic (Criteria 1C)), it is not possible to use ICD9 queries because ICD9 codes do not incorporate quantitative information. We need to extract this information from the notes for detailed descriptions. MedLEE is capable of recognizing quantitative measurements and encoding them in a structured output as shown below in Figure 2.
Figure 2.
Example MedLEE Output
3. Concept Encoding Granularity
Granularity is a common challenge for both ICD9-based diagnoses and MedLEE-processed notes. Inappropriate uses of terms that are too general can cause false positives in queries over both data sources. For example, the general ICD9 codes “cerebral vascular disease” and “cerebral vascular accident” were used to encode diagnoses for ischemic stroke, hemorrhage stroke or carotid occlusion. In our sample set 2, one false-positive patient was included by the ICD9 query, but proved to be ineligible because the “hemorrhage” condition was specified in his or her note only. Similarly, in discharge summaries, clinicians often used the general term “stroke” in the patients’ past medical histories without specifying whether the stroke was ischemic or hemorrhage. If no further details were provided, we had to search for a more specific ICD9 diagnosis that may specify the type of stroke for the patient. However, often no specific information was available. In our sample set 1, 14% of the patients had ICD9 diagnoses for “Hemiplegia”. However, hemiplegia can be caused by different etiological reasons; stroke is only one cause. Including this ICD9 diagnosis in the query could cause false positives and lower the query’s accuracy. NLP-processed notes are a good data source that supplements etiological information.
4. Accuracy of Temporal Information
Accuracy of temporal information is identified as a common challenge for both ICD9 codes and NLP-processed notes. Eligibility criteria usually have temporal constraints on clinical phenotypes. In our case study, we needed to find patients with cerebral vascular disease within the past 5 years, but not within the past 8 weeks. Although every ICD9 diagnosis code has a primary diagnosis time, this time stamp does not necessarily indicate the disease onset time, but the diagnosis time. If a patient had stroke in the past and now has paralysis, diagnosis for stroke may be given using a general term “cerebral vascular disease” for current admission. In this scenario, the primary diagnosis time is current, but the disease may have started a long time prior to the admission date. Similarly, narrative patient reports may state “stroke in the past” but did not specify when it started. Our CDW would capture accurate temporal information of diseases only if patients were admitted every time a disease occurred and if the disease was diagnosed and documented. Temporality assessment among clinical phenotypes in narrative clinical notes is still an active but challenging research area8.
Conclusion
This study is an early step toward identifying an optimal combination of structured (e.g., ICD9 codes) and NLP-processed unstructured data (e.g., discharge summaries) for clinical trials pre-screening. We conclude that NLP-processed notes are valuable data sources for clinical trial pre-screening by providing past medical histories as well as more specific details about diseases that are unavailable in ICD9 codes.
We will extend this analysis to multiple types of clinical notes and other clinical trials with different diseases to assess the generalizability of our findings. In this study we did not utilize multiple notes of one patient and therefore did not obtain as comprehensive patient information as possible for the NLP-based query. In the future, we plan on processing multiple records for one patient in order to obtain more comprehensive information; however, the retrieval may be more complicated then because there will be a larger possibility that some of the information will be inconsistent, and a more complex process may be needed to resolve inconsistencies.
Acknowledgments
We thank Drs John Ennever and David Wajngurt for reviewing our ICD9 codes and queries and Dr. Adam Wilcox for providing access to the CDW. This work is supported by grants LM007659, LM00865, and LM06910 from NLM and UL1 RR024156 from the National Center for Research Resources (NCRR).
References
- 1.Sinackevich N, Tassignon J-P. Speeding the Critical Path. Applied Clinical Trials. 2004 [Google Scholar]
- 2.Aronsky D, Haug PJ, Lagor C, Dean NC. 2005. Accuracy of Administrative Data for Identifying Patients With Pneumonia; pp. 319–328. [DOI] [PubMed] [Google Scholar]
- 3.Embi PJ, Jain A, Clark J, Bizjack S, Hornung R, et al. Effect of a Clinical Trial Alert System on Physician Participation in Trial Recruitment. Arch Intern Med. 2005;165:2272–2277. doi: 10.1001/archinte.165.19.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiszman M, Chapman W, Aronsky D, Evans R, Haug P. Automatic Detection of Acute Bacterial Pneumonia from Chest X-ray Reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gundlapalli AV, South BR, Phansalkar S, Kinney AY, Shen S, et al. Application of Natural Language Processing to VA Electronic Health Records to Identify Phenotypic Characteristics for Clinical and Research Purposes. Proc of 2008 AMIA Summit on Translational Bioinformatics. 2008:36–40. [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. JAMIA. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UMLS http://www.nlm.nih.gov/research/umls/, cited July 10, 2008.
- 8.Zhou L, Parsons S, Hripcsak G. The Evaluation of a Temporal Reasoning System in Processing Clinical Discharge Summaries. JAMIA. 2008;15(1):99–106. doi: 10.1197/jamia.M2467. [DOI] [PMC free article] [PubMed] [Google Scholar]