Abstract
Domain reference ontologies are being developed to serve as generalizable and reusable sources designed to support any application specific to the domain. The challenge is how to develop ways to derive or adapt pertinent portions of reference ontologies into application ontologies. In this paper we demonstrate how a subset of anatomy relevant to the domain of radiology can be derived from an anatomy reference ontology, the Foundational Model of Anatomy (FMA) Ontology, to create an application ontology that is robust and expressive enough to incorporate and accommodate all salient anatomical knowledge necessary to support existing and emerging systems for managing anatomical information related to radiology. The principles underlying this work are applicable to domains beyond radiology, so our results could be extended to other areas of biomedicine in the future.
Introduction
The application of ontology in support of biomedical research and the practice of medicine is widely recognized. There is an explosion of independent but largely uncoordinated efforts in building ontologies in the different biomedical domains, and most of these projects are lightweight or end use-specific ontologies, aptly called application ontologies because they only meet circumscribed needs and are not equipped to scale, extend or generalize to other applications. On the other hand there are ontologies that aim to comprehensively represent domains of basic biomedical science such as anatomy, physiology and pathology and which are reusable and generalizable to meet the needs of any application requiring structured information for the particular domain. However these so-called reference ontologies are often too large and complex for end users and application developers to access, process and utilize. There is therefore a need for methods to create modular and manageable application ontologies by piecing together components derived from multiple sources, especially from domain reference ontologies which can support all sorts of applications that pertain to the corresponding domains. The approach is gaining increasing acceptance because it can serve two practical purposes: 1) reference ontologies provide a principled and robust framework on which to build applications specific to the biomedical realities under the purview of a particular field1, and 2) because of this underlying ontological framework, the derivations facilitate and promote integration, interoperability and reuse of knowledge among application ontologies.
Derivation of application ontologies
There are at least two methods for deriving an application ontology from a reference ontology, one by extracting the entire ontology as a subset of one or more reference sources, and the other by augmenting an existing terminology project or lightweight ontology with ontological elements from one or more reference ontologies. In this study we explore the latter approach, and demonstrate how we can use this approach to augment the RadLex radiology terminology (http://www.rsna.org/radlex/). Our approach entails exploiting the disciplined and sound ontological framework of an anatomy reference ontology, the Foundational Model of Anatomy (FMA) ontology2 and incorporating subsets of the FMA into the organizational structure of the lexicon. The result is an application ontology dedicated to radiological anatomy that is both robust and sufficiently expressive to capture and accommodate salient anatomical information needed for radiology-related tasks. In the process of creating this application ontology we gain valuable insights into the requirements and operations for developing generalizable methodologies for programmatically generating any ontology subset or application “view” from any source ontology.
Materials and Methods
RadLex: Standard Radiology Lexicon
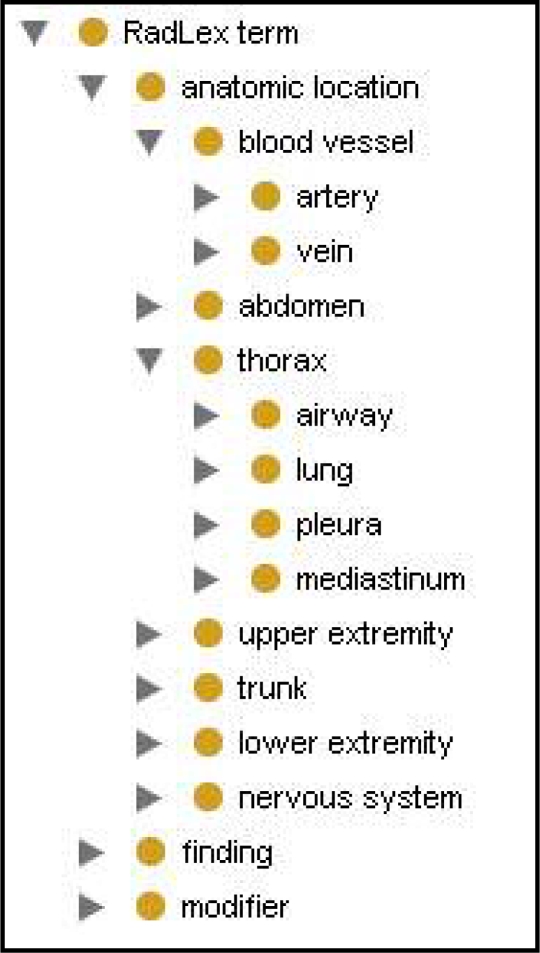
The Radiological Society of North America (RSNA) developed a publicly available terminology, RadLex, to provide a uniform standard for all radiology-related information3. RadLex encompasses many complex domains, ranging from basic sciences to imaging technologies and acquisition. The lexicon is organized into a subsumption hierarchy with RadLex term as the root; it subsumes over 7400 terms organized in 9 main categories or types; anatomic location being but one among others such as treatment, uncertainty and image quality. Compiled by committee members from different radiology subdomains, the resulting terminology artifact is a mix- and –match collection of terms and relations adapted from a variety of sources. While the focus of RadLex is on providing controlled terminology, its hierarchical structure points to its evolutionary direction to ontology. Marwede et al. describes the ontological challenges to the current organizational structure of RadLex due to the fact that it does not yet have a principled ontological framework4. Viewed from an ontological perspective, RadLex has the following limitations that could be resolved by adopting the framework of a sound reference ontology such as the FMA: 1) being term-oriented, RadLex currently ignores the entities to which its terms project; 2) the lack of a taxonomy grounded in biomedical reality; 3) the ambiguity and mixing of relations (such as is_a, part_of, contained_in) represented by the links between the nodes of the term hierarchy (Figure 1). Our test case relates to assessing how a portion of a reference ontology, such as the FMA, can be adopted to lend ontological rigor to RadLex in resolving the challenges enumerated above.
Figure 1.
RadLex hierarchy of mixed relations in Protégé5; for example, artery is_a blood vessel and lung part_of thorax.
Foundational Model of Anatomy (FMA) Ontology
The Foundational Model of Anatomy Ontology is an open source reference ontology that represents the spatio-structural description of the entities and the relations which constitute and form the structural phenotype of the human organism at all biologically salient levels of granularity2 (http://fma.biostr.washington.edu). Based on a theory that explicitly defines anatomy and its content, it provides a framework that can incorporate and accommodate all entities under the purview of the anatomy domain. It is implemented as a computable artifact6 and is primarily intended for developers of computable terminologies and application ontologies in clinical medicine and biomedical research. Since the FMA is both broader and more fine-grained than extant anatomy texts or terminologies (e.g., it has as its high level nodes such types as Material anatomical entity and Immaterial anatomical entity, and such leaf types as Left third rib, none of which are to be found in other sources), its merits are primarily valued by ontologists, whereas practitioners of clinical medicine and of biomedical research and education find it not entirely consistent with their tradition-based knowledge of anatomy. Therefore, the benefits the FMA offers to such end users should be realized through particular derivative application ontologies which reconcile the view of anatomy prevalent, for example, among radiologists or anatomy teachers with its ontological representation in the FMA.
We illustrate in the following sections how the theory of anatomy proposed by the FMA can enrich and enhance the taxonomy and the relations network of RadLex. Here we cover only certain portions of RadLex’s anatomy, and concentrate only on select anatomical areas as proof of concept.
Results
Clarification of semantics
The term-centric approach adopted in RadLex generates some confusion in the semantics intended for its terms. Terms relating to anatomy are represented in the RadLex category Anatomic location, which subsumes the same terms used by other disciplines of biomedicine that refer to anatomical entities that actually exist in reality. However when a RadLex term like heart is used as a label by the radiologist, it is an annotation of an area in a concrete radiological image that corresponds to the actual heart of a particular patient. Thus we are dealing with a term given two entirely different meanings, as a radiology image entity and as a real anatomical entity. It is therefore necessary to clarify what the RadLex anatomical terms are meant to designate in order to assure consistency in the representation of anatomy across all domains. We propose that RadLex should reserve its collection of anatomical terms to refer to anatomical reality, and then create a separate ontology for the image findings representing that reality. We therefore renamed anatomic location as the FMA root term Anatomical entityto reflect this agreement in the meaning.
Derivation of FMA-RadLex
An anatomy subset from the FMA can be derived either by obtaining an entire copy of the FMA and pruning the ontology down to the required specifications (de novo construction) or by mapping the existing terminology project to the FMA, carving out the ontology around the mappings and finally incorporating the derivatives into the target project. The latter method was applied in constructing the anatomy application ontology for RadLex. Although the operations were carried out manually in this project, current efforts are underway to implement both methods programmatically.
Anatomy taxonomy
The backbone of a sound ontology is a class or type subsumption hierarchy that propagates transitively through is_a relation the inheritance of properties and attributes that explicitly define a type and its subtypes. This assures unambiguous assignment of any radiology-related anatomical entity in the taxonomy. We applied this principled approach to RadLex by first mapping high level RadLex terms to the corresponding FMA terms, and then importing their corresponding FMA supertypes into the RadLex taxonomy. The following highest level RadLex anatomical structures, blood vessel, head, neck, abdomen, thorax, upper extremity, trunk, lower extremity, spine and nervous system all mapped to their FMA counterparts, which in turn provided their respective supertypes: Organ part (blood vessel), Cardinal body part (head, upper extremity and lower extremity), Cardinal body part subdivision (neck, trunk, thorax, abdomen), Organ system (nervous system) and Organ system subdivision (spine). Other terms at different levels of the RadLex tree mapped to the other FMA supertypes such as Organ (heart), Anatomical compartment (mediastinum), Anatomical cluster (inguinal canal), Portion of tissue (ependyma), Anatomical set (meninges), Anatomical space (peritoneal cavity), Anatomical surface (pleural surface), and Anatomical line (soleal line).
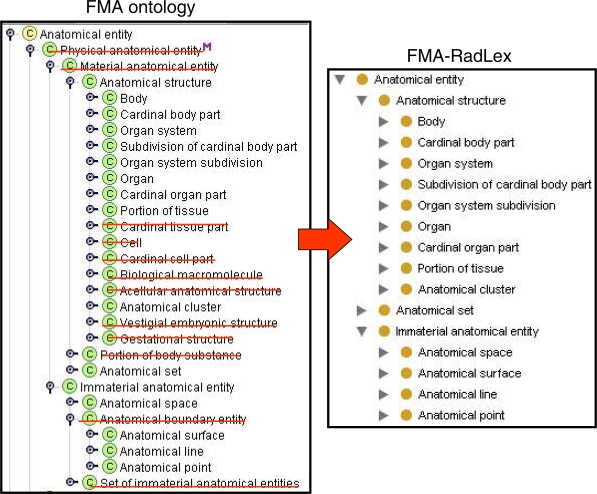
We harmonized the RadLex anatomy taxonomy in line with the top level types of the FMA by incorporating as well the highest level parents of the imported supertypes, namely; Anatomical structure which subsumes 3-D objects that have inherent shape, e.g. body, organ system, and organ, and Immaterial anatomical entity which encompasses types that have no mass property, such as Anatomical space, e.g lumen of artery, Anatomical surface e.g. surface of heart, Anatomical line, e.g. mid-clavicular line, and Anatomical point, e.g. apex of heart, (Figure 2, right).
Figure 2.
FMA-RadLex (right) derived from the FMA (left) by deletion (strikeouts) and addition of links as shown by the is_a link of Anatomical structure, which was deleted from Material anatomical entity but added to Anatomical entity.
To construct the same ontology via the de novo approach in an automated system, the operation would involve a series of deletion and addition of links (Figure 2, left). For example, in our model anatomical types representing microscopic entities which are not relevant to radiology such as Cell, Cardinal cell part, Biological macromolecule, Cardinal tissue part, are deleted from Anatomical structure. Removing unwanted anatomical types as intervening nodes in the tree and then splicing the remaining nodes in order to achieve the taxonomy specificity required by RadLex are carried out with the same deletion/addition operations. For example, the is_a link of the class Anatomical structure is deleted from Material anatomical entity and then added directly to Anatomical entity as this is a valid inferred relation, but now is made explicit consequent to deleting the intervening types. Both Physical anatomical entity and Material anatomical entity are then deleted from the taxonomy. This operation can be carried out in all levels of the hierarchical tree. Figure 2 displays how the high level FMA taxonomy is pruned down to RadLex’s requirements.
We retained most of the existing structural and non-structural relations in RadLex and assured that the separate implementation of the is_a and part_of relations in the select areas we examined conform to the definitions set forth in the OBO Relation Ontology (RO)7. As a result FMA-RadLex retains the traditional view of anatomy familiar to radiologists while endowing the application ontology with a sound ontological structure.
Discussion
The development of domain reference ontologies is an ongoing initiative aimed at promoting the integration and interoperability of the different and disparate biomedical data and systems. The main purpose of domain ontologies is to serve as the primary source for content and ontological framework from which modular or lightweight ontologies dedicated to specific applications for the particular domain can be derived and potentially can be shared with other application ontologies
We have demonstrated we can use a reference ontology to enhance and augment an existing application ontology, adding a robust semantic framework. Specifically, we augmented RadLex with a subset of FMA to enhance it with a semantic framework for structuring its domain knowledge—an improvement in its content that will enable it to provide the semantics for associating and correlating diverse radiology-related data. The RadLex anatomy application ontology was constructed by importing high level anatomical types from the FMA and enforcing throughout selected test areas is_a and part_of relations as separate hierarchies. RadLex anatomical entities of all sorts can now be properly re-assigned in the taxonomy according to their properties and attributes made explicit by the derived framework of the FMA.
Although our focus has been on RadLex and an application ontology for radiology, we believe the principles underlying our work are extensible and generalizable to other domains and application ontologies. Ultimately, a broader range of ontologies will benefit from reuse and harmonization with existing, robust reference ontologies. Our methods could provide a basis for creating or enhancing application ontologies.
Conclusion
While the mapping of reference and application ontologies to one another should optimally be accomplished by automated methods, our work on prototype ontologies shows that such correlations require ontological changes that must be effected before machine-based methods can be usefully implemented.
Acknowledgments
This work has been supported by the NIH grant HL087706.
Reference:
- 1.Menzel C. In: Reference Ontologies -Application Ontologies: Either/Or or Both/And? KI2003. Smith B, editor. Hamburg; Germany: 2003. [Google Scholar]
- 2.Rosse C, Mejino JLV. The Foundational Model of Anatomy Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. New York: Springer; 2007. pp. 59–117. [Google Scholar]
- 3.Langlotz CP. RadLex 2006. a new method for indexing online educational materials. Radiographics. 26:1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 4.Marwede D, Fielding M, Kahn T. Proc AMIA 2007. Chicago. IL: 2007. RadiO: A Prototype Application Ontology for Radiology Reporting Tasks; pp. 513–517. [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin DL. Creating and curating a terminology for Radiology: Ontology Modeling and Analysis. J Digit Imaging. 2007 doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noy NF, Mejino JLV, Musen MA, Rosse C. Pushing the envelope: challenges in frame-based representation of human anatomy. Data & Knowledge Engineering. 2004;48:335–359. [Google Scholar]
- 7.Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector A, Rosse C. Relations in biomedical ontologies. Genome Biology. 2005;6(5):R46.15. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]