Abstract
Many clinical reports contain family history, which is valuable information for clinical decision support and research. We developed a simple natural language processing algorithm to identify and extract family histories. The algorithm was tested on a set of discharge summaries and outpatient clinic notes. The precision and recall of extracting all diagnoses were 85.12% and 86.93%, respectively. The precision and recall of differentiating family history from patient history diagnoses were 96.30% and 92.86%, respectively. Both the precision and recall of exact family member assignment were 92.31%.
Introduction
A number of natural language processing (NLP) applications have been developed to extract key findings such as past and present diagnoses for point-of-care decision support as well as clinical research [1–5]. As part of the National Center for Biomedical Computing, Informatics for Integrating Biology & the Bedside (I2B2) [6], we developed an open-source and modularized natural language processing system: the Health Information Text Extraction (HITEx) System [7]. HITEx is a suite of open source NLP tools, written in Java, which builds on top of the General Architecture for Text Engineering (GATE) framework [8].
We have used HITEx to parse discharge summaries and outpatient visit notes [7]. One challenge which we encountered was that family history is sometimes mixed with a patient’s own history and diagnoses in the same section, paragraph, or sentence. In order not to report a patient’s family history as the person’s own diagnosis, it is necessary for us to differentiate the two types of information.
Also, while family histories may be considered false positives in the context of diagnosis extraction, they provide valuable, and sometime critical information for patient care and scientific research [9]. Breast cancer risk predication models, for example, often include family history of breast cancer as a key variable and as a surrogate for genetic information [10]. Therefore, it is not only necessary to distinguish family history from personal history, but also to capture the details of family history (e.g., grandmother and mother with hypertension, versus a cousin with hypertension).
To identify and extract family history information, we have developed a simple rule-based algorithm. For evaluation, this algorithm was applied to 350 sentences which were randomly selected from a set of 2,000 discharge summaries and outpatient clinic notes of the Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH).
Methods
The family history extraction process consists of three main steps: pre-processing, family member and diagnosis concept identification, and family member/patient assignment.
Pre-processing
First, clinical reports are split into sections (e.g., diagnosis, history, and medication), and section headings are coded using a locally developed taxonomy. Second, content of each section is tokenized and split into sentences. Third, noun phrases are extracted after part-of-speech processing. Fourth, noun phrases are mapped to Unified Medical Language System (UMLS) concepts [11].
In this study, all the pre-processing tasks are conducted using the existing HITEx components.
Family member and diagnosis concept identification
Because our main interest is to assign various diagnoses to the correct person (a family member or the patient), UMLS concepts that fall into these two categories (diagnosis and family member) are tagged as such.
Family member concepts are mainly identified by one UMLS semantic type: family group (T099).
We define diagnosis as a concept which belongs to one or more of 8 UMLS semantic types:
congenital abnormality (T019)
acquired abnormality (T020)
injury or poisoning (T037)
disease or syndrome (T047)
mental or behavioral dysfunction (T048)
cell or molecular dysfunction (T049)
anatomical abnormality (T190)
neoplastic process (T191).
Family member/patient assignment
In the last step, a set of rules is used to associate the diagnosis or group(s) of diagnoses with the most relevant family member or group(s) of family members. Besides family members and diagnoses, the rules employ 3 other high level annotations:
Conjunction – tokens that may indicate the end of a family history-related phrase, but may also be a part of such phrase. Examples of conjunctions include “,” (comma) and “and”.
Sentence boundary – tokens that identify the sentence boundaries, for example period (“.”).
Patient possession – a token or group of tokens that indicates with a high probability that the sentence describes patient, not family history diagnoses. For example: “patient had” or “patient has”.
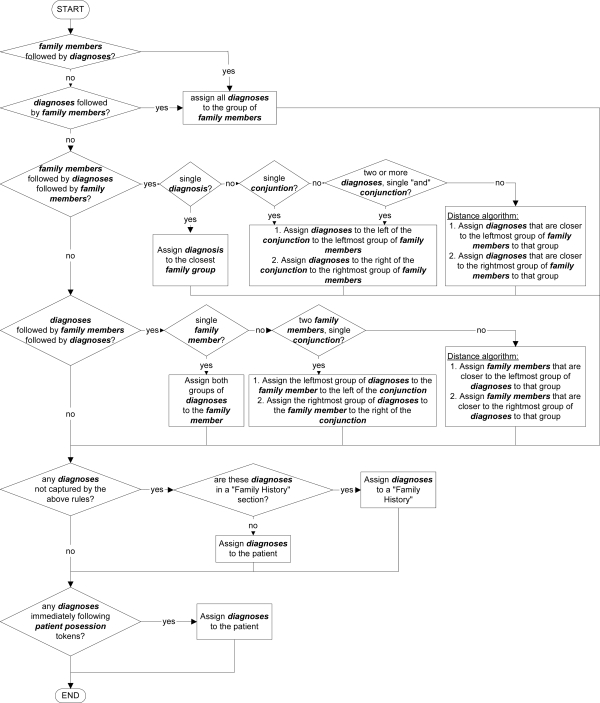
Tokens or concepts which are not classified as family member, diagnosis, conjunction, sentence boundary or patient, are ignored by the association rules. The rules are represented in Figure 1.
Figure 1.
The association rules used to assign diagnoses to family members.
Evaluation
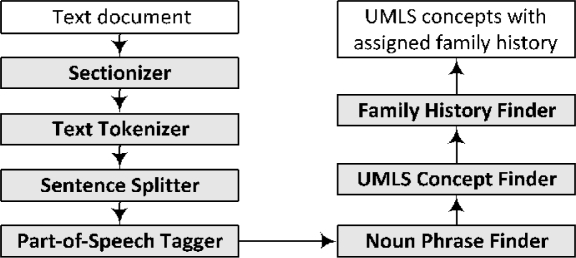
The family history identification and extraction algorithm was implemented in Java and as a GATE module. For evaluation, this module was used along with existing HITEx modules to form a family history extraction pipeline, shown in Figure 2.
Figure 2.
The pipeline for family history extraction.
A total of 2000 reports were randomly selected from the Partners Research Patient Data Registry (RPDR) [12]: 500 discharge summaries from Brigham and Women’s Hospital (BWH), 500 discharge summaries from Massachusetts General Hospital (MGH), 500 outpatient notes from BWH, and 500 outpatient notes from MGH. We included these two types of reports from two different hospitals to provide better coverage of different writing styles.
The reports were parsed by the family history extraction pipeline application. The application identified UMLS concepts that are diagnoses and assigned each concept to either patient or one or more family members. In this evaluation, only sections with history-related titles (e.g., “history”, “family history”, “history of present illness”, or “social history”) were used. (As a part of HITEx, we have identified more than 1000 section headers and mapped them to section categories, according to an internally developed taxonomy. Dozens of sections were categorized as history-related).
We randomly selected 350 sentences from the reports. A clinician (nurse) who is also an author on this paper performed the following review tasks: (1) identified diagnoses in each sentence; (2) decided whether each diagnosis was related to the patient or patient’s family; and (3) if a diagnosis was family related, decided which family members were involved. The sentences were presented along with the names of the document sections to provide the reviewer with some contextual information. In order to assess the inter-rater agreement, a second clinician (physician) who is not an author on this paper was asked to perform the same task on 100 sentences randomly selected from the 350 sentences. Both clinicians are familiar with the UMLS and were asked to consider concepts of the 8 UMLS semantic types that we previously described as diagnoses.
The diagnoses identified by the first reviewer were treated as the gold standard. We first calculated the precision and recall of the HITEx in extracting all diagnoses. For diagnoses identified by both the gold standard and HITEx, the precision and recall of the family history diagnosis identification were calculated. For those diagnoses that were identified by both the gold standard and HITEx as family history, the precision and recall of the specific family member assignment was calculated. Similarly, the diagnoses and family history statuses identified by the second reviewer were also compared to the gold standard, to provide us with a sense of inter-rater agreement.
For the purpose of evaluation, partial match (e.g. “old infarct of the right frontal region” and “old infarct”) with the gold standard was considered to be a match in the identification of diagnoses. We made this decision because the UMLS sometimes does not have an exact match of the concept (e.g. “old infarct of the right frontal region”) that the clinician identified. To simplify the analysis and review process, negation, temporal and other modifiers were not taken into account. The abilities of algorithm to distinguish patient history and family history and to assign diagnoses to a family member are independent of other modifiers. For example, correctly identified negated family history diagnosis is still a family history diagnosis (or absence thereof).
Results
In the 350 evaluated sentences, 375 diagnoses were identified by the gold standard and 383 concepts were found by HITEx. The gold standard and HITEx agreed on 326 concepts. Among these 326 diagnoses, the reviewer identified 28 family history related diagnoses, and HITEx identified 27. The gold standard and HITEx agreed on 26 concepts. Finally, out of the 26 agreed upon family history, 24 were given the same family member assignment by the gold standard and HITEx.
The HITEx’s precision and recall of identifying diagnoses were not very high (Table 1). This is consistent with our previously reported evaluation of HITEx’s ability to extract principal diagnosis, co-morbidity and smoking status [7], and is comparable to the results from a number of previous studies in the literature [1–3, 5]. Some of HITEx’s errors were caused by wrong part of speech tagging and noun phrase extraction, aggressive stemming, wrong disambiguation, etc. Majority of the HITEx’s errors in extracting diagnosis, however, were caused by the difference between what the HITEx/UMLS and the human reviewer considered to be a diagnosis. The boundaries between the findings and diagnoses as well as between the signs and symptoms and diagnoses are not always clear. The two human reviewers also disagreed sometimes about what is a diagnosis and what is not. For example, while the first reviewer considered “postmenopausal” and “unable to void” diagnoses, the other reviewer did not.
Table 1.
Precision, recall and F-measure for diagnoses and family history identification
| Precision | Recall | F-measure | |
|---|---|---|---|
| Diagnoses | 0.8512 | 0.8693 | 0.8602 |
| Family History | 0.9630 | 0.9286 | 0.9455 |
| Specific Family Member | 0.9231 | 0.9231 | 0.9231 |
When diagnoses were correctly identified, HITEx performed well in assigning diagnoses to patients or family members (Table 1). It achieved 96.30% precision and 92.86% recall in detecting family history diagnoses, and 92.31% precision and 92.31% recall in specific family member assignment. We found that errors in family history assignment were likely to occur in those sentences that were complex, with multiple groups of diagnoses or multiple groups of family members, e.g., “The cardiac risk factors included post-menopausal, hypertension, diabetes mellitus, questionable cholesterol, smoking, but no family history”.
When we used the first reviewer as the gold standard, the precision, recall and F-measure of the second reviewer were not very impressive (Table 2). In fact, the F-measure of the second rater was slightly lower than that of the HITEx. If we used the second reviewer as the gold standard and tested the first review against it, it would result in similar imperfect results. This reflects the significant inter-rate disagreement in defining diagnosis. On other hand, the two raters agreed perfectly on family history status and specific family member assignment.
Table 2.
The precision, recall and F-measure of the second reviewer in diagnoses and family history identification, when using the first reviewer as the gold standard.
| Precision | Recall | F-measure | |
|---|---|---|---|
| Diagnoses | 0.9063 | 0.7838 | 0.8406 |
| Family History | 1.0000 | 1.0000 | 1.0000 |
| Specific Family Member | 1.0000 | 1.0000 | 1.0000 |
Discussion
Family history is an important type of clinical information for patient care as well as scientific research. This paper presents a new algorithm for identifying and extracting family histories from free-text clinical reports.
The algorithm was evaluated using a set of discharge summaries and outpatient notes. First, the algorithm extracted all diagnoses (both patient and family). It achieved 85.12% in precision and 86.93% in recall. Second, it differentiated family history from patient history. At this step, the algorithm demonstrated a good ability to detect family history diagnoses (96.30% precision and 92.86% recall). Third, the algorithm assigned the diagnoses related to family history to specific family members, and achieved 92.31% precision and 92.31% recall. In this evaluation we didn’t examine the negation status or temporal status of diagnoses, which could be extracted by adding HITEx Negation Finder or Temporal Finder modules to the pipeline. The achieved performance is adequate for many information retrieval and secondary analysis tasks; however, the extracted family histories should not be relied upon as the sole data source in clinical practice.
A related prior study by Friedlin et al [13] reported high accuracy rate (sensitivity = 93% and positive predictive value = 97%) in the extraction of family histories. Friedlin’s study differs from ours in several respects: 1) it focused on sections clearly labeled as family history, thus did not need to differentiate family from patient history; 2) it classified family members as primary, secondary or unknown relatives, while we assign diagnoses to exact family members (e.g., father, mother, or sister); 3) although Friedlin’s algorithm was only described very briefly, it appears to be somewhat different from ours as we consider the use of conjunction words and symbols.
The prevalence of family histories in clinical records is relatively low (6.48%). To objectively evaluate the system’s ability to pick up family histories, we have to randomly select and review a large number of sentences that do not contain family histories. To acquire a number of family histories sufficient for a thorough evaluation, a manual review of thousands of sentences would be required, for which we did not have enough resources.
In the BWH and MGH’s discharge summaries and outpatient notes which we have analyzed, family history information is not always documented under sections clearly labeled as “family history”. We suspect this may not be a unique problem of the BWH and MGH reports. The ability to distinguish family history from patient history could help to reduce false positives when extracting a patient’s past and present diagnoses from such reports. It could provide valuable family history information for data mining and hypothesis testing.
A significant limitation of the algorithm is that it is error-prone when handling complex and ambiguous sentences. It also does not have the ability to resolve co-reference. In the evaluation, the human reviewers were provided with individual sentences along with the associated section headings, but no other context. In addition, we used one clinician’s review as the gold standard in the evaluation and found significant disagreement between the gold standard clinician and a second clinician in terms of diagnosis identification. Ideally, we would recruit more clinicians and create a more reliable gold standard by consensus.
We have incorporated the family history module into HITEx. For future work, we plan to further test and refine the module.
Conclusion
Family history information found in clinical reports is valuable for many applications such as breast cancer risk prediction. Correct identification and extraction of family history presents a challenge for NLP systems, given the heterogeneity of clinical reports. We have developed and tested a simple, rule-based algorithm to extract family history from clinical reports. The algorithm was moderately successful in extracting all diagnoses. It showed good ability in differentiating family history from patient history. It also achieved high accuracy in exact family member assignment.
Acknowledgments
This research was funded by NIH grant number U54 LM008748. We thank Drs. Shawn Murphy, Ross Lazarus, Susanne Churchill and Isaac Kohane for their advice and collaboration.
References
- 1.Chapman WW, Haug PJ. Comparing expert systems for identifying chest x-ray reports that support pneumonia. Proc AMIA Symp. 1999:216–20. [PMC free article] [PubMed] [Google Scholar]
- 2.Divita G, Tse T, Roth L.Failure analysis of MetaMap Transfer (MMTx) Medinfo 200411(Pt 2): 763–7. [PubMed] [Google Scholar]
- 3.Friedman C, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haug PJ, et al. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp. 1997:814–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Taira RK, Soderland SG. A statistical natural language processor for medical reports. Proc AMIA Symp. 1999:970–4. [PMC free article] [PubMed] [Google Scholar]
- 6.I2B2: Informatics for Integrating Biology & the Bedside[http://www.i2b2.org]2007
- 7.Zeng QT, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham H, et al. he TIPSTER Text Program (Phase III) 6 Month Workshop. Morgan Kaufmann; California: 1997. GATE – a TIPSTER-based General Architecture for Text Engineering. [Google Scholar]
- 9.Guttmacher AE, Collins FS, Carmona RH. The family history–more important than ever.[see comment] New England Journal of Medicine. 2004;351(22):2333–6. doi: 10.1056/NEJMsb042979. [DOI] [PubMed] [Google Scholar]
- 10.Gail MH, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually[see comment] Journal of the National Cancer Institute. 1989;81(24):1879–86. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 11.Humphreys BL, et al. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nalichowski R, et al. Calculating the benefits of a research patient data repository. AMIA Annu Symp Proc. 2006:1044. [PMC free article] [PubMed] [Google Scholar]
- 13.Friedlin J, McDonald CJ. Using a natural language processing system to extract and code family history data from admission reports. AMIA Annu Symp Proc. 2006:925. [PMC free article] [PubMed] [Google Scholar]