Abstract
We describe an application (“Medline Publications”) written for the Facebook platform that allows users to maintain and publish a list of their own Medline-indexed publications, as well as easily access their contacts’ lists. The system is semi-automatic in that it interfaces directly with the National Library of Medicine’s PubMed database to find and retrieve citation data. Furthermore, the system has the capability to present the user with sets of other users with similar publication profiles. As of July 2008, Medline Publications has attracted approximately 759 users, 624 of which have listed a total of 5,193 unique publications.
Introduction
In recent years, there has been a great deal of discussion regarding the problem of scientific collaboration, and it now seems clear that the days of researchers being able to restrict themselves to one narrow field of study are rapidly drawing to a close.1,2,3 In order to conduct truly translational research, we must work together with other researchers outside of our own fields of expertise— sometimes very far outside.4 The question, of course, is: how are we to identify, locate, and contact researchers outside of our respective fields or institutions who might be working on complementary projects? The volume of published research is such that keeping up with the latest work in our own field is a challenge, to say the least, and most research institutions are set up in such a way that their various research groups are unintentionally “siloed” from one another.
Fortunately, a second trend has been developing contemporaneously with this increased recognition of the importance of collaboration: social networking websites. Once the exclusive realm of the undergraduate student, sites such as Facebook (Palo Alto, CA) have been gaining increasingly wide acceptance among academics of all ages. These sites are designed to help their users find and maintain contact with one another, and also to identify users who share common interests or backgrounds.
Although the intended use of these features has historically been social in nature, there is no reason why they might not also prove useful for professional or scientific purposes.
This paper describes the “Medline Publications” (MP) Facebook application. MP leverages the Facebook platform to assist researchers in finding colleagues who may be working on similar or complementary projects. MP does this in three ways:
Enabling users to list their Medline-indexed publications on their “profile” page, thereby making them visible to other users;
Automatically displaying all of a user’s “friends”’ publications, thereby helping users to keep abreast of their colleagues’ work;
Connecting the user with other users who have similar publication profiles, thereby exposing the researcher to new potential collaborators.
Background
There are a variety of existing systems that act as specialized or enhanced interfaces to the National Library of Medicine’s Medline database of biomedical publications. Many of these systems have features that assist their users in identifying sets of related publications, or tools for suggesting topics based on a set of publications.5—10 Existing systems, however, typically model the problem from either a “publication-centered” or “topic-centered” point of view.11 Our system takes an “author-centered” approach, which we feel fits well with the holistic and collaborative nature of translational research as well as with the user-centric focus of most social networking websites.
There have been several attempts to use social networking technologies to facilitate scientific communication and collaboration, some of which have been commercial in nature (Community of Science, Nature Network) and others institution-specific.12,13,14 Irrespective of these systems’ relative advantages or disadvantages, one fact is clear: each such system that an individual participates in represents one more username and password to remember, one more website to check each morning, and one more online “profile” to remember to update.
The excessive cognitive load caused by participating in too many social networking systems has become a common enough phenomenon to warrant its own name: “social networking fatigue”.15 We believe that MP represents the first attempt to build scientific social networking into an existing and widely-used system. Given that many of our potential users already have Facebook accounts, our system’s barriers to entry are significantly lower than those of other scientific collaboration systems. Users need only to check one or two boxes to install and begin to use our system, and are spared the repetitive and seemingly endless process of entering form after form of personal information common to most new web applications.
System Architecture
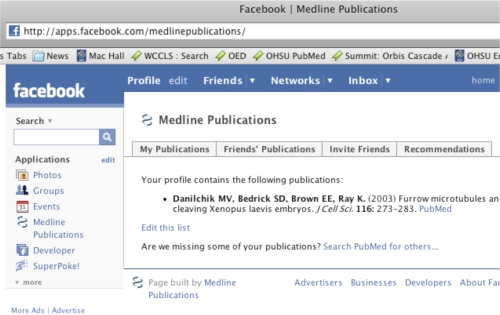
Applications written for the Facebook platform can seamlessly embed themselves into the standard Facebook user interface (see figure 1). Under the most common design pattern, this is accomplished by allowing the Facebook website to act as a proxy between the user and the third-party application. From the perspectives of both the user and the third-party application, all interactions that take place appear to be directly from and with Facebook. As a result of this architecture, developing a Facebook application is simultaneously easier and more difficult than traditional web application development. On the one hand, Facebook manages many of the infrastructure-level details associated with any web application: user authentication, session management, etc. On the other hand, many standard web programming techniques become much more complicated once a proxy layer is put in place, and it takes some time and patience to become accustomed to the particulars of working with the Facebook platform.
Figure 1.
The Facebook API allows third-party applications to embed themselves relatively seamlessly within the larger Facebook user interface.
MP is written in the Ruby programming language, and is built on top of the Ruby on Rails framework. It also makes extensive use of the BioRuby library,16 as well as the GNU Scientific Library.17 MP uses the National Library of Medicine’s Entrez E-Utils18 to integrate with the NLM’s PubMed database. MP also uses a PostgreSQL relational database to store user and publication data.
System Features
MP enables a user to build and manage a list of their Medline-indexed publications, and to make that list available to their contacts in the Facebook system. Unlike many publication management programs, which often require their users to manually enter the citations they wish to list, MP automates as much of the process as possible by integrating directly with PubMed. When a user first “installs” MP, our system uses Facebook’s Application Programming Interface (API) to obtain the user’s name, which it then uses to dynamically build a PubMed query. MP then uses the Entrez E-Utils to execute this query and retrieve a set of citations that (hopefully) includes the user’s own publications. MP then presents this list to the user, who then selects which publications they wish to add to their profile.
While this straightforward and simplistic approach does generally work, we discovered early on that it has several important limitations. Its efficacy depends heavily on two factors: how completely and accurately the user’s Facebook name matches the name under which they publish, and how common their name is in PubMed’s index. In practice, we have found that the vast majority of our users have registered with Facebook using their real, full name. However, there have been several users who reported difficulties in finding their publications due to name changes (from maiden to married names, for example). These cases have been relatively few and far between. The second limiting factor, however, presents a much more serious challenge: our name-based approach completely fails users whose names have high document frequencies in Medline (i.e., whose names or initials are quite common).
We considered several possible solutions to this problem, all of which involved including more information— academic affiliation, geographic location, etc.— in the dynamically-generated PubMed query. Unfortunately, limitations inherent to the Facebook platform prevented us from being able to reliably obtain these data from our users’ profiles. Many users simply never provide Facebook with this information, and among those users who do there is no way to tell how accurate it might be. Furthermore, as our user base increased, we discovered a third limiting factor: many of our users had published articles as part of a consortium, or under the name of their institute. Since the users’ names did not appear anywhere in these publications’ Medline records, our name-based query strategy would always fail to discover these citations.
Our intention had originally been for this part of system to be entirely automated. Upon examining the challenges presented by these limiting factors, however, we chose to include some manual publication-curation features. Users can now manually enter lists of Entrez PMID keys, or enter a custom query of their own devising. These tools have proven to be both popular and effective, and allow our users fine-grained control over their publication lists while still protecting them from the tedium of manually entering citations into the system.
Of course, the whole point of a social networking application is that users do not exist in a vacuum, but rather that they are part of a large inter-connected social graph. Facebook’s APIs allow third-party developers to access their users’ social graphs and interact with its various nodes. One way that MP uses these APIs is to provide users with easy access to the publication lists of any of their connections that are also MP users.
MP offers another social feature: a recommendation engine. This component of our system is intended to help users discover researchers or papers that they may not otherwise have encountered. As previously mentioned, our recommendations are author-centric rather than publication- or topic-centric. This means that we are truly matching users to other users, rather than directly to particular publications or to topic groups.
Our system’s algorithm uses NLM Medical Subject Heading (MeSH) terms as the basic unit of analysis. All Medline-indexed publications have a set of MeSH terms assigned to them at their time of entry, and our system maintains an index of users and their publications (and therefore their MeSH terms). Our assumption is that users working on complementary topics will have more MeSH terms in common than users who are working on completely unrelated topics.
Recommendation Algorithm
Our system builds and maintains a list of the MeSH terms that each of our users’ publications have been assigned by the Medline indexers, and computes frequency counts for each term. We then construct an n-dimensional term vector v for each user, where n is the size of the set of unique MeSH terms used by all of our users. For any given MeSH term t, vt contains the number of papers that the user published which were assigned t as a MeSH term. Given v, we can find similar term vectors (i.e., similar users) by calculating the angle between v and each of our users’ term vectors. The smaller the resulting angle, the more similar the users are.
Unfortunately, this naïve approach to calculating similarity turned out to be unacceptably slow, due in part to the large total number of MeSH terms used by our user base. Fortunately, we are far from the first programmers to attempt a recommendation system, and there exist several different straightforward approaches to the problem. The algorithm we employ is very similar to that used by classical Latent Semantic Indexing.19
Under our final algorithm, we combine the term vectors into an n × m matrix, where m is equal to our number of users. We then take the Singular Value Decomposition (SVD) of this matrix, which (among other things) enables us to approximate our original n × m matrix with an arbitrarily smaller one by using the first g eigenvalues of one of the components of the SVD. The fidelity of the approximation depends on how many eigenvalues we choose to retain; in practice, we have found that the first 30 eigenvalues are typically sufficient. Comparing hundreds of 12,000-dimensional vectors is time-consuming and unwieldy; comparing hundreds of 30-dimensional vectors is nearly instantaneous. Given a novel term vector, we simply project it into the lower-dimensional space derived from the SVD of our large matrix, compute angle distances, and pick the smallest ones. Another advantage to this SVD-based approach is that we can easily store and reuse the output of the expensive parts of the calculation, which improves our overall system performance greatly.
We have applied several modifications to the basic methodology described above. First and foremost, we exclude from consideration the MeSH terms with the highest document frequency (i.e., the most commonly used MeSH terms). For our current user base, we exclude “Humans”, “Animals”, “Female”, and “Male” from our recommendation analysis. Secondly, we normalize each MeSH term’s frequency count to between 0 and 1.0 across our entire user base. This helps to balance our recommendations: some users’ publication profiles were such that one particular MeSH term was overpowering their other MeSH terms to an undesirable degree.
The recommendation system is very much a work in progress, and represents one of our major future areas of work. We are currently planning several different evaluations that will help us to tune and improve the recommendation engine’s performance.
User Base
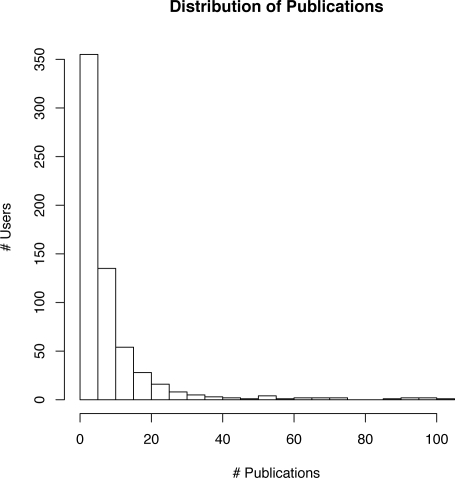
As of July 2008, approximately 759 Facebook users have added MP to their accounts. Of these, 624 have added a total of 5,193 unique publications to their profiles. Interestingly, there are a total of 5,414 publications currently in the database, which indicates some level of interconnectedness amongst our users (consult JE Andrews’ 2003 paper20 for a thorough discussion of the co-authorship network phenomenon). The distribution of publications has a definite skew (see figure 2), and ranges from 1 to 105. While the mean number of publications per user is 8.7, the median is only 5 and (standard deviation: ≈13).
Figure 2.
Distribution of publications among users. Note the highly skewed distribution and “long tail”.
The vast majority of our users (490,) have between 1 and 10 publications listed, and most (355) between 1 and 5. It is tempting to explain this finding as being the result of the demographics of Facebook as a whole: while it is not possible to positively determine our users’ ages, it seems plausible to suppose that younger researchers are over-represented within our application’s user base. That said, our application’s publication distribution generally follows the pattern predicted by Lotka’s law (a special case of Zipf’s law stating that, in any given field, the number of authors making n contributions is generally equal to 1/n2 of those making one contribution21). We are therefore hesitant to ascribe our skewed distribution to any particular demographic attributes of our user base.
Geographic location is similarly difficult to determine reliably using Facebook’s APIs. We are, however, able to obtain very coarsely grained location information, and can report that our user base extends throughout the United States, in both the private and the public sector. Furthermore, we have users in several European and Latin American countries, as well as in Australia.
System Limitations
Our system is clearly in its infancy, and has several important limitations. First and foremost among these is that it is only able to handle publications that are indexed in Medline. While this is a relatively minor limitation in our biomedical context, there are many potential users who do academic research in other fields. Several of these potential users have already requested that MP support other repositories such as the ACM Portal, Eric,22 HCI Bibliography,23 or IEEE Xplore. At the moment, support for non-Medline repositories is not on our roadmap; due to the fundamental design of MP, it would be a decidedly non-trivial undertaking to support other repositories. However, should enough users request such a feature, it might be worth investing the time.
In spite of this and other limitations, MP is currently quite functional, and has (anecdotally) proved professionally useful to several users. For example, one early beta-tester (DFS) unexpectedly met one of his “matches” at the recent HIMSS conference in Orlando, FL. After an initial awkward moment (think “online informatics predator”), they had a pleasant discussion regarding uses of clinical decision support for medication ordering.
Future development of the Medline Publication application
First and foremost, we wish to conduct formal evaluations of the system’s usability and functionality. This will involve engaging with our user base to identify missing features, and to optimize our recommendation algorithm. Luckily, the fact that our application runs within the larger context of Facebook means that we have a variety of tools at our disposal for identifying, coordinating, and following up with possible research subjects.
A second axis of future research lies within our user base itself. Our application represents a window into the social and professional lives of several hundred biomedical researchers at all career stages and in several countries. We know what they have published and with whom, and, for many of them, we know at least something about their academic or professional affiliations. Furthermore, we have access to their social graphs, which could help us discover useful social patterns within our user base. This data set is a potentially valuable source of information about how modern researchers carry out their work. One of our goals going forward is to explore ways to mine this rich vein of data while still respecting and protecting the privacy of our users.
Conclusion
In many ways, our seemingly simple reference management application finds itself to be, in essence, a laboratory for exploring many of today’s most compelling research questions: the power and promise of social networking; the temptations and pitfalls of dual purpose data; the challenges of enabling translational research. By examining these issues within the controlled and relatively small environment of a Facebook application, perhaps we will discover useful principles and techniques for use in the wider world.
Acknowledgments
We gratefully acknowledge the beta-testing help of David Dorr, Adam Wright, and Joshua Richardson. This work was funded in part by NLM Training Grant T15LM007088.
References
- 1.Zerhouni EA. US biomedical research: basic, translational, and clinical sciences. JAMA. 2005 Sep;294(11):1352–8. doi: 10.1001/jama.294.11.1352. [DOI] [PubMed] [Google Scholar]
- 2.Reece EA. A clarion call for translational and collaborative research. Am J Obstet Gynecol. 2006 May;194(6):1507–9. doi: 10.1016/j.ajog.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Sung NS, Crowley WF, Genel M, Salber P, Sandy L, Sherwood LM, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003 Mar;289(10):1278–87. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan B, Brennan PF, Dowling AF, Friedman CP, Peel V. Toward an informatics research agenda: key people and organizational issues. JAMIA. 2001 Dec;8(3):235–41. doi: 10.1136/jamia.2001.0080235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas SM, Montelione GT, Gerstein M. Pub- Net: a flexible system for visualizing literature derived networks. Genome Biol. 2005 Jan;6(9):R80. doi: 10.1186/gb-2005-6-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannidis JPA, Bernstein J, Boffetta P, Danesh J, Dolan S, Hartge P, et al. A network of investigator networks in human genome epidemiology. Am J Epidemiol. 2005 Aug;162(4):302–4. doi: 10.1093/aje/kwi201. [DOI] [PubMed] [Google Scholar]
- 7.Newman ME. The structure of scientific collaboration networks. Proc Natl Acad Sci USA. 2001 Jan;98(2):404–9. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smalheiser NR, Torvik VI, Bischoff-Grethe A, et al. Collaborative development of the Arrowsmith two node search interface designed for laboratory investigators. J Biomed Discov Collab. 2006 Jan;1:8. doi: 10.1186/1747-5333-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smalheiser N, Zhou W, Torvik V. Anne O’Tate: A tool to support user-driven summarization, drill-down and browsing of PubMed search results. J Biomed Discov Collab. 2008 Feb;3(1):2. doi: 10.1186/1747-5333-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu W, Yesupriya A, Wulf A, et al. An automatic method to generate domain-specific investigator networks using PubMed abstracts. BMC medical informatics and decision making. 2007 Dec;7:17. doi: 10.1186/1472-6947-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Synnestvedt MB, Chen C, Holmes JH. CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annual Symposium proceedings. 2005 Jan;:724–8. [PMC free article] [PubMed] [Google Scholar]
- 12.Schleyer T, Spallek H, Butler B, Kelleher C, Johnson S. Online Communities for Translational Research. AMIA Spring Symposium. 2007 [Google Scholar]
- 13.Cohen D. Facebook for scientists? BMJ. 2007 Aug;335(7616):401 EP. [Google Scholar]
- 14.Johnson C. A survey of current research on online communities of practice. The Internet and Higher Education. 2001 Jan;4(1):45–60. [Google Scholar]
- 15.Lee E. Social sites becoming too much of a good thing. San Francisco Chronicle. 2006 Nov 2;:A–1. [Google Scholar]
- 16.Goto N, Nakao M, Kawashima S, Katayama T. BioRuby: open-source bioinformatics library. Genome Informatics. 2003 Jan;:14, 629–630. [Google Scholar]
- 17.Free Software Foundation GNU Scientific Library. http://www.gnu.org/software/gsl/
- 18.National Library of Medicine Entrez E-Utils. http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html [DOI] [PubMed]
- 19.Berry M, Dumais S, O’Brien G. Using Linear Algebra for Intelligent Information Retrieval. SIAM Review. 1995 Jan;37(4):573–595. [Google Scholar]
- 20.Andrews JE. An author co-citation analysis of medical informatics. JMLA. 2003 Jan;91(1):47–56. [PMC free article] [PubMed] [Google Scholar]
- 21.Lotka A. The frequency distribution of scientific productivity Journal of the Washington. Academy of Sciences. 1926 Jan;16(12):317–32. [Google Scholar]
- 22.Institute of Education Studies US Dept. of Education ERIC: Education Resources Information Center. http://www.eric.ed.gov
- 23.Perlman G. The HCI Bibliography Project. SIGCHI Bulletin. 1991;23(3):15–20. [Google Scholar]