Abstract
Objective
: This study evaluated natural language processing methods to extract clinical data from free text in surgical reports related to cardiac pacing and defibrillation in order to populate a registry.
Methods
: The information extraction system that we have developed is a name entity recognition system based on GATE using regular expressions. 232 reports were analyzed. For each report, we performed manual abstraction, we collected the information stored in the registry, and we performed information extraction with our system. Sensitivity, positive predictive value and accuracy were used to evaluate our method.
Results
: Our system extracted information, including numeric data, text and combination of numbers and strings, with a high sensitivity (>90%) and very high predictive positive value (>95%). It featured a precision that was higher than the precision of the original registry database populated by manual input.
Conclusion
This tool based on GATE open source components provides a robust approach to extracting information from documents related to a specific narrow domain such as pacemaker reports. Further evaluation is needed for application to broader domains.
Introduction
With more than 500,000 pacemaker implantations worldwide per year, cardiac resynchronization therapy alone or combined with an implantable defibrillator is a highly promoted medical treatment. The number of patients that underwent permanent pacemaker implantation has increased due to novel indications and socio-economic factors [1]. Device failures, as well as increasing global costs, led to the establishment of national and international registries to achieve quality assurance and improve patient safety [2–4]. As many others, the French national registry is a multi-centric electronic repository that collects information for each device implantation [5], including information about devices (e.g., serial number), clinical contexts (e.g., disease), and procedures (e.g., pacing mode) Data entry in registries relies on electronic forms and manual data input. It is independent from clinical follow-up and therefore remains labor intensive and costly, and exhaustiveness is an important and unsolved issue. On the other hand, most clinical data are stored in free text and Natural Language Processing (NLP) may be used to extract information from text [6] and avoid multiple entry of the same information.
Several recent NLP systems have shown promising results in extracting information from medical narratives [7–10]. For example, to extract information from pathology reports, which contain poor narration and tabular data (such as pacemaker implantation reports), a preprocessor was integrated to MedLEE [7,11,16]. In a recent paper, Turchin used regular expressions (a metalanguage which describes string search patterns), to extract numeric data form free-text [12].
The objective of this paper is to study whether automated information extraction from free-text clinical reports may be used to help populate a pacemaker registry database. More precisely, we developed a system based on a limited set of rules and regular expressions to extract information related to the patients, the medical devices, and the procedures. The information extracted by our system was then compared to the information extracted by an expert and to the data stored in the registry.
Methods
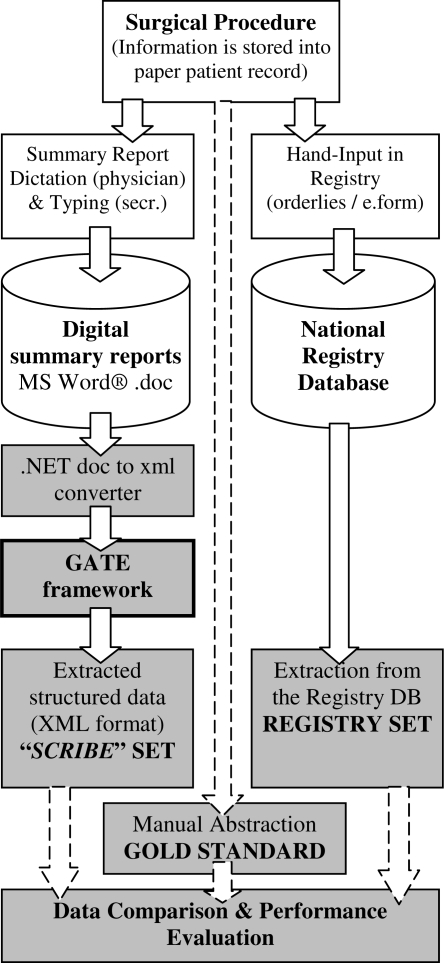
This study compares the information extracted from medical narratives with the structured data stored in a national registry (Fig. 1).
Figure 1.
Study Workflow (grey) based on Clinical (summary reports) and Registry Workflow
Corpus
We built a corpus containing all the reports produced at the Department of Cardiology, Rennes University Hospital for patients that were implanted with a pacemaker or a defibrillator during year 2005. A total of seven cardiologists dictated those documents and all documents had approximately the same structure and contained the same items. All the reports were stored in Microsoft Word® 97/2000 format. This corpus is referred as the discharge summary set or ‘SCRIBE’ set (Fig 1).
For each SCRIBE report, we extracted the corresponding dataset from the French National Registry of Pacemakers. The records in this database contain data manually entered by nurses using an online electronic form. This set is referred as the registry set.
Discharge summaries and registry records were manually checked for patient linkage and the data present within a single source were excluded from analysis.
Data Categories
A pacemaker expert (cardiologist) defined a list of 6 categories of relevant data from both sources:
Patient name (text)
Cardiologist name (text)
Device model (mixed text and number string, limited number of values)
Manufacturer name (brand name) of the implanted pacemaker pulse generator (mixed text and number string, limited number of values)
Pulse generator Serial number (mixed text and number string, unlimited number of values)
Device Stimulation Mode (NASPE international nomenclature [13], composed of up to five letters codes - e.g., AAI, DDDR, DDDRV).
Data Extraction Method
The information extraction system that we have developed is based on GATE (General Architecture for Text Engineering) [14], which is an open-source NLP framework. GATE modules are called CREOLE modules, CREOLE standing for Collection of REusable Objects for Language Engineering and include tokenisers, sentence splitters, and gazetteers. Among these modules, we used three components:
a non-language-specific Tokeniser
a Lexicon matcher (Gazetteer)
JAPE grammar, used to support regular expressions matching. JAPE grammar is a java compatible language used in GATE to apply rules to text annotations. Annotations can be made by other CREOLE components (e.g., Gazetteer), but also by the grammar itself.
Algorithm:
We randomly selected four reports from the SCRIBE dataset. These reports were manually analyzed to create the rules in JAPE grammar, using regular expressions. The rules were based on the syntactic and/or semantic co-occurrence patterns found in these four documents and on domain knowledge. For example, the possible values for Device Stimulation Mode include AAI, DDDR, DDDRV, and a matching pattern can be easily created; alternatively, many named entities were found nested in surrounding fixed words, which can be used as “hook” words in rules.
Two additional steps were necessary to use GATE in SCRIBE annotation workflow. First, Word® documents were converted into xml documents using .NET technology to be compatible with the GATE format. Second, the xml GATE files were parsed to extract the strings assigned to each category.
Evaluation
For each procedure mentioned in the corpus, we performed manual abstraction from the patient paper record (PR). Manual abstraction was then compared to the data extracted using SCRIBE, and to the data stored in the Registry database, to determine the status of the extracted items: True Positive (TP, correct information), False Positive (FP, wrong information), or False Negative (FN, missing data). Every FP was classified either human correctable or un-correctable, depending on the ability for the expert to infer the correct data from the extracted data (i.e. misspelled manufacturer name was correctable whereas incorrect serial number was not).
We measured sensitivity (Se = TP/[TP+FN]), positive predictive value (PPV = TP/[TP+FP]) and accuracy (Acc = TP/Total) values and compared them for SCRIBE and Registry extractions.
Results
287 SCRIBE clinical reports were obtained. 248 records were stored in the registry (year 2005). A total of 303 procedures were present in at least one source. 55 were only present in the SCRIBE set, whereas 16 were found in the registry but had no corresponding SCRIBE report.
Therefore, a total of 232 unique free-text files linked to registry entries were analyzed. Comparing to gold standard (PR), the sensitivity with SCRIBE was higher than the Registry for 4 categories and lower for 2. The positive predictive value of all 6 categories was higher with SCRIBE than in the registry. Accuracy was higher with SCRIBE for 4 categories out of 6. It was lower for Stimulation Mode and equivalent for Pulse Generator Manufacturer Name. The results are summarized in Table 1, with details in Table 2.
Table 1.
Sensitivity, Positive Predictive Value and Accuracy (SCRIBE vs. registry)
| Sensitivity
|
Positive predictive value
|
Accuracy
|
||||
|---|---|---|---|---|---|---|
| SCRIBE | Registry | SCRIBE | Registry | SCRIBE | Registry | |
| Patient Name | 100.0 | 100.0 | 99.1 | 93.5 | 99.1 | 93.5 |
| Cardiologist Name | 100.0 | 100.0 | 99.1 | 19.0 | 99.1 | 19.0 |
| Pulse Generator Manufacturer Name | 93.1 | 99.5 | 99.5 | 93.1 | 92.7 | 92.7 |
| Pulse Generator Model Name | 99.6 | 98.7 | 98.3 | 97.4 | 97.8 | 96.1 |
| Serial Number | 92.4 | 98.9 | 96.7 | 81.7 | 89.7 | 81.0 |
| Stimulation Mode | 56.0 | 100.0 | 100.0 | 80.2 | 56.0 | 80.2 |
| Mean rates for all six categories | 90.2 | 99.5 | 98.8 | 77.5 | 89.1 | 89.8 |
Table 2.
Number and kinds of errors (false positives and false negatives) for both workflows
| Item Category | # TP in both | SCRIBE # FP | Registry # FP | SCRIBE # FN | Registry # FN |
|---|---|---|---|---|---|
| Patient Name | 217 | 2 | 15 | 0 | 0 |
| Cardiologist Name | 43 | 2 | 188 | 0 | 0 |
| PG Manuf. Name | 198 | 1 (correctable) | 16 (9 correctable) | 16 missing values | 1 |
| PG Model Name | 81 | 4 (3 correctable) | 6 (4 correctable) | 1 | 3 |
| Serial Number | 164 | 7 (4 correctable) | 42 (32 single character error) | 5 missing values (files) + 12 failures of the rule | 2 |
| Mode | 84 | 0 | 46 wrong values | 102 missing values (files) | 0 |
Discussion
This study evaluates the feasibility and performance of a named entity recognition system based upon GATE and relying mostly on regular expressions for automated abstraction of selected information from free-text summary reports, in comparison to manual input in an electronic form. A limitation of this study is that the reference standard used in the evaluation step was created by a single domain expert. SCRIBE software achieved accuracy rates that are equal or superior to manual input by healthcare professionals for all categories except one (89 to 99.1 vs. 19 to 96.1), and higher positive predictive value for all categories. Sensitivity was generally higher in registry data, but equal for 3 categories. Other authors reported similar results in similar contexts, namely with ad hoc methods applied to narrow domains. Regular expressions achieved 90–98% sensitivity and comparable specificity on arterial blood pressure numeric data, but lower accuracy than SCRIBE [12]. Barrows [15] found that an ad hoc pattern matching method used upon ophthalmology visit notes had a better recall than a standard version of MedLEE, and 98% precision (vs. 100% for MedLEE). However, the combination of MedLEE and a pre-processor applied to pathology reports achieved rates equivalent to SCRIBE [16]. In [17], the authors reported 100% positive predictive value in the extracting of numeric data after using postprocessing algorithms. As in our approach, sensitivity was lower than positive predictive rate.
Errors belong to four categories:
Missing values (FN) in text reports represented 123 out of 139 FN cases for SCRIBE.
Typing errors (e.g., five occurrences of “5” instead of “S” in the registry) are responsible for 42 errors in the registry whereas only few typing errors occurred with SCRIBE (clerical personnel).
For the Cardiologist Name, a default value existed in the registry electronic form. This item was almost never correctly modified by health professionals, which caused 188 FP errors in the registry. For Stimulation Mode, 46 wrong values in the registry came from the fact that the correct mode could not be represented in NASPEE nomenclature, as this mode (AAI Safe R) is a modern combination of two modes (“AAI” and “DDD”), which could not be represented as a 5 letter code). This is a classic shortcoming of rigid systems.
12 mistakes were due to grammar failure (for example, “0” replacing “O” caused a grammar rule to mismatch). One regular expression in the SCRIBE algorithm is responsible for all these errors. They are due to the presence of free-text comments in twelve reports, which modified the sentence about the serial number. Because of the small training set, such cases were not detected.
We used GATE as a development platform with three major benefits:
GATE is an open source framework, within which many multi-language NLP modules are already implemented and can be reused to save development resources.
Although our approach relies on term matching and on a regular expression based grammar, GATE possesses the capability to handle a more complex medical language processing system. HITEx, a NLP tool designed in Boston Harvard Medical School uses GATE to process medical narratives inthe field of asthma with promising results [18].
The graphical development environment and the edition of the rules needed no preliminary programming skills, and it was possible to train a domain expert to create the regular expressions. The whole process was reduced to less than one month for pacemaker implantation reports, which is tremendously faster than the conception of more complex NLP systems.
Recent studies have explored different algorithms for automated extraction of data and especially numeric patterns [17], and a recent publication uses ad hoc regular expressions with interesting results in term of performance and savings in design time [12]. Our method shares several advantages and shortcomings of these techniques, but differs in some ways:
Grammar rules were successfully developed directly by a domain specialist, and only very few rules (14 rules) were necessary to extract relevant data.
Our approach specifically addressed a corpus of similar documents. These documents contained free-text sequences but hardly long narrative parts and these may need more complex NLP systems to be processed. Also, the system did not have to manage negative sentences, which need additional algorithms, e.g. [19, 20]. Generalizability to other fields is also questionable, since the domain studied was narrow, and variability in document is low. These features might be necessary conditions to obtain high performance despite a low number of rules.
This study was conducted on the basis of an existing workflow consisting in one data input for the clinical reports, and another one for the registry. We noticed that 55 records were missing in the registry. Compared to the 303 procedures performed during 2005, the summary report corpus reached an exhaustiveness rate of 95%, when registry contained data for only 82% implantations, despite the legal constraint for data submission. Moreover, the registry included both undetected and uncorrectable mistakes (more than 85% of the records). Therefore, in this study we have shown that the text corpus based approach performs better than current manual coding to populate the registry.
Conclusion
This method extracted information from free-text reports in the field of pacemaker implantation with adequate accuracy, and required only open source resources and few weeks of domain specific development. This approach could potentially replace tedious manual input of data to populate registries.
Acknowledgments
This work was supported in part by a grant from Medtronic and Boston Scientific.
The authors thank Pierre Zweigenbaum and Vincent Claveau for their comments.
References
- 1.Apkon M, Singhaviranon P. Impact of an electronic information system on physician workflow and data collection in the intensive care unit. Intensive Care Med. 2001;27:122–130. doi: 10.1007/s001340000777. [DOI] [PubMed] [Google Scholar]
- 2.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 3.Barrows RC, Busuioc M, Friedman C. Limited parsing of notational text visit notes: ad-hoc vs. NLP approaches. Proc AMIA Symp. 2000:51–55. [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–288. [PMC free article] [PubMed] [Google Scholar]
- 6.Mendonça Eneida A, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform. 2005;38:314–321. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Taira RK, Soderland SG. A statistical natural language processor for medical reports. Proc AMIA Symp. 1999:970–974. [PMC free article] [PubMed] [Google Scholar]
- 8.Tange HJ, de Hasman A, F P, Schouten HC. Medical narratives in electronic medical records. Int J Med Inform. 1997;46:7–29. doi: 10.1016/s1386-5056(97)00048-8. [DOI] [PubMed] [Google Scholar]
- 9.Turchin A, Kolatkar NS, Grant RW, Makhni ML, Pendergrass EC, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc. 2006;13:691–695. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voorham J, Denig P. Computerized extraction of information on the quality of diabetes care from free text in electronic patient records of general practitioners. J Am Med Inform Assoc. 2007 doi: 10.1197/jamia.M2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Hua, Friedman C. Facilitating research in pathology using natural language processing. AMIA Annu Symp Proc. 2003:1057. [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Qing T, Goryachev S, Weiss S, Sordo M, Murphy SN, Ross L. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham H, Humphreys K, Gaizauskas R, Wilks Y. GATE – a TIPSTER-based General Architecture for Text Engineering in he TIPSTER Text Program (Phase III) 6 Month Workshop. Morgan Kaufmann; California: 1997. [Google Scholar]
- 14.Cunningham H, Maynard D, Bontcheva K, Tablan V. GATE: A Framework and Graphical Development Environment for Robust NLP Tools and Applications. Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL'02); Philadelphia. July 2002. [Google Scholar]
- 15.Honigman B, Lee J, Rothschild J, et al. Using computerized data to identify adverse drug events in outpatients. J Am Med Inform Assoc. 2001;8(3):254–66. doi: 10.1136/jamia.2001.0080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12(4):448–57. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essin DJ. Intelligent processing of loosely structured documents as a strategy for organizing electronic health care records. Methods Inf Med. 1993 Aug;32(4):265–8. [PubMed] [Google Scholar]
- 18.Mutalik PG, Deshpande A, Nadkarni PM. Use of general purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc. 2001;8(6):598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]