Abstract
As Electronic Healthcare Records become more prevalent, there is an increasing need to ensure unambiguous data capture, interpretation, and exchange within and across heterogeneous applications. To address this need, a common, uniform, and comprehensive approach for representing clinical information is essential. At Partners HealthCare System, we are investigating the development and implementation of enterprise-wide information models to specify the representation of clinical information to support semantic interoperability. This paper summarizes our early experiences in: (1) defining a process for information model development, (2) reviewing and comparing existing healthcare information models, (3) identifying requirements for representation of laboratory and clinical observations, and (4) exploring linkages to existing terminology and data standards. These initial findings provide insight to the various challenges ahead and guidance on next steps for adoption of information models at our organization.
INTRODUCTION
The value of healthcare information interoperability and reuse is widely recognized1,2. Disparate systems and applications store clinical information in different formats and data structures, and support different types of access, querying, and update mechanisms. Furthermore, critical clinical knowledge such as decision support logic and documentation templates are hard-coded and embedded into clinical information systems. This can lead to high costs of implementing interoperable systems on one hand, and supporting maintenance and change of clinical knowledge on the other. As Partners HealthCare System (PHS) moves towards a Service-Oriented Architecture (SOA) to achieve flexibility, agility, and interoperability, there is a need for a common representation and standards to specify the structure and semantics of clinical information for unambiguous data capture, interpretation, and exchange across the enterprise.
In response to these needs, we are investigating the development and implementation of information models to specify the representation of clinical information to support semantic interoperability across the enterprise. Our overall goals include studying evolving information modeling efforts (e.g., Intermountain Health Care’s detailed clinical models3–6, HL7 Reference Information Model [RIM]7, and openEHR8) to determine if any could be adopted or adapted to meet our requirements, ensuring a tight coupling with Terminology Services, and defining the role of a runtime Information Model Service. In the initial phase of the project, the main objectives were to identify a systematic process for model development, gain a better understanding of information models and related efforts, investigate the requirements for representing laboratory and clinical observations, examine the use of terminology and other standards, and gain insight to the various challenges. In this paper, we provide a summary of initial experiences and findings with respect to these objectives.
BACKGROUND
Clinical systems should be able to represent and reason with information in a consistent and standardized manner. The interoperability of clinical systems will be limited if different conceptualizations and representations are adopted. Of concern is the capability to represent the same information in different ways across systems. For example, information could be represented in a pre-coordinated fashion (e.g., single name-value pair for “left patellar deep tendon reflex intensity is 2+”) or post-coordinated style (three name-value pairs for “deep tendon reflex intensity is 2+”, “body part is patella”, and “laterality is left”)4. Another example demonstrates the variety of ways for capturing results of lung auscultation where one representation could be finding-focused (e.g., “wheezing was found in the right upper lobe”) while another may be location-focused (e.g., “the right upper lobe has wheezing …”)4. In these cases, a formal representation of the information and use of standardized terminologies could enable systems to recognize the equivalent representations while allowing for flexibility and extensibility to accommodate the variations in use and presentation3–5
An information model specifies the representation of information, typically entities, their relationships, and constraints in a given domain. Information models can be viewed as technology-independent specifications that provide a single common representation of information, and can support the different needs of applications through mapping or transformations. Several efforts describe the need for coordinated creation and adoption of models and standard terminologies to support primary and secondary uses of data (e.g., in the longitudinal EHR, real-time patient-specific decision support, sharing of data internally and externally, data aggregation, and research)5,6.
MODEL DEVELOPMENT PROCESS
Our process of developing and implementing information models is iterative and incremental. The key phases we have identified are as follows (where some are analogous to those in the software development process and others are focused on issues specific to information models):
Review of Existing Models – analyze and compare existing healthcare information modeling efforts and their respective models
Model Creation and Evolution – use an iterative process for creating and evolving models based on exemplars, use cases, and requirements
Model Alignment – link elements of the model with various healthcare standards including terminologies, data types, and units
Model Validation – test the model with additional examples and use cases to determine if it supports the various requirements
Model Implementation – identify and implement aspects of the model in the context of an application and technology platform
Model Evaluation – after implementation, determine if the model meets the needs of the applications
At each phase, gaps and weaknesses in the model (e.g., mis-alignments and inconsistencies) and lack of support for key requirements (e.g., functional, business, and application) are determined. This results in the next iteration of enhancing the model.
The following sections report on our methods and early results related to the first three phases: review and comparison of existing models, requirements for creating an initial information model for laboratory and clinical observations (Observation Model), and alignment to existing standards.
REVIEW OF EXISTING MODELS
To identify existing or evolving healthcare-related information models in the field, we examined related work by standards bodies, academic groups, organizations, healthcare institutions, and industry in the United States and internationally. Based on this review, an initial set of information models and terminologies (and their underlying models) were selected for further investigation. The information models included HL7 RIM, openEHR, IHC detailed clinical models, and previous modeling efforts at our organization; the terminologies included LOINC9, SNOMED CT10, ICNP11, and CCC12. Each of the information models and terminologies was reviewed and initial lists of key features were created to reflect what to consider for model development. Table 1 summarizes the features along with a description or examples for the information models reviewed.
Table 1.
Base Set of Key Features of Information Models
| Feature | Description or Example(s) |
|---|---|
| Primary Focus | e.g., data exchange, EHR modeling |
| Domain(s) Modeled | e.g., specific or various domains |
| Multi-Level Modeling | e.g., metamodel, information model, data model |
| Formalisms Used | e.g., XML, UML |
| Linkage to Standards | e.g., LOINC, SNOMED CT, HL7 |
| Constraints | Support for constraints or rules |
| Compositionality | Support for post-coordination or composition of information |
| Change management | Existence of mechanisms for change management and versioning |
| Tools | Tools used, adopted, or developed for various aspects of the modeling process |
| Status | e.g., under development, needs extensions, in production use |
INITIAL OBSERVATION MODEL
One of the objectives of the project was to determine if any of the existing models reviewed in the previous phase could be used or adapted in order to meet local requirements. As a prerequisite to this, we examined the types of content and data that need to be represented for laboratory and clinical observations. Our initial approach was bottom-up where we used a set of exemplars, use cases, and understanding of various requirements (e.g., business, functional, and application) to drive an initial structure for observations. In this section, two of the examples (Blood Pressure and Hemoglobin A1c) are used to highlight the features of observations. The general use cases include supporting clinical decision support, clinical documentation, and knowledge management activities across the enterprise.
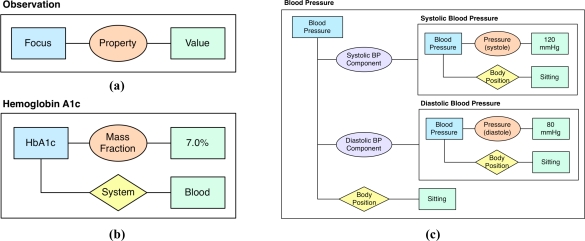
Based on analysis of the examples, we identified a base set of entities, relationships, and constraints for representing observations (Observation Model). Each observation may be viewed as a Focus-Property-Value triplet where focus is the clinical phenomenon being observed, property is the feature or property being measured, recorded, or observed, and value is the value of the feature or property (Figure 1a). For instance, a systolic blood pressure observation could be represented as: focus is blood pressure, property is pressure of systole, and value is 120 mmHg.
Figure 1.
Observation Model. An Observation represented as a Focus-Property-Value triplet (a); Hemoglobin A1c represented as an atomic observation with a context property of “System” (b); Blood Pressure represented as a compound observation consisting of two atomic observations as components for systolic and diastolic blood pressure. In this example, each observation has a context property of “Body Position” with the value “Sitting”. A constraint may be defined that specifies that the values of these context properties for the components must equal the value for the main compound observation (c). (Focus = blue rectangle, Observed Property = orange oval, Component Property = purple oval, Context Property = yellow diamond, Value = green rectangle)
In addition to the core Focus-Property-Value triplet, an observation may be associated with a set of contextual information and constraints that are specific to a particular type of observation and need to be defined accordingly. This may include information that refines the meaning of the observation or provides additional information (referred to as “qualifiers” by some efforts10). For example, systolic blood pressure can be qualified with a body position such as “sitting” where this context property would attach to the focus of the observation. Constraints are another feature that specify restrictions to elements or relationships between the elements of observations such as the properties and values (e.g., a constraint could limit the values that the context property of position can have for a blood pressure observation).
Observations may be distinguished as either atomic or compound. An atomic observation measures a single indivisible property (e.g., single laboratory test or diastolic blood pressure) and may be associated with zero or more context properties and constraints (Figure 1b). A compound observation consists of at least two or more component observations (atomic or compound) where the compound observation and each of its components may be associated with zero or more context properties and constraints. For example, blood pressure is a compound observation consisting of two atomic observations as components, systolic blood pressure and diastolic blood pressure (Figure 1c). In the former, the property is specialized as a component property and in the latter case as an observed property.
ALIGNMENT WITH STANDARDS
One of the key features of information models is linkage to existing standards. We have initially examined alignments with the following terminology and data standards given their current use at our institution:
LOINC9: a database of universal identifiers for laboratory and other clinical observations. Each LOINC record corresponds to an observation and includes six major axes for: component/analyte, kind of property, time aspect, system/sample, type of scale, and type of method.
SNOMED CT10: a multi-axial, hierarchical classification system for healthcare consisting of concepts, hierarchies to organize the concepts, and relationships to link the concepts. Among the 19 top-level hierarchies are observable entity for representing a question or procedure that can produce an answer or a result, clinical finding for representing the result of a clinical observation, assessment, or judgment, and linkage concept to assert or construct relationships.
HL7 Version 3 Data Types7: define the meaning of values that can be assigned to a data element. The specification attempts to define all the data types needed for healthcare information exchange including Physical Quantity (PQ) and a range of coded types (e.g., Coded Descriptor [CD] and Coded With Equivalents [CE]). PQ includes a value and unit where units can be taken from the Unified Code for Units of Measure (UCUM)13.
Using the exemplars and initial representation of observations from the previous phase, we examined terminology alignment at two levels: (1) mapping of the information model to the terminology model and (2) identifying concepts using the Regenstrief LOINC Mapping Assistant (RELMA) for LOINC and CliniClue browser for SNOMED CT. Table 2 summarizes the initial findings for alignment of the Observation Model (Figure 1a) to the terminology models and data types; Table 3 presents identification of concepts for blood pressure in SNOMED CT to determine which may be used in the information model (e.g., in Figure 1c).
Table 2.
Observation Model Alignment to Standards. This table depicts how each element in the Observation Model maps to the structure of LOINC and SNOMED, and what HL7 data types could apply.
| Element | LOINC (Axis/Axes) | SNOMED CT (Hierarchy) | HL7 V3 Data Types |
|---|---|---|---|
| Focus | Component | Observable entity | Coded type |
| Property | Property | Coded type | |
| Value | Scale | Clinical finding | PQ, Coded type |
| Context | Method, System | Linkage concept | Coded type |
Table 3.
Examples of Concept Mapping.This table depicts several concepts related to blood pressure (out of over 150) in SNOMED CT.
| ID | Description (Preferred) | Top-Level Hierarchy |
|---|---|---|
| 75367002 | blood pressure | Observable entity |
| 271649006 | systolic blood pressure | Observable entity |
| 271650006 | diastolic blood pressure | Observable entity |
| 392570002 | blood pressure finding | Clinical finding |
| 271605009 | position of body and posture | Observable entity |
| 246273001 | patient position | Linkage concept |
| 9851009 | body position finding | Clinical finding |
| 33586001 | sitting position | Clinical finding |
DISCUSSION
One of the key drivers of investigating the creation of an information model for laboratory and clinical observations is that observations about a patient drive the basic care activities such as assessment, diagnosis, planning, treatment, and evaluation. Uses that are of particular interest at this time include nursing assessments (e.g., identifying the observations that were used to make judgments for standardized assessment tools), clinical documentation (e.g., capturing and representing structured observations within clinical notes), and clinical decision support (e.g., capturing data in a uniform way for logic and rules). Given the wide range of uses and heterogeneous applications across the enterprise, there is a need for a common information model to enable applications to represent, exchange, and reason with observations in a consistent and standardized manner.
Challenges
Through the examples used to study the requirements for observations and the interface between information models and standard terminologies, we encountered several issues that are consistent with reported findings by other groups. These challenges include:
Multiple equivalent representations of information: There may be a variety of ways to represent the same information. Several pre-coordinated and post-coordinated representations could be possible and information may be represented at different levels of granularity; with respect to the Observation Model, there may be various possibilities for what the focus or properties could be. One strategy is to itemize the alternatives, select one based on requirements, and allow for transformations as needed.
Lack of concept coverage in the terminologies: There may be no concepts (or concepts are at a different level of granularity) in the terminology for elements in the information model. In these cases, recommendations could be made to the organization responsible for maintaining the terminology or local codes created to handle these gaps.
Multiple mappings from the information model to the terminologies: Some observations relate to hundreds of concepts. Finding appropriate concepts to align with an information model is labor-intensive and time-consuming. Both pre-coordinated and post-coordinated options may be available; in the case of another example for tobacco use history, many pre-coordinated SNOMED CT concepts exist. For example, “moderate cigarette smoker (10–19 cigs/day)” and “ex-heavy cigarette smoker (20–39/day)” include information about status, method, and frequency of tobacco use.
Development of appropriate post-coordination strategies: There is currently limited documentation of strategies for post-coordination in SNOMED CT. An information model specifies the relationships among component concepts and constructs a framework of how to compose them (e.g., a series of fields), while terminologies may provide possible values for these fields with standard codes. Projects such as HL7 TermInfo7 are aiming to develop approaches for using standard terminologies such as SNOMED CT.
Change management: A strategy will be needed for propagation of SNOMED CT changes to the alignment mappings. SNOMED CT continues to change at a rapid pace, including content and hierarchy changes. Change management is a significant barrier to adoption. A large effort is needed to map updates to legacy systems.
Complex semantic relationships: An interesting example was found for the SNOMED CT interprets relationship. For the blood pressure example, “blood pressure finding (finding)” interprets “blood pressure (observable entity)”, which falls under feature of entity. With another example of heart murmur14, “heart murmur finding (finding)” does not interpret “heart murmur (observable entity)” that falls under feature of entity and instead interprets “turbulent blood flow (observable entity)”, which is a function. This relationship for heart murmur varies from that for blood pressure where the latter finding interprets a feature of entity while the former interprets a system function. Such differences may increase the difficulty of semantic structure mapping between SNOMED CT and the information model.
Next Steps
As part of this work, we have described six key phases for information model development. The presented work covers aspects of the first three phases: review of existing models, model creation and evolution, and model alignment. Given the iterative and incremental nature of the model development process, next steps include building upon the findings from these three phases as well as delving into model validation, implementation, and evaluation.
Review and analysis of existing information models and terminologies revealed a base set of key features that may be used to guide model development. The key features that were considered in this first phase of the project and will continue to be evolved include: use of existing formalisms (UML with plans to explore XML, OWL, and RDF), linkage to standards (LOINC, SNOMED CT, HL7, etc.), multi-level modeling, support for constraints, and support for compositional construction.
The initial Observation Model defines a core construct for laboratory and clinical observations (i.e., Focus-Property-Value triplet), differentiates atomic and compound observations, and seeks to accommodate contextual information such as qualifiers. There is a range of other contexts such as modifiers (e.g., negation or family history) and temporal information; these are also essential aspects of observations and will be addressed in subsequent phases of the project. In addition, we have primarily focused on the “facts” related to observations; other information about the “act” (e.g., timestamps) and “participants” (e.g., author) also need to be included and will similarly be incorporated in future iterations of the model.
With a basic understanding of observations, a next step includes identifying how best to formalize the Observation Model; this may involve determining if any existing or evolving information models may be used or adapted, or if the strategy should be to align or harmonize the Observation Model with these models. A thorough evaluation of the models that were reviewed and continued in-depth review of other identified efforts (e.g., HL7 Clinical Statements7, CEN/TC 251 and ENV/EN 136061, and ISO/TC 21515) is needed to determine the approach that best meets the requirements.
CONCLUSION
In this paper, we have described our early efforts and experiences in: (1) exploring existing work related to healthcare information models that offers insights on essential features and successes, (2) designing an information model for observations (Observation Model) to be generalizable, flexible, and extensible, and (3) studying how an Observation Model would align to industry-wide standards such as LOINC, SNOMED CT, and HL7 V3 data types. The initial findings provide insight to the various challenges ahead and guidance on next steps for adoption of information models at our organization.
References
- 1.Eichelberg M, Aden T, Diesmeier J. A survey and analysis of electronic healthcare record standards. ACM Computing Surveys. 2005;37(4):277–315. [Google Scholar]
- 2.Goossen WT. Intelligent semantic interoperability: Integrating knowledge, terminology and information models to support stroke care. Stud Health Technol Inform. 2006;122:435–9. [PubMed] [Google Scholar]
- 3.Evans DA, Cimino JJ, Hersh WR, Huff SM, Bell DS. Toward a medical-concept representation language. The Canon Group J Am Med Inform Assoc. 1994 May-Jun;1(3):207–17. doi: 10.1136/jamia.1994.95236153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyle JF, Mori AR, Huff SM. Standards for detailed clinical models as the basis for medical data exchange and decision support. Int J Med Inform. 2003 Mar;:69, 2–3, 157–74. doi: 10.1016/s1386-5056(02)00103-x. [DOI] [PubMed] [Google Scholar]
- 5.Huff SM. Ontologies, vocabularies, and data models. In: Greenes RA, editor. Clinical decision support the road ahead. Elsevier; 2006. pp. 307–324. [Google Scholar]
- 6.Huff SM, Rocha RA, Bray BE, Warner HR, Haug PJ. An event model of medical information representation. J Am Med Inform Assoc. 1995 Mar-Apr;2(2):116–34. doi: 10.1136/jamia.1995.95261905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.http://www.hl7.org/v3ballot/html/welcome/environment/index.htm
- 8.http://www.openehr.org/
- 9.http://www.regenstrief.org/medinformatics/loinc/
- 10.http://www.snomed.org/index.html
- 11.http://www.icn.ch/icnp.htm
- 12.http://www.sabacare.com/index.html
- 13.http://aurora.rg.iupui.edu/UCUM/
- 14.Green JM, Wilcke JR, Abbott J, Rees LP. Development and evaluation of methods for structured recording of heart murmur findings using SNOMED-CT post-coordination. J Am Med Inform Assoc. 2006 May-Jun;13(3):321–33. doi: 10.1197/jamia.M1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.http://isotc.iso.org/