Abstract
Translating evidence into clinical practice is a complex process that depends on the availability of evidence, the environment into which the research evidence is translated, and the system that facilitates the translation. This paper presents InfoBot, a system designed for automatic delivery of patient-specific information from evidence-based resources. A prototype system has been implemented to support development of individualized patient care plans. The prototype explores possibilities to automatically extract patients’ problems from the interdisciplinary team notes and query evidence-based resources using the extracted terms. Using 4,335 de-identified interdisciplinary team notes for 525 patients, the system automatically extracted biomedical terminology from 4,219 notes and linked resources to 260 patient records. Sixty of those records (15 each for Pediatrics, Oncology & Hematology, Medical & Surgical, and Behavioral Health units) have been selected for an ongoing evaluation of the quality of automatically proactively delivered evidence and its usefulness in development of care plans.
Background
Documented plans of care, individualized for each patient could provide a means for interdisciplinary teams to put evidence-based research into practice and ensure quality and continuity of care. Translation of evidence into healthcare practice is viewed as a function of three critical elements: 1) the level and nature of the evidence; 2) the environment (context) into which the evidence is introduced; and 3) the methods of facilitating the evidence delivery [1].
The nature of evidence-based practice (EBP) is defined as the integration of best available research evidence with clinical expertise and patient values [2, 3]. Many resources provide detailed descriptions of the steps in active search for evidence-based decision support: 1) asking searchable, answerable clinical questions, 2) searching for the best evidence; 3) critically appraising the evidence; 4) implementing the applicable evidence; and 5) evaluating the outcomes [4, 5].
When applied to nursing, this model of EBP requires the nurses to proactively pull the resources devoting some additional time to question formulation and search for evidence, most probably, on a computer. This activity might not be seen as valuable as spending time with a patient [6]. Interviews with 50 nurses revealed that not all wanted access to evidence-based information, and those who did wanted the up-to-date information to be available when and where needed [6]. These findings are in agreement with a systematic review of clinical decision support systems that identifies the following key features of a successful system: 1) computer generated decision support is provided automatically and is integrated in the workflow; 2) support is delivered at the time and location of decision making 3) recommendations are actionable; and 4) the system does not depend on clinicians’ initiative for use [7].
EBP is enabled by the availability of adequate resources. As the EBP resources are abundant [8], they require judicious use at every step of the EBP process in a specific context [9].
The context in which evidence is being introduced is viewed as the organizational environment defined by the prevailing culture, human relations expressed through leadership, and monitoring and evaluation mechanisms [10]. Based on the observation that an essential additional component of the context is uncertainty caused by patients’ status, complexity of teamwork, unpredictability of nurses’ work, and changing management [11], we propose developing a tool that can be adjusted locally depending on the organizational goals and types of evidence suitable for the environment.
The existing systems that facilitate evidence delivery through linking evidence to a patient’s record either provide access to a pre-populated evidence base using a set of carefully selected topics [12], or require some user interaction in the evidence search step [13]. Our goals were to test whether the search step could be automated and the evidence for dynamically generated topics could be retrieved automatically, reducing clinicians’ cognitive burden and demands on their time.
Although each critical element of translating evidence into nursing practice has been studied separately, to our knowledge, we present the first attempt to develop a system that automatically initiates retrieval of available evidence and can be tuned to address the selection and delivery of patient-specific evidence in a health care environment.
Methods
The complexity of our task is best addressed using the spiral model of system development [14]. The spiral model allows designing the whole system but implementing only the highest priority features in an initial prototype. Several iterations of design refinements and prototyping based on users’ feedback allow developing an evaluated and tested final system.
System design
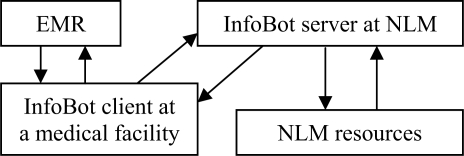
The overall design of the InfoBot system is shown in Figure 1. The system consists of two modules one of which (InfoBot client) interacts with an Electronic Medical Record (EMR). The second module (InfoBot server) resides at NLM and provides information from resources requested by the InfoBot client.
Figure 1.
InfoBot system design
The evidence search and delivery process is triggered by a new entry in one of the monitored EMR fields. The text entered into the field is extracted from the EMR into the InfoBot client and sent to the NLM server in a request to identify clinical terminology and deliver information from evidence-based resources pre-selected by the health care organization and stored in the InfoBot client as a set of rules
We use the NLM MetaMap service [15] to identify the Unified Medical Language System® (UMLS®) Metathesaurus® concepts in the EMR text [16]. The next steps in the InfoBot server pipeline depend on the preferences of a specific health care organization encoded in a set of rules selected from the InfoBot pick list during the InfoBot client installation at the health care facility. In general, the identified clinical terms and the additional UMLS information related to the terms are used to query the selected resources.
NLM resources used in the initial implementation include definitions of biomedical terms available in the UMLS, MEDLINE® citations and links to the full-text articles, MedlinePlus® articles [17], ClinicalTrials.gov [18], and Nursing Standards of Practice and Procedure Guidelines provided by the NIH Clinical Center.
Information retrieved from these resources is stored in the InfoBot database. This information can be displayed by request at the healthcare institution via Web pages dynamically generated by the InfoBot server. The system can also return the information to the healthcare institution in XML format.
Prototype implementation
The initial prototype presented in this paper supports the end-to-end information flow and implements the core functionality of the future clinical decision support system. The goal of this prototype is to test the feasibility of the system and the soundness of its architecture.
EMR and InfoBot client requests
Rather than attempting a direct interface with the National Institutes of Health's Clinical Research Information System (CRIS) at this early development stage, we extracted the following fields from CRIS: Clinical Center unit, Chief Complaint, Clinical Trial protocol number, and Interdisciplinary problems, goals and interventions. The interdisciplinary problems, goals and interventions fields consist of two parts: 1) a controlled vocabulary term that categorizes the problem, for example, Patient/Family Education Goals, Metabolic/Endocrine Problem, Safety Interventions, etc; and a free text part that describes the actual problem, for example, the following safety problem description: “high falls risk secondary to impaired vision”.
The extracted fields were stored in a database representing an EMR. The EMR-InfoBot client interaction was simulated by sending a “new record” request with information from one of the restored EMR records to the client. The EMR request contains an anonymized patient unique identifier, the identifier of the set of rules to be applied to the record (RuleSet), and the value entered into the field, for example, the clinical protocol number or the interdisciplinary problem text.
Upon receiving information from the EMR, the InfoBot client registers the request, the patient identifier, and the RuleSet in its internal table and sends this information and the extracted text to the InfoBot server that also stores this information in its internal tables monitored by a work distribution manager that ensures the RuleSet is applied and the request is processed in the pipeline.
InfoBot server functions
The following InfoBot client requests selected in a study session with the NIH Clinical Center evidence-based practice educators are currently supported by the InfoBot server: 1) identifying biomedical terms in free text; 2) linking those terms to UMLS definitions, 3) linking to the NIH Clinical Center Nursing Standards of Practice and Procedure Guidelines; 4) searching PubMed for Cochrane reviews; 5) linking to MedlinePlus and other patient education articles, and 6) linking a Clinical Trial protocol number to information about the trial in the ClinicalTrials.gov database. In practice, requests two through five depend on the information obtained in the first request and are combined into a pipeline defined in the RuleSets.
The key step in the InfoBot pipeline is the identification of the biomedical terms in the free text entered into the EMR fields. InfoBot text processing starts with expansion of acronyms and abbreviations that are unambiguous within the context of the Clinical Center, but could be mapped to irrelevant terms or not recognized by MetaMap. For example, decub is expanded to decubitus ulcer. Next, spelling in each of the free text snippets is checked using the NCBI ESpell E-Utility [19].
The expanded corrected text is submitted to the MetaMap API by the InfoBot MetaMap wrapper module. The MetaMap results are post-processed in this module to identify and extract biomedical single-word and multi-word terms that are recognized as UMLS concepts and belong to the following general semantic groups: problems, interventions, medications, anatomy, and findings. In addition to the extracted text, the module extracts the concepts’ unique identifiers, semantic groups, and preferred UMLS names from the MetaMap results. If information is extracted, it is stored in the response being formed by the server and the response status continues to be in process. If no concepts are identified in the corrected text, the server responds to the InfoBot client providing this information and the distribution manager marks the request as finished. The extracted information triggers retrieval of definitions, Standards of Practice, and related articles.
The definition retrieval module uses the unique concept identifiers to retrieve the UMLS definitions. The definitions are indexed with their identifiers and stored in the InfoBot database table. The definition retrieval module queries the table and adds available definitions to the InfoBot server response.
The Standards of Practice retrieval module uses preferred concept names to retrieve the Nursing Standards of Practice and Procedure Guidelines. Retrieval of these resources is supported by indices stored in the InfoBot server database. The Standards of Practice and Procedure Guidelines were indexed manually by an experienced medical indexer.
The patient education materials retrieval module also uses preferred concept names to retrieve MedlinePlus articles. Those articles were indexed with the UMLS preferred concept names identified in their titles using MetaMap. The index was manually edited to exclude terms irrelevant to the document and stored in the InfoBot server database. Both Standards of Practice and Patient Education Materials indices link the preferred names assigned to a document to the document URL (at the Clinical Center Nursing Intranet and MedlinePlus respectively).
The MEDLINE retrieval module uses phrases extracted from the text to retrieve Cochrane reviews. The module formulates queries and sends requests to the Repository for Informed Decision Making (RIDeM), which was developed to extract and store elements of a clinical scenario from MEDLINE citations. RIDeM determines the clinical task supported by the publication and the potential level of evidence. It automatically extracts the problem(s), the intervention(s), the number and characteristics of patients, and the patient-oriented outcomes of the study from the abstract of the publication [20].
The queries formulated by the MEDLINE retrieval module contain the search preferences encoded in the RuleSet (for example, searching for up to ten Cochrane reviews using PubMed E-Utilities [19]) and the query terms in the form of a well-formed clinical question. The latter is possible because of the semantic groups associated with the identified biomedical terms and is used to filter out retrieval results in which the studied problem differs from the one submitted in the request. Our current query formulation strategy is a Boolean AND of the identified elements of the well-formed clinical question. RIDeM response to the InfoBot request contains an ordered list of the titles, extractive summaries, and links to PubMed citations and full-text articles. The InfoBot MEDLINE retrieval module parses the received information and adds it to the response.
The Protocol retrieval module satisfies requests to link to a clinical trial protocol by a look-up in the InfoBot server table that stores a URL of the study details submitted to the ClinicaTrials.gov registry [18]. The Protocol retrieval module searches the table that indexes each URL using the corresponding clinical trial identifier.
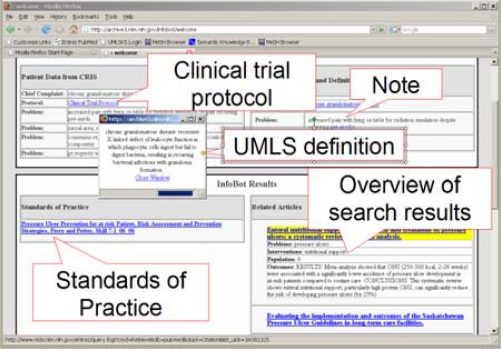
When all modules involved in the RuleSet pipeline are done, the InfoBot server response becomes available to the InfoBot client. The InfoBot client Web page with an augmented patient record created for the evaluation of the quality of evidence is shown in Figure 2. The following section details the creation of this record.
Figure 2.
A prototype display of an augmented interdisciplinary team record
Augmenting a record: a worked example
The record for a patient with the Chief Complaint of chronic granulomatous disease is built incrementally. The first request to the InfoBot server contains the chief complaint, which is mapped to the UMLS concept Granulomatous Disease, Chronic (identifier: C0018203; semantic group: problem) by the MetaMap wrapper module. This successful mapping initiates the pipeline: the concept identifier retrieves the definition of the term and the preferred name retrieves the MedlinePlus article. The extracted biomedical term (in this case the whole Chief Complaint entry), the definition and the link to the MedlinePlus article become available to the InfoBot client. When the patient’s record is accessed in the prototype interface, the text in the Chief Complaint field is hyperlinked to the definition of the term (displayed in a small window in the upper left corner of Figure 2). The title of the MedlinePlus article (linked to the article) is added to the Related Articles area in the lower right corner of the interface. The next request is issued to link the Clinical Trial Protocol field to the ClinicalTrials.gov entry.
Of the four interdisciplinary problem notes subsequently added to the patient’s record, two contain terms that trigger the RuleSets and augment the record, although many more terms are initially identified in the text. For example, the following concepts are found in the text “increased pain with lying on table for radiation simulation despite receiving pre-meds”: lying (Supine Position, C0038846, Organism Attribute); table (C0039224, Manufactured Object); and pre-meds (Premedication, C0033045, intervention). Of these, the term pre-meds is processed further to retrieve its definition. We hope the timely delivered definition “Preliminary administration of a drug preceding a diagnostic, therapeutic, or surgical procedure. The commonest types of premedication are antibiotics (ANTIBIOTIC PROPHYLAXIS) and anti-anxiety agents. It does not include PREANESTHETIC MEDICATION” will clarify why pre-meds do not alleviate pain and will help with an appropriate plan of care.
The term decubitus ulcer identified in the subsequently added problem text triggers the successful completion of the whole pipeline in addition to retrieval of its definition: a practice guideline on assessment and prevention of ulcers and two Cochrane reviews, one on benefits of following guidelines and one on nutritional support for patients at risk of developing pressure ulcers are added to the record.
Results and discussion
Using 4,335 de-identified interdisciplinary team notes for 525 patients, the system automatically extracted biomedical terminology from 4,219 notes. The types of terms that triggered search for evidence and numbers of patient records and interdisciplinary notes containing the extracted terms (see Table 1) allowed the system to subsequently link at least one resource (other than the chief complaint definition and the clinical trial protocol) to 260 of the 525 patient records. There are several reasons for not finding evidence for all patients: 1) 65 records contained no notes; 2) some of the notes were not informative enough, for example, a Research Participation Problem described as “lack of knowledge”; 3) we applied very strict rules to trigger term selection assuming that it is better not to flood the record with too general and potentially irrelevant information that could be retrieved using terms pain, discussion, admission, etc.
Table 1.
Clinical terms automatically identified in the interdisciplinary team notes
| Clinical term type | Total notes | Total patients |
|---|---|---|
| problems | 709 | 68 |
| interventions | 445 | 54 |
| medications | 68 | 6 |
| anatomy | 223 | 24 |
| findings | 946 | 108 |
Conclusions and Future Work
The positive comments obtained in the ongoing evaluation of the prototype indicate the fully automated evidence-based practice support system is feasible and has a potential to proactively support development of an evidence-based plan of care. The proposed general system architecture is sound, but the methods of query term selection and query generation need to be refined. The implementation of the next system prototype will be based on the results of the ongoing evaluation of the quality and usefulness of the retrieved evidence resources.
Acknowledgments
Authors would like to thank Susanne Humphrey for manual indexing of the Nursing Standards of Practice and Procedure Guidelines, Joseph Chow for his contribution to InfoBot implementation.
References
- 1.Kitson A, Harvey G, McCormack B. Enabling the implementation of evidence based practice: a conceptual framework. Qual Health Care. 1998 Sep;7(3):149–58. doi: 10.1136/qshc.7.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiCenso A, Guyatt G, Ciliska D, editors. St Louis, Missouri: Elsevier Mosby; 2005. Evidence-based nursing: a guide to clinical practice. [Google Scholar]
- 3.Rycroft-Malone J, Seers K, Titchen A, Harvey G, Kitson A, McCormack B. What counts as evidence in evidence-based practice? J Adv Nurs. 2004 Jul;47(1):81–90. doi: 10.1111/j.1365-2648.2004.03068.x. [DOI] [PubMed] [Google Scholar]
- 4.Fineout-Overholt E, Melnyk BM, Schultz A. Transforming health care from the inside out: advancing evidence-based practice in the 21st century. J Prof Nurs. 2005 Nov-Dec;21(6):335–44. doi: 10.1016/j.profnurs.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Brady N, Lewin L. Evidence-based practice in nursing: bridging the gap between research and practice. J Pediatr Health Care. 2007 Jan-Feb;21(1):53–6. doi: 10.1016/j.pedhc.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Bond CS. Nurses and computers. An international perspective on nurses’ requirements. Medinfo. 2007;12(1):228–32. [PubMed] [Google Scholar]
- 7.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005 Apr 2;330(7494):765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oermann MH. Internet resources for evidence-based practice in nursing. Plast Surg Nurs. 2007 Jan-Mar;27(1):37–9. doi: 10.1097/01.PSN.0000264160.68623.0a. [DOI] [PubMed] [Google Scholar]
- 9.Rutledge DN. Resources for assisting nurses to use evidence as a basis for home care nursing practice. Home Health Care Management & Practice. 17(4):273–280. [Google Scholar]
- 10.Kitson AL, Rycroft-Malone J, Harvey G, McCormack B, Seers K, Titchen A. Evaluating the successful implementation of evidence into practice using the PARiHS framework: theoretical and practical challenges. Implement Sci. 2008 Jan 7;3:1. doi: 10.1186/1748-5908-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott SD, Estabrooks CA, Allen M, Pollock C. A context of uncertainty: how context shapes nurses' research utilization behaviors. Qual Health Res. 2008 Mar;18(3):347–57. doi: 10.1177/1049732307313354. [DOI] [PubMed] [Google Scholar]
- 12.Bakken S, Currie LM, Lee NJ, Roberts WD, Collins SA, Cimino JJ. Integrating evidence into clinical information systems for nursing decision support. Int J Med Inform. 2007 Sep 27; doi: 10.1016/j.ijmedinf.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhadad N, Kan MY, Klavans JL, McKeown KR. Customization in a unified framework for summarizing medical literature. Artif Intell Med. 2005 Feb;33(2):179–98. doi: 10.1016/j.artmed.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 14.Boehm B. A Spiral Model of Software Development and Enhancement. IEEE Computer. 1988 May;21(5):61–72. [Google Scholar]
- 15.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MedlinePlus. 2008 Available at http://www.nlm.nih.gov/medlineplus/ Accessed February292008
- 18.ClinicalTrials.gov. 2008 Available at: http://clinicaltrials.gov Accessed February 292008
- 19.Entrez Programming Utilities. 2008http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.htmlAccessed February 292008
- 20.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]