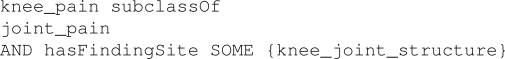Abstract
The ability to model, share and re-use value sets across multiple medical information systems is an important requirement. However, generating value sets semi-automatically from a terminology service is still an unresolved issue, in part due to the lack of linkage to clinical context patterns that provide the constraints in defining a concept domain and invocation of value sets extraction. Towards this goal, we develop and evaluate an approach for context-driven automatic value sets extraction based on a formal terminology model. The crux of the technique is to identify and define the context patterns from various domains of discourse and leverage them for value set extraction using two complementary ideas based on (i) local terms provided by the Subject Matter Experts (extensional) and (ii) semantic definition of the concepts in coding schemes (intensional). A prototype was implemented based on SNOMED CT rendered in the LexGrid terminology model and a preliminary evaluation is presented.
Introduction
In the context of terminologies and coding schemes, a value set is an uniquely identifiable set of valid values that can be resolved at a given point in time to an exact set (collection) of codes. Value sets exist to provide possible values for data elements (i.e., variables) that can be present, for example, in an electronic health record. The main objective of modeling value sets is to specify a concept domain with certain slots or attributes of interest such that the attribute-values can be obtained from one or more terminologies of interest. An example of a concept domain could be “world countries”, and the representative value set will include countries such as USA and UK.
Typically, value sets can be drawn from pre-existing coding schemes such as SNOMED CT or ICD by constraining the value selection based on logical expressions (e.g., all sub-concepts of the concept colon_cancer). Although useful in practice, this process can be manually intensive, ad-hoc, and in some cases, inadequate, thereby warranting the development of techniques for (semi-) automatic extraction of value sets. However, generating value sets (semi-) automatically from a terminology service is still an unresolved issue, in part due to the lack of (i) linkage to clinical context patterns that act as constraints in defining a concept domain, (ii) techniques for automatically analyzing membership of values to a particular concept domain, and (iii) approaches based on formal languages such as the Web Ontology Language (OWL1).
To address some of these requirements, we propose a novel approach for context-driven value sets extraction and evaluation called LexValueSets. The crux of LexValueSets is to render the semantics of a concept domain using a formal model that takes into consideration various context patterns (e.g., location, time duration), specified typically by subject matter experts (SMEs), to drive the development of two complementary techniques for value sets extraction: extensional and intensional. The extensional approach comprises of an explicitly enumerated set of local terms, provided initially by SMEs, that correspond to an initial list of values for different slots of the concept domain, and are used for automatically extracting concepts from a particular terminology or a coding scheme.2 For example, given a concept domain pain in humans, the set of local terms for a slot location would comprise of hand, hip and other anatomical structures. The intensional technique, on the other hand, leverages the computable semantic definition of a concept domain to automatically identify relevant concepts for filling the slots. For example, the SNOMED CT concept “661005 jaw region structure” can be used to fill the location slot of pain since it is a finding site for the SNOMED CT concept “274667000 jaw pain”.
We have developed a prototype implementing these techniques for value set extraction based on graph traversal techniques. In particular, we have adopted the LexGrid terminology model3 for rendering terminologies as well as defining the semantics and context patterns of concept domains. We have also performed preliminary evaluation based on SNOMED CT and our initial investigation has shown satisfactory results as well as provided insights for further work.
Related Work
Alan Rector introduced representing value partitions and value sets using OWL in [1]. The objective was to model the descriptive features (a.k.a qualities, attributes or modifiers) of classes (or concepts, in general) as properties whose ranges specify the constraints on the values that the property can take on. These values can be defined either as partitions of classes or enumerations of individuals. However, [1] does not provide any specific mechanism or implementation for generating the value sets. This limitation was addressed by Rector et al. in [2], where the authors model concept domains as well as terminologies in OWL, and demonstrate how relevant value sets can be extracted to bind them to electronic health records and messages. Our work is motivated by [2] as we leverage a formal model for representing concept domains and terminologies. Additionally, we introduce two complementary techniques for automatic value sets extraction and provide preliminary results based on use cases from SNOMED CT. Even though existing software, such as Apelon DTS4 and openEHR5, provide techniques for (semi-) automatic value sets extraction, they only allow identifying relevant values based on lexical matching and provide no mechanism for exploiting semantic relationships between the concepts for value set extraction.
Another set of work which is very closely related to our effort is in the area of ontology querying. The main objective of such techniques is to provide the ability to identify relevant concepts from a particular ontology based on a search criteria (e.g., user-specified keywords). Power-Aqua [3] allows users to ask questions using (controlled) natural language constructs, which are then translated into a set of logical queries for deriving a set of RDF6 triples. These triples essentially contain the ontological concepts relevant to the user query. A similar approach is proposed in the PANTO framework [4] where the user questions are converted into SPARQL7 queries for querying ontologies. PANTO also enables advanced querying with negations, superlatives and comparatives. On a slightly different note, Alani et al. [5] proposed leveraging Wikipedia for identifying important concepts that represent a particular domain. Specifically, given a particular domain name (e.g., anatomy), the technique will first identify important terms from Wikipedia that are relevant to the domain (e.g., hands, brain, bones) and apply them to search concepts within ontologies. Although we do not provide the ability to convert natural language queries to logical queries for value sets extraction, it is of our interest to explore such an approach in the future.
Methods
Extensional Value Set Extraction
Traditionally, approaches for value set extraction have focused primarily on employing SMEs for manually selecting a set of values for a particular concept domain which is often tedious and cumbersome. Consequently, the ability to (semi-) automate parts of the processing is required, and an important aspect to achieve this automation is to explicitly and unambiguously represent the concept domains.
Our extensional value sets extraction technique attempts to address this requirement based on the following model:
The technique assumes that the concept domain under consideration has been defined using a formal model such as OWL that can be represented within the LexGrid environment. For example, Figure 1 shows the definition of the pain concept domain introduced earlier using the Manchester OWL syntax8.
As part of the concept domain definition, the technique also assumes that a preliminary list of local terms (i.e., keywords used in general by SMEs) for various slots are provided. For example, in Figure 1, the duration slot is defined with two local terms. Similarly, the location slot can be filled with local terms (not shown in Figure 1).
Once the concept domain definition and local terms are provided, the final step is to initiate the value set extraction process based on a target terminology.
Figure 1.
Manchester OWL syntax representation of the pain concept domain
The crux of the final step comprises of two major elements: (a) first, the local terms are used to perform LexGrid queries to identify terminological concepts that are lexically related, and (b) second, the identified terminological concepts are used to extract semantically related concepts by traversing the terminology hierarchy to form the value set. To perform the lexical similarity between local terms and concept definitions, we leverage a set of lexical matching algorithms (e.g., “sounds-like” or double metaphone, stemmed words querying) implemented in the LexGrid API. For example, if HAND is a local term for consideration and is used to search for lexically related concepts in a terminology, such that the match is “exact”, the concept hand from Figure 2 will be selected. Once the lexical search returns candidate concept(s) from the target terminology, the next step traverses the concept relationships in the terminology to determine semantically related concepts. Within the scope of our work, we say that concepts A and B are semantically related (denoted by A ∼Sem B) if either A ≡ B (A is equivalent to B) or A ⊑ B (A is a sub-concept of B) such that neither A nor B correspond to OWL:Thing. Thus, if the concept hand is selected from Figure 2 corresponding to the local term hand (step (a)), the traversal technique (step (b)) will extract the concepts left_hand and right_hand since both semantically relate to hand.
Figure 2.
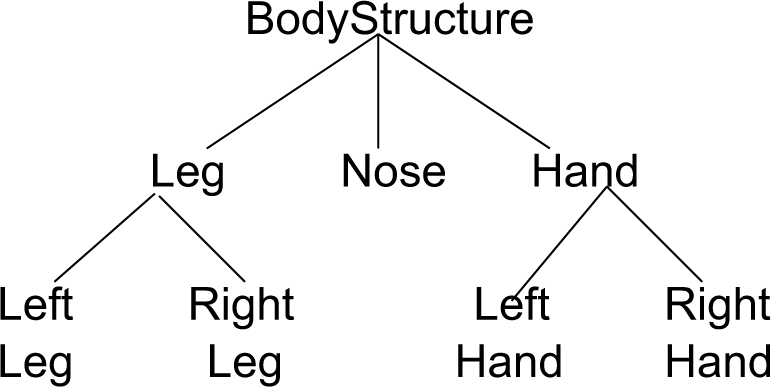
A sample is-a hierarchy
Algorithm 1.
Identifying Semantically Related Concepts via Hierarchy Traversal
| 1: | procedure HierarchyTraversal(T, x, RS, VSet) |
| 2: | if ((T = ∅) Or (x = ∅) Or (RS = ∅) Or (x ≁SemRS)) then |
| 3: | return null |
| 4: | end if |
| 5: | if (∃x′ ∈ T.((x′ ⊑ x) ∧ (x′ ∉ VSet))) then |
| 6: | VSet := VSet ∪ {x′} |
| 7: | if (∀x″ ∈ T.((x Assoc x″) ∧ (x″ ~SemRS)) then |
| 8: | VSet := VSet ∪ {x″} |
| 9: | end if |
| 10: | HierarchyTraversal(T, x′, RS, VSet) |
| 11: | else |
| 12: | if (∀x″ ∈ T.((x Assoc x″) ∧ (x″~SemRS)) then |
| 13: | VSet := VSet ∪ {x″} |
| 14: | end if |
| 15: | end if |
| 16: | return VSet |
| 17: | end procedure |
Algorithm 1 shows the pseudo code for the traversal technique illustrated above. The procedure HIERARCHY-TRAVERSAL takes as input the target terminology T, the terminological concept x, the range RS of the slot S to which x belongs, and the value set VSet (initially empty) which is being extracted. The procedure assumes that T can be represented as a directed acyclic graph and does a variant of depth first traversal (DFT) starting from the concept x (which acts as a node in T). At first, it checks whether T, or RS, or x are null or x is not semantically related to RS —if either of these conditions hold, the execution halts (lines 2–4). On the other hand, if none of these conditions hold, the procedure checks for sub-concepts of x, such that if a sub-concept x′ exists, it is added to VSet (lines 5–6). Additionally, if x′ is associated to any other concept x″ (apart from the sub-concept or equivalence association), such that x″ is semantically related to RS, x″ is added to VSet (lines 7–8). A similar step to extract associated concepts is also done if x is a leaf node in the hierarchy (lines 12–13). The procedure is recursively invoked until all the hierarchies originating at x have been explored.
Intensional Value Set Extraction
In the context of terminologies, intensional value sets are defined by a computable expression that can be resolved to an exact list of codes. For example, an intensional value set definition could be defined as, “All SNOMED CT concepts that are sub-concepts of the SNOMED CT concept
Diabetes_Mellitus”. In LexValueSets, we leverage this notion along with the computable semantic definition of a concept domain to automatically (a) identify candidate terminological concepts that are semantically related to the concept domain, and (b) extract association-values of those candidate concepts to fill the concept domain slots.
In particular for step (a), the technique identifies candidate concepts by analyzing their semantic relatedness to the concept domain based on its semantic definition. For example, consider a concept knee_pain as shown in Figure 3. It can be regarded as semantically related to the pain concept domain (Figure 1) assuming that (i) joint_pain ⊑ pain, (ii) hasLocation ≡ hasFindingSite, and (iii) knee_joint_structure ⊑ body_structure. Once such concepts are identified from a target ontology, step (b) extracts values from those associations that correspond to the appropriate slots in the concept domain, and uses them as the starting point for traversing the terminology (based on Algorithm 1) to determine additional semantically related concepts. For instance, assuming that hasLocation ≡ hasFindingSite, the concept knee_joint_structure (and its sub-concepts obtained after traversal) can be used to fill the location slot of the pain concept domain. Arguably, an important aspect of the approach is that the semantic definitions of the concept domain and terminological concepts can be interpreted uniformly either by assuming that they are based on the same underlying representational model and/or the required concept-mappings have been pre-defined.
Figure 3.
Manchester OWL syntax representation of the knee_pain concept
Results
Prototype Implementation
To evaluate our proposal, we have implemented both the extensional and intensional value sets extraction approaches based on the clinical context information defined in a real Mayo Clinic clinical element model (i.e., a structure that defines some related set of data) for pain in humans. The 20070731 version of SNOMED CT of UMLS (version 2007AC) was used as the target terminology and the extraction algorithms were built using Mayo’s LexGrid API. In this model, a concept domain called pain was defined with slots such as location, duration, pain scale value, and others. For each slot, a list of locally defined terms was used as the picklist (see Figure 1 for a snapshot of the model).
For the extensional approach, we considered all the local terms from the slot hasLocation as keywords and executed the lexical algorithms against SNOMED CT concepts rendered within the LexGrid environment. For this pilot study, we defined three types of value sets: the first value set (VS-EA) contains all the matching results (filtered only for the anatomical concepts of SNOMED CT) obtained directly from the lexical match, the second value set (VS-EB) contains additional child concepts obtained via traversing the hierarchy of concepts contained in VS-EA, and the third value set (VS-EC) contains all concepts from VS-EB and additional concepts obtained through traversing the target concepts of all associations (apart from the sub-concept or equivalence associations) for each concept in VS-EB.
For the intensional approach, we leveraged SME-defined mappings between the concepts and properties in the clinical element model and SNOMED CT. For example, we considered that the pain concept domain corresponds to SNOMED CT concept “22253000 pain” and the slot hasLocation corresponds to the association “363698007 finding site”. Based on this premise, our technique first identifies all the sub-concepts of “22253000 pain” in SNOMED CT and extracts the target concepts of the association “363698007 finding site” as candidates for the value set (VS-IA) corresponding to the location slot. Furthermore, all the children of concepts contained in VS-IA are used to create another value set (VS-IB). And finally, using all concepts in VS-IB, additional associations (apart from the sub-concept or equivalence associations) are traversed to identify semantically related concepts for creating the value set VS-IC.
To evaluate the techniques, we compared the overlap between the value sets extracted by both the approaches and provide a preliminary analysis in the following. Additionally, we evaluated the usefulness of the value sets through a questionnaire to the SMEs at Mayo Clinic.
Preliminary Evaluation
To evaluate our implementation, 103 local terms corresponding to the slot hasLocation for the pain concept domain were used as query terms in the extensional approach. 777 pain concepts in SNOMED CT were identified and used in the intensional approach. The total number of concepts in the Body_Structure (concept ID: 123037004) branch of SNOMED CT is 28646. All the experiments were executed on a Windows XP SP2 notebook with Intel Core2 2.00 GHz CPU and 2GB RAM.
Based on the implementation of the extensional and intensional techniques, 858 anatomical concepts were extracted for VS-EA, 17128 for VS-EB, 25635 for VS-EC, 217 for VS-IA, 24404 for VS-IB, and 26712 for VS-IC. The number of concepts in both VS-EC and VS-IC is close to the total number of concepts in the Body_Structure branch of SNOMED CT.
Table 1 shows the number of overlapping concepts between the value sets. Specifically, the number of overlapping concepts between VS-EA and VS-IA is 23 (accounting for about 3% of the concepts in VS-EA), whereas the number of overlapping concepts between VS-EA and VS-IB is 811, accounting for about 93% of the concepts in VS-EA. This result indicates that most concepts in VS-EA are more granular (i.e., closer to the leaf nodes in the SNOMED CT hierarchy) than those identified in VS-IA that are derived by the intensional approach. The number of overlapping concepts between VS-EC and VS-IC is 25606, accounting for about 99.9% of VS-EC and 95.8% of VS-IC. This result indicates that the coverage of the two value sets for both the approaches, once hierarchy traversal is employed, is almost same.
Table 1.
Overlap between extensional and intensional value sets
| Intensional Approach | |||
|---|---|---|---|
| VS-IA(n=217) | VS-IB (n=24404) | VS-IC (n=26712) | |
| Extensional Approach | |||
| VS-EA (n=858) | 23 | 811 | 829 |
| VS-EB (n=17128) | 136 | 17056 | 17099 |
| VS-EC (n=25635) | 215 | 24156 | 25606 |
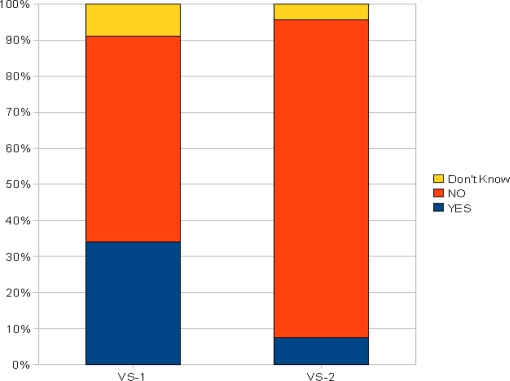
Furthermore, in this pilot study, we considered overlapping concepts between VS-EA and VS-IB since they could potentially capture a set of concepts that are useful to support the process of “manual mapping” (i.e., identifying the mappings of local terms to the SNOMED CT concepts) done by SMEs. In particular, we randomly selected (i) 100 concepts from the 811 overlapping concepts between VS-EA and VS-IB (value set VS-1), and (ii) 100 concepts from VS-IB that do not overlap with VS-EA (value set VS-2) as control groups. An evaluation question was designed as follows: “Which concepts in VS-1 and VS-2 are an appropriate value for the slot hasLocation of the pain concept domain?”. Two nosologists participated in the evaluation. Figure 4 shows the results—approximately 35% of concepts in VS-1 were considered appropriate as opposed to only 7% of concepts in VS-2 (p<0.001, X2 test). The result confirmed our hypothesis that the value set extracted from local term triggered approach (i.e., extensional) is, in general, more useful than the intensional approach for supporting SMEs’ manual mapping tasks.
Figure 4.
Nosologists Evaluation
Discussion
New techniques for automatic extraction of value set proved beneficial for SMEs in determining an initial list of appropriate values. In particular, the values extracted by obtaining more granular concepts (via hierarchy traversal) in the extensional approach yielded better results, thereby asserting that, in general, traversing via associations could be useful in refining the value sets. However, this does not necessarily imply that our intensional approach is not a viable strategy—the results shown in Figure 4 are based on a random selection of 100 concepts out of 23593 concepts, and hence, it is possible that this set of selected concepts did not meet SMEs’ requirements. This in turn calls for more rigorous cross-validation of the results. Additionally, our current evaluation is a lacking a measurement of how much SMEs’ effort and time was reduced by using LexValueSets as opposed to manually modeling value sets; we plan to conduct such a study in the future. Furthermore, in the current implementation, we applied simple lexical matching techniques for identifying lexically related concepts. Consequently, it is of our interest to explore more advanced lexical matching algorithms [6–8] and compare them for optimizing the extraction process. For further analysis, we are also considering aggregating the granular concepts obtained in the extensional approach to higher-level concepts and comparing them with those obtained via the intensional approach.
As mentioned earlier, an integral aspect of our intensional value sets extraction algorithm is the ability to determine semantic correspondences between the concepts and associations in a terminology with that of the concept domain (e.g., is hasLocation hasFindingSite). At present, such an analysis is facilitated in parts by manual mapping done by SMEs. In the future, we plan to explore semi-automated techniques for specifying mappings between concepts and associations [9]. Additionally, we intend to leverage existing ontology reasoners such as Pellet [10] in identifying semantically related concepts. On a slightly different aspect, our extraction techniques do not take into consideration various issues about management and governance of value sets. For example, the existing LexValueSets implementation cannot enable automatic percolation of a change in a value set whenever there is a change in the terminology version. We believe this will be an important requirement since many terminologies (e.g., Gene Ontology) are updated frequently. Finally, our current implementation for value sets extraction considers only one target terminology or coding scheme at a time, and we intend to enable consideration of multiple terminologies simultaneously.
Conclusion
The ability to model, share and re-use value sets across multiple medical information systems is an important requirement. Traditionally, such value sets have been constructed manually by subject matter experts (SMEs) making the entire process cumbersome, time consuming, and in some cases, inadequate. In this paper, we have shown how (semi-) automatic context-driven value sets extraction techniques can be applied for generating an initial list of appropriate values that can be evaluated by SMEs. In particular, we developed two complementary approaches for automatic value sets extraction where one leverages a set of locally defined terms provided by SMEs, corresponding to the values for the concept domain properties or attributes, to trigger the value set extraction process (extensional approach), and the other leverages a computable semantic definition of the concept domain to select a set of terminological concepts (intensional approach). Both the techniques are based on determining lexical and semantical relatedness between concepts and leverage well-studied graph traversal techniques. We also evaluate the feasibility of our approach based on use cases from SNOMED CT and provide preliminary evaluation results.
Acknowledgments
The authors would like to thank De-bra Albrecht and Donna Ihrke for their assistance and participation in numerous discussions leading to this work.
Footnotes
In practice, the local terms correspond to a set of keywords used by SMEs within the context of a particular application that are manually mapped to concepts in coding schemes.
References
- [1].Rector A.Representing Specified Values in OWL: “Value partitions” and “Value sets”In: W3C Working Group Note, http://www.w3.org/TR/swbp-specified-values/; 2005
- [2].Rector A, Qamar R, Marley T. Binding Ontologies and Coding Systems to Electronic Health Records and Messages. 2nd International Workshop on Formal Biomedical Knowledge Representation CEUR WS Proceedings; 2006. pp. 11–19. [Google Scholar]
- [3].Lopez V, Motta E, Uren VS.PowerAqua: Fishing the Semantic WebIn: 3rd European Semantic Web Conference. LNCS 4011, Springer-Verlag; 2006393–410. [Google Scholar]
- [4].Wang C, Xiong M, Zhou Q, Yu Y.PANTO: A Portable Natural Language Interface to Ontologies In: 4th European Semantic Web Conference. LNCS 4519, Springer-Verlag; 2007473–487. [Google Scholar]
- [5].Alani H, Noy N, Shah N, Shadbolt N, Musen M.Searching Ontologies Based on Content: Experiments in the Biomedical Domain In: 4th International Conference on Knowledge Capture.ACM Press; 200755–62. [Google Scholar]
- [6].Kondrak G.N-Gram Similarity and Distance In: 12th International Conference on String Processing and Information Retrieval. LNCS 3772, Springer-Verlag; 2005115–126. [Google Scholar]
- [7].Porter MF. An Algorithm for Suffix Stripping. In: Readings in Information Retrieval. Morgan Kaufmann Publishers Inc; 1997. pp. 313–316. [Google Scholar]
- [8].Alvarez MA, Lim S.A Graph Modeling of Semantic Similarity between Words In: 1st IEEE International Conference on Semantic Computing.IEEE CS Press; 2007355–362. [Google Scholar]
- [9].Shvaiko P, Euzenat J.A Survey of Schema-Based Matching Approaches In: Journal of Data Semantics IV LNCS 3730, Springer-Verlag; 2005146–171. [Google Scholar]
- [10].Sirin E, Parsia B, Grau BC, Kalyanpur A, Katz Y. Pellet: A Practical OWL-DL Reasoner. Journal of Web Semantics. 2007;5(2):51–53. [Google Scholar]