Abstract
Automated document classification can be a valuable tool for enhancing the efficiency of creating and updating systematic reviews (SRs) for evidence-based medicine. One way document classification can help is in performing work prioritization: given a set of documents, order them such that the most likely useful documents appear first. We evaluated several alternate classification feature systems including unigram, n-gram, MeSH, and natural language processing (NLP) feature sets for their usefulness on 15 SR tasks, using the area under the receiver operating curve as a measure of goodness. We also examined the impact of topic-specific training data compared to general SR inclusion data. The best feature set used a combination of n-gram and MeSH features. NLP-based features were not found to improve performance. Furthermore, topic-specific training data usually provides a significant performance gain over more general SR training.
Introduction
Systematic reviews (SRs) are an essential component in the practice of Evidence-based Medicine (EBM), providing recommendations for medical treatment, diagnosis, prognosis, and etiology based on the best available biomedical evidence. Because new information constantly becomes available, medicine is continually changing, and SRs must undergo periodic updates. Currently, experts in EBM manually review thousands of articles on specific classes of drugs in order to synthesize treatment recommendations to direct the standard of practice and continually improve the standard and cost-effectiveness of clinical care.1 The Cochrane Collaboration estimates that at least 10,000 total SRs are needed to cover most health care problems, with less than half of this number completed after 10 years of concerted effort by the EBM community. New trials are currently published at a rate of more than 15,000 per year, making the need for improved efficiency in preparing and updating reviews urgent.2
By applying automated document classification techniques early in the process, at the stage of identifying and screening the possibly relevant literature, the workload of the systematic reviewers at these early stages can be reduced. Automated document classification can help by identifying the most promising documents, reducing the human workload, and allowing more time to be spent on the more analytical parts of the task including: conducting more detailed analyses, writing more complete reports, and producing a greater number of complete reviews more quickly.
When performing systematic reviews, the review topics are periodically updated. This creates an opportunity to use the article inclusion/exclusion decisions made previously as a training set within a machine learning framework. This training set can be used to create a classifier that prioritizes work for future review updates. This concept of work prioritization for document-intensive tasks has several attractive features. First, by reviewing the most likely important documents before other documents the human reviewers or curators are more likely to be able to “get up to speed” on the current developments within a domain more quickly. Also, while reviewers or curators can certainly read all the documents in a collection given sufficient time, many tasks have practical limitations, and it is expedient to read first the documents that are most likely useful. If time or resource limitations prevent reading all documents within a collection, reviewing documents in order of most likely importance allows a more confident decision to be made about when the document review can be considered complete.
Most commonly in bioinformatics text classification research, the performance of a given system on a given task is measured by precision and recall, which are then combined into a single F-measure.3 These metrics measure the ability of a classifier to make correct predictions identifying which documents are positive (desired for the given task), and which are negative (should be ignored for the given task). Precision is the fraction of correct positive predictions, recall is the fraction of true positive documents correctly prediction, and the F-measure is the geometric mean of the two.
While the precision, recall, and F-measure metrics are common and useful, they are not optimal or even relevant for all tasks. These measures are focused on the ability of a classification system to make binary predictions on the documents of interest. If the task is not to separate documents into positive and negative groups, but instead to prioritize which documents should be reviewed first and which later, then precision, recall, and F-measure do not provide sufficient information. Furthermore, the binary prediction measures are heavily influenced by the prevalence of positive cases, and therefore it is difficult to use these measures to make meaningful comparisons on the performance of a system across tasks with different class prevalence.
Given a set of documents, where some are positive and some are negative for a given task, a good measure of the quality of a specific document ordering is the area under the receiver operating curve (ROC), usually abbreviated “AUC”. This measure is the area under the curve traced out by graphing the true positive rate against the false positive rate, at all thresholds of sensitivity for a given document classification system, where 1.0 is a perfect score and 0.50 is equivalent to a random ordering. The AUC is a good measure of the quality of work prioritization for a given classifier because it is equivalent to the probability of a randomly chosen positive sample being ranked higher than a randomly chosen negative sample.4 It is independent of class prevalence and therefore is a good overall measure to use when the false positive/negative cost tradeoff is not known in advance or when comparing performance on tasks with different class prevalence.
In this work we use AUC as a consistent metric to compare alternative methods for applying machine learning techniques to automated work prioritization of systematic review topics. First we investigate the best set of features to be used from among available feature types including n-gram, manual annotation, and natural language processing (NLP) derived features. Other biomedical document classification research has found that features derived from manual annotation, such as MeSH terms, and NLP-based conceptual features, such as those produced by MetaMap5, improved classification performance.6–9 We sought to determine whether these types of features improved SR classification as well.
Second, using the best feature set found, we compare the performance obtained using SR topic specific training data versus non-topic specific training data. Work of other investigators, such as Aphinyanaphongs6, 7, has shown that high quality EBM articles can be predicted with good accuracy irrespective of the particular biomedical subject domain of the article. Since the task of selecting high quality EBM articles is similar to a non-topic specific version of selecting articles for inclusion in a SR, we examined the level of performance improvement obtained when training on data specific to an individual topic compared to training on SR inclusion decision data not specific to that SR topic.
Methods
Evaluation
To evaluate the alternative classification feature sets (described below) we applied each of them in various combinations to the fifteen biomedical document triage topics that we have previously used in our work on the use of automated document classification for drug-related systematic review update.10 We term each unique combination of feature sets a feature system.
The test collection contains the titles, abstracts, and MeSH terms for over 8000 documents that have been judged by experts for inclusion in 15 different systematic drug reviews. Each review comprises between 300 and 3500 journal article judgments. The 15 review topics represent a wide range of drugs, diseases, and medical knowledge, and also appear to have a wide range of difficulty in terms of machine learning tasks. The collection is publicly available.11
Each feature system was evaluated by performing five repetitions of two-way cross-validation with stratification to keep the ratio of positive to negative samples consistent between training and test splits. Each two-way cross-validation was randomly and independently split, and each feature system was subject to the exact same sequence of splits. Therefore each measurement consisted of 10 measurements of the AUC, which were then averaged to create an average AUC score for each system for each topic.12 Systems were then compared across the set of 15 SR topics using a non-parametric rank-based repeated measures statistical analysis with post-hoc paired Wilcoxon tests.13 Systems were divided into statistically significant (α = 0.05) ranked performance groups, where differences between systems within a rank group are not statistically significant. Systems placed into lower rank groups are statistically distinct from the overall best performing system in each of the higher rank groups.
We also compared topic specific training data versus non-topic specific training data using the best overall feature system found in the first set of experiments. For each of the 15 SR topics, we compared cross-validation (intra-topic training) on the topic to training on the data samples from the remaining 14 topics (inter-topic training) prior to classification on the given topic. To improve comparability, we used the same sequence of splits for both intra- and inter-topic training, scoring AUC on a one half of the given topic data at a time, repeating the splits five times, and averaging the results. Results were compared topic-by-topic for statistically significant differences using the t-test with α = 0.05.
Feature Systems
Algorithms based on support vector machines have been consistently shown to work well on biomedical text with large numbers of sparse features and are one of the most popular methods for classifying biomedical text.8, 14 We used the SVMLight14 implementation of SVM at default settings to compare feature systems including: unigram based features, n-gram based features of token length 1–4, annotation based features using MeSH terms, and UMLS (Unified Medical Language System) CUI-based features extracted using Metamap by applying the MMTx 2.4.C distribution. All features were represented as binary vectors where the element of a feature vector was set to 1 if that feature was included in a given document and 0 otherwise. Test samples were ranked for AUC computation based on the SVM signed-margin distance. We compared systems using feature types both singly and in combination.
Unigram (ABTITLE) and n-gram based features (NGRAM12, NGRAM13, and NGRAM14) were extracted from the text of the title and abstract of the MEDLINE record for each of the articles in the dataset using the StandardAnalyzer in Lucene15 as a tokenizer, which also applies a small stop-word list. N-grams consisted of from one to four contiguous tokens not separated by punctuation.
For features based on MeSH terms (MESH), we included features representing primary terms, main terms and subheadings separately, as well as complete MeSH terms as given in the MEDLINE record. For the Metamap features, we evaluated systems using two types of UMLS CUI-based features. The first type (MMTXFIRST) used just the first and highest scoring UMLS term identified for each phrase. The second type used all of the high scoring UMLS terms matched to a given phrase in the text (MMTXALL). All features of the given types were input to the SVM classifier. In order to limit the number of possible combinations and interactions to a manageable set, neither stemming nor feature selection was performed.
Results
Average AUC for each of the 13 compared feature systems over each of the 15 tasks are shown in Table 1. The mean average AUC for each system across all topics is also shown at the bottom of the table. The table separates groups of systems into statistically indistinguishable equal rank groups according to the results of the RMEQ analysis (α = 0.05 significance).
Table 1.
Comparison of mean cross-validation AUC across all 15 tasks for various combinations of feature sets.
| Rank Group 1 System Features | Rank Group 2 System Features | |||||
|---|---|---|---|---|---|---|
| TASK | ABTITLE+MESH+NGRAMS12 | ABTITLE+MESH+MMTXFIRST | ABTITLE+MESH | ABTITLE+NGRAMS13 | ABTITLE+NGRAMS14 | ABTITLE+NGRAMS12 |
| ACEInhibitors | 0.9462 | 0.9362 | 0.9378 | 0.9465 | 0.9465 | 0.9478 |
| ADHD | 0.9244 | 0.9054 | 0.9262 | 0.9155 | 0.9125 | 0.9203 |
| Antihistamines | 0.7228 | 0.6913 | 0.7048 | 0.7185 | 0.7289 | 0.7060 |
| AtypicalAntipsychotics | 0.8180 | 0.8045 | 0.8013 | 0.8256 | 0.8254 | 0.8164 |
| BetaBlockers | 0.8911 | 0.8679 | 0.8721 | 0.8690 | 0.8675 | 0.8704 |
| CalciumChannelBlockers | 0.8738 | 0.8408 | 0.8419 | 0.8397 | 0.8431 | 0.8349 |
| Estrogens | 0.8876 | 0.8692 | 0.8753 | 0.8502 | 0.8456 | 0.8526 |
| NSAIDS | 0.9515 | 0.9383 | 0.9412 | 0.9538 | 0.9546 | 0.9496 |
| Opioids | 0.8972 | 0.9086 | 0.8996 | 0.9017 | 0.8994 | 0.9029 |
| OralHypoglycemics | 0.7815 | 0.7603 | 0.7753 | 0.7530 | 0.7478 | 0.7598 |
| ProtonPumpInhibitors | 0.8600 | 0.8360 | 0.8429 | 0.8573 | 0.8589 | 0.8546 |
| SkeletalMuscleRelaxants | 0.7383 | 0.6851 | 0.6594 | 0.5988 | 0.5974 | 0.5962 |
| Statins | 0.9007 | 0.8704 | 0.8785 | 0.8617 | 0.8617 | 0.8555 |
| Triptans | 0.9097 | 0.9204 | 0.9112 | 0.9054 | 0.9009 | 0.9076 |
| UrinaryIncontinence | 0.8905 | 0.8878 | 0.8825 | 0.8784 | 0.8766 | 0.8782 |
| MEAN | 0.8662 | 0.8481 | 0.8500 | 0.8450 | 0.8445 | 0.8435 |
The overall top scoring system includes features based on unigrams, MeSH terms, and n-grams of length 2 (ABTITLE+MESH+NGRAM12) with a mean average AUC of 0.8662 overall. The other system in the top rank group incorporated MMTXFIRST features instead of n-gram features (ABTITLE+MESH+MMTXFIRST), had a mean average AUC of 0.8481, and was statistically indistinguishable from the top ranked system.
The systems combining unigram-based features with either MeSH or higher order n-gram based features performed equivalently and comprised rank group 2. There were no significant differences between using n-gram features of lengths up to two, three or four. Rank group 3 includes the ABTITLE+MESH+MMTXALL system, as well as the system that used only MESH-based features. Rank group 4 shows equivalent performance between the ABTITLE+MMTXFIRST and ABTITLE system, implying there was no significant performance gain by adding MMTXFIRST features to unigram based features. The ABTITLE+MMTXALL system placed alone in the fifth rank group, performing worse than the ABTITLE+MMTXFIRST system. Systems consisting solely of MMTx features, MMTXFIRST and MMTXALL, finished in the lowest rank group with mean average AUCs of 0.7423 and 0.7333.
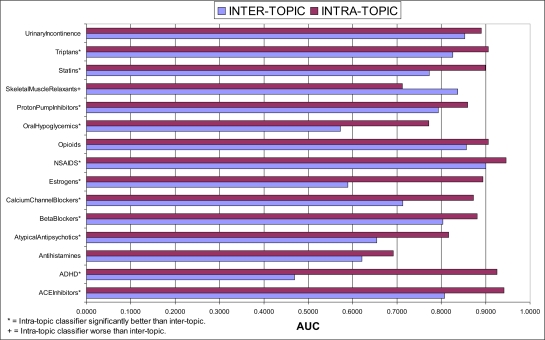
Figure 1 presents the results of comparing training on inter- versus intra-topic documents. Intra-topic performance is significantly better than inter-topic on 11 of the 15 topics, and about equal for 3 topics. For 9 of the topics seeing an improvement, the difference is greater than 0.10, and for some topics, such as ADHD, Estrogens, and OralHypoglycemics, the inter-topic results are not much better than random, while the intra-topic results are quite good. Interestingly, for one topic, SkeletalMuscleRelaxants, the inter-topic AUC is actually much better than the intra-topic performance.
Figure 1.
Comparison of mean AUC between inter- and intra-topic training for all 15 topics.
Discussion
Several useful conclusions can be made from these results in terms of optimizing AUC for SR review document prioritization. First and foremost, there was no advantage to be gained using the MMTx derived UMLS CUI features over other feature types. While a system using these features was included in the top rank group, the ABTITLE+MESH+NGRAMS12 system performed just as well, if not better. Since extracting UMLS CUI features with MMTx is a computationally and time-intensive operation, and extracting n-grams is fast and simple, n-gram based features, in combination with MeSH terms, are to be preferred. Also, while inclusion of n-gram features was helpful in achieving maximum performance, there was no increased benefit in going from 2-gram to 3- or 4-gram length features.
This result contrasts somewhat with that of Yetisgen-Yildiz and Pratt, who found that including MMTx-derived features improved results over their text only system when applied MEDLINE abstracts. However, they did not include n-gram features in their work.9 In fact, in only this one case (ABTITLE+MESH+MMTXFIRST) did adding MMTx-based features to a system (ABTITLE+MESH) improve that system’s rank group, and as noted, this improvement was not more than that derived from adding simple n-grams. For the other feature systems, adding MMTX-based features did not show an improvement. The ABTITLE+MESH system performed significantly better than the ABTITLE+MESH+MMTXALL system. We were hoping that MMTx-derived features would be helpful in situations where MeSH terms were not available. This was not the case, as the ABTITLE system performed about the same as the ABTITLE+MMTXFIRST system, implying that adding the MMTXFIRST features did not improve performance to the unigram-only system. Taken alone, it did not matter whether MMTXFIRST or MMTXALL features were used, as both MMTX-only systems performed poorly. However the MMTXFIRST features were beneficial in combination with other features (ABTITLE+MESH+MMTXFIRST), while the MMTXALL features were somewhat disruptive to classification task performance.
MeSH-based features were essential for top performance. Removal of these features from a system resulted in a system of lower performance. For example, removing MeSH features from the best ABTITLE+MESH+NGRAMS12 system resulted in the ABTITLE+NGRAMS12 system being ranked one group lower. Removing MeSH features from the ABTITLE+MESH system resulted in the ABTITLE system ranking two groups lower.
The results shown in Figure 1 make it clear that training on data specific to a SR topic almost always achieves performance greater than training on more general systematic review inclusion data, and is sometimes essential for good performance. Therefore topic-specific training should be done when data is available, such as when updating a previous SR. However, there are two circumstances where training on general SR inclusion data may be warranted. First, when no topic-specific training data is available general training on SR inclusion data may be sufficient. Across the 15 studied topics the mean average AUC was 0.7380 when trained on inter-topic documents, as compared to 0.8610 for intra-topic. Some topics, such as Triptans, did well with inter-topic training (average AUC = 0.8262), although not as well as with intra-topic. This level of performance may be acceptable for some uses. Secondly, for one topic, SkeletalMuscleRelaxants, the intra-topic performance was poor. An examination of the dataset shows the likely reason for this. There are only 9 positive documents out of 1643 for this topic. This is the lowest absolute number of positive documents across the 15 topics and evidently was not enough positive data to adequately characterize the task. The general SR training data more adequately described the parameter space for inclusion in this SR.
Conclusion
In this work we have shown that automated work prioritization can be accomplished with high performance across a range of drug-related SR topics with a straightforward feature set. Extension to non-drug related SRs is an area for future work. Unlike in previous work, we found that NLP-based features do not provide a significant advantage over simpler feature types. We have also shown that topic-specific training data is required for best performance. However, there are circumstances where the performance achieved using classifiers trained on general (inter-topic) training data may be useful or necessary. To allow focus on the feature system issues, stemming, feature selection, and feature weighting were not included. Also, it is presently unknown what level of performance is necessary in order to provide adequate value to the SR process. Furthermore, prospective studies must be conducted to determine what level of performance can be achieved on future document collections. Further work will investigate these and related issues.
| Rank Group 3 System Features | Rank Group 4 System Features | Rank Group 5 System Features | Rank Group 6 System Features | |||||
|---|---|---|---|---|---|---|---|---|
| TASK | ABTITLE+MESH+MMTXALL | MESH | ABTITLE+MMTXFIRST | ABTITLE | ABTITLE+MMTXALL | MMTXFIRST | MMTXALL | |
| ACEInhibitors | 0.9329 | 0.9206 | 0.9312 | 0.9356 | 0.9260 | 0.7614 | 0.7917 | |
| ADHD | 0.8924 | 0.9286 | 0.9001 | 0.9193 | 0.8831 | 0.7215 | 0.6543 | |
| Antihistamines | 0.6912 | 0.6733 | 0.6585 | 0.6803 | 0.6627 | 0.5528 | 0.5668 | |
| AtypicalAntipsychotics | 0.8004 | 0.7781 | 0.7915 | 0.7839 | 0.7882 | 0.7569 | 0.7574 | |
| BetaBlockers | 0.8534 | 0.8479 | 0.8429 | 0.8388 | 0.8275 | 0.7468 | 0.7307 | |
| CalciumChannelBlockers | 0.8357 | 0.8352 | 0.8049 | 0.8015 | 0.8014 | 0.7294 | 0.7577 | |
| Estrogens | 0.8506 | 0.8729 | 0.8345 | 0.8339 | 0.8122 | 0.7155 | 0.6718 | |
| NSAIDS | 0.9402 | 0.9095 | 0.9298 | 0.9355 | 0.9318 | 0.8289 | 0.8767 | |
| Opioids | 0.9035 | 0.8822 | 0.9120 | 0.8984 | 0.9032 | 0.8246 | 0.8061 | |
| OralHypoglycemics | 0.7554 | 0.7260 | 0.7369 | 0.7519 | 0.7348 | 0.6650 | 0.6902 | |
| ProtonPumpInhibitors | 0.8154 | 0.8404 | 0.8224 | 0.8291 | 0.8022 | 0.7586 | 0.7525 | |
| SkeletalMuscleRelaxants | 0.6748 | 0.6642 | 0.6046 | 0.5734 | 0.5995 | 0.6071 | 0.5360 | |
| Statins | 0.8649 | 0.8500 | 0.8301 | 0.8319 | 0.8259 | 0.7552 | 0.7487 | |
| Triptans | 0.9192 | 0.8423 | 0.9121 | 0.9008 | 0.9098 | 0.8623 | 0.8269 | |
| UrinaryIncontinence | 0.8811 | 0.8493 | 0.8753 | 0.8678 | 0.8655 | 0.8492 | 0.8321 | |
| MEAN | 0.8407 | 0.8280 | 0.8258 | 0.8255 | 0.8183 | 0.7423 | 0.7333 | |
Acknowledgments
This work was supported by grants 1R01LM009501-01 from the National Library of Medicine and ITR-0325160 from the National Science Foundation.
References
- 1.Mulrow C, Cook D. Systematic Reviews: Synthesis of Best Evidence for Health Care Decisions. Philadelphia, PA: The American College of Physicians; 1998. [Google Scholar]
- 2.Mallett S, Clarke M. How many Cochrane reviews are needed to cover existing evidence on the effects of health care interventions? ACP J Club. 2003;139(1):A11. [PubMed] [Google Scholar]
- 3.Cohen AM, Hersh W. A survey of current work in biomedical text mining. Briefings in Bioinformatics. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 4.Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform. 2005;38(5):404–15. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 6.Aphinyanaphongs Y, Statnikov A, Aliferis CF. A comparison of citation metrics to machine learning filters for the identification of high quality MEDLINE documents. J Am Med Inform Assoc. 2006;13(4):446–55. doi: 10.1197/jamia.M2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12(2):207–16. doi: 10.1197/jamia.M1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersh WR, Cohen AM, Yang J, Bhupatiraju RT, Roberts P, Hearst M. TREC 2005 genomics track overview. Proceedings of the Fourteenth Text Retrieval Conference - TREC 2005; 2005; Gaithersburg, MD. 2005. [Google Scholar]
- 9.Yetisgen-Yildiz M, Pratt W. The effect of feature representation on MEDLINE document classification. AMIA Annu Symp Proc. 2005:849–53. [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification. JAMIA. 2006;13(2):206–219. doi: 10.1197/jamia.M1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen AM.Systematic Drug Class Review Gold Standard Data, http://davinci.ohsu.edu/~cohenaa/systematic-drug-class-review-data.html, accessed Feb 19 2008.
- 12.Fawcett T. ROC Graphs: Notes and Practical Considerations for Researchers. Palo Alto, CA: HP Labs; 2004. [Google Scholar]
- 13.Cohen AM, McWeeney SK.RMEQ: A tool for computing equivalence groups in repeated measures studies Linking Literature, Information and Knowledge for Biology Proceedings of the BioLINK2008 Workshop2008;Toronto, ON2008 [Google Scholar]
- 14.Joachims T. Text categorization with support vector machines: learning with many relevant features. Proceedings of the 10th European Conference on Machine Learning; 1998. pp. 137–142. [Google Scholar]
- 15.Apache Software Foundation, Lucene, 2008http://lucene.apache.org/, accessed January 31, 2008.