Abstract
Auditing large, rapidly evolving terminological systems is still a challenge. In the case of RxNorm, a standardized nomenclature for clinical drugs, we argue that quality assurance processes can benefit from the recently released application programming interface (API) provided by RxNav. We demonstrate the usefulness of the API by performing a systematic comparison of alternative paths in the RxNorm graph, over several thousands of drug entities. This study revealed potential errors in RxNorm, currently under review. The results also prompted us to modify the implementation of RxNav to navigate the RxNorm graph more accurately. The RxNorm web services API used in this experiment is robust and fast.
Introduction
Auditing relations in biomedical terminologies generally requires the development of complex ad hoc programs [1, 2]. Terminological systems such as the Unified Medical Language System (UMLS), SNOMED CT and RxNorm are published as relational tables. Traversing graphs of relations in these systems typically requires multiple queries to the database to be integrated into specific programs.
In the past few years, programming interfaces have been developed for the UMLS [3, 4] and RxNorm [5], as well as for generic terminology services, such as the HL7 Common Terminology Services [6] and their implementation through LexGrid [7]. Such application programming interfaces (APIs) consist of a set of functions that can be embedded in programs (e.g., to get all the synonyms of a given concept), allowing users to manipulate the terminology programmatically without having to perform low-level queries against a database. One popular form of APIs is web services, a collection of protocols (e.g., Simple Object Access Protocol or SOAP) and standards (e.g., XML) for interchanging data between applications [8]. Users of the web services can use a variety of languages such as Java and Perl to invoke the web services.
The Web Services API recently released for RxNorm provides various functions for exploring the relations among drug entities in RxNorm. For this reason, it appears to be suitable for testing the consistency of the relations represented in RxNorm. The objective of this paper is to introduce to readers the functionality of the RxNorm API and demonstrate its usefulness as a Quality Assurance tool in verifying the structure and contents of the RxNorm data set.
Background
RxNorm is a standardized nomenclature for clinical drugs developed by the National Library of Medicine [9, 10]. The RxNorm data set is organized around concepts with normalized drug names which can include information about ingredients, strengths and dose forms. RxNorm uses “term types” (listed in Table 1 below) to distinguish among these various kinds of drug entities.
Table 1.
RxNorm Term Types
| Term Type | Example |
|---|---|
| Ingredient | Cetirizine |
| Precise ingredient | Cetirizine Dihydrochloride |
| Brand name | Zyrtec |
| Clinical drug component | Cetirizine 5 MG |
| Branded drug component | Cetirizine 5 MG [Zyrtec] |
| Clinical drug name | Cetirizine 5 MG Oral Tab- |
| Branded drug name | Zyrtec 5 MG Oral Tablet |
| Clinical drug form | Cetirizine Oral Tablet |
| Branded drug form | Cetirizine Oral Tablet [Zyrtec] |
| Dose form | Oral Tablet |
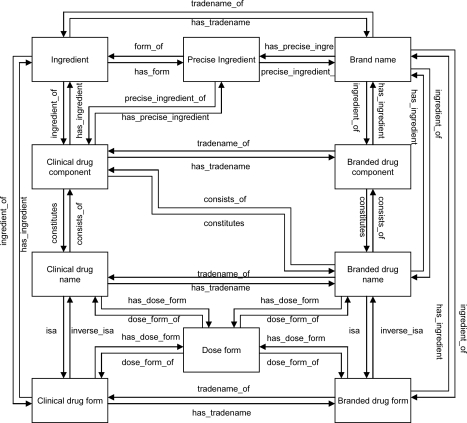
The RxNorm drug entities are related to each other by a well-defined set of named relationships. For example, brand name concepts are related to branded drug component concepts by the relationships ingredient_of and has_ingredient. Figure 1 shows the relationships between the various kinds of drug entities.
Figure 1.
Relations among RxNorm entities
RxNorm Web Services API
A browser called RxNav1 was developed in 2004 to access the RxNorm data set and display graphically all related concepts and the relations between them. RxNav uses web services to access the RxNorm data. In early 2008, the web services that access the RxNorm data were enhanced and made available publicly. The current API comprises functions for resolving drug names and codes into RxNorm identifiers, for accessing the properties of drug concepts (including their relations to other drug concepts), as well as various housekeeping functions. The complete list of functions of the API is displayed in Annex 1. In addition, a description of the API in the Web Service Definition Language (WSDL) is available at http://mor.nlm.nih.gov/download/RxNormDBService.wsdl.
Quality assurance in RxNav
RxNorm data have the structure of a graph. As shown in Figure 1, RxNorm relations are often purposely redundant. For example, given an ingredient, to get the related clinical drug names the following paths could be taken:
Path 1:
Get the clinical drug components of the ingredients using the ingredient_of relationship.
Get the clinical drug names of the clinical drug components using the consists_of relationship.
Path 2:
Get the clinical drug forms of the ingredients using the ingredient_of relationship.
Get the clinical drug names of the clinical drug forms using the inverse_isa relationship.
In terms of quality assurance, one major concern is that the traversal implemented in RxNav for linking two kinds of drug entities (e.g., ingredient and clinical drug) may not yield the same results as alternate paths (e.g., paths 1 and 2 above).
In this study, we use functions from the RxNorm API to assess the consistency of traversal of the RxNorm graph when using several alternate paths.
Methods
In selecting alternate relationship paths to compare, the paths actually implemented in the RxNav application were first examined. For historical reasons, the most direct path between two kinds of drug entities was not always used. For example, as shown in Figure 1, when starting with a brand name, it is possible to get the related branded drug names directly by using the ingredient_of relationship. However, the RxNav application actually gets the branded drug forms from the brand name with the ingredient_of relationship and then uses those branded drug forms with the inverse_isa relationship to retrieve the branded drug names.
For the study, four sets of paths were chosen, based on the fact that the path used in the RxNav application was not a direct path, but that a direct path did exist. So both the indirect path used in the application and the direct path not used were selected for comparison. In addition to comparing direct and indirect paths, we also wanted to compare several indirect paths. To this end, we added a second indirect path to one of the sets. Table 2 below shows the paths which were selected – the relations between the term types are omitted.
Table 2.
Paths tested
| Set Id | Path taken |
|---|---|
| Set 1 direct | Brand name → branded drug name |
| Set 1 indirect | Brand name → branded drug form → branded drug name |
| Set 2 direct | Branded drug form → clinical drug form |
| Set 2 indirect | Branded drug form → branded drug name → clinical drug name → clinical drug form |
| Set 3 direct | Ingredient → brand name |
| Set 3 indirect | Ingredient → clinical drug component → branded drug name → branded drug form → brand name |
| Set 4 direct | Clinical drug form → ingredient |
| Set 4 indirect 1 | Clinical drug form → clinical drug name → branded drug name → clinical drug component → ingredient |
| Set 4 indirect 2 | Clinical drug form → clinical drug name → clinical drug component → ingredient |
To test the data, a Java program was created to use the RxNorm API functions. The program takes as input a file of RxNorm identifiers and reads command line parameters to determine which API functions to call. The returns from the API calls are printed to a file.
For the direct paths, the API function getRelated-ByRelationship is used. For example, from the brand name Zyrtec (RxCUI = 58930), this function returns five branded drug names, including Zyrtec 10 MG Chewable Tablet (541030) when called with the relationship ingredient_of as parameter.
For the indirect paths, the API function getRelatedByType is used since this is the function called by the RxNav application and reflects the indirect path listed in Table 2. For example, from the brand name Zyrtec, this function also returns five branded drug names when called with the term type “SBD” (for branded drug name) as parameter.
Also, getRelatedByRelationship was used in the analysis phase to test segments of the indirect path to determine the source of the differences between the direct and indirect paths.
The paths were tested using all RxNorm concepts of the starting term type for the set. The March 2008 version of the RxNorm data set was used. This included 3,460 ingredients, 9,716 brand names, 11,346 branded drug forms and 8,154 clinical drug forms.
Results
Table 3 shows the results of the paths tested in Table 2. The second column of the table indicates the number of concepts that were tested of the starting term type. For example, in set 1, 9,716 brand name concepts were tested. The third column indicates how many of those start concepts led to 1 or more target concepts from the path taken. The fourth column indicates how many concepts were found at the final term type in the path.
Table 3.
Path Results
| Set Id | Start concepts | Start # found | # target concepts |
|---|---|---|---|
| Set 1 direct | 9,716 | 9,696 | 14,499 |
| Set 1 indirect | 9,716 | 9,696 | 14,499 |
| Set 2 direct | 11,346 | 11,346 | 11,346 |
| Set 2 indirect | 11,346 | 11,312 | 11,312 |
| Set 3 direct | 3,460 | 1,710 | 16,508 |
| Set 3 indirect | 3,460 | 1,701 | 16,360 |
| Set 4 direct | 8,154 | 8,154 | 12,436 |
| Set 4 indirect1 | 8,154 | 4,020 | 5,790 |
| Set 4 indirect2 | 8,154 | 8,094 | 12,340 |
Discussion
Findings
In all cases, the direct path yielded at least as many results as the indirect path and only in set 1 did the direct and indirect paths produce exactly the same results.
The results of set 1 (retrieving branded drug names starting with brand names) did reveal that 20 brand names have no currently related branded drug names. An example is the brand name Centrax. Upon further investigation it was discovered that a branded drug name originally existed for Centrax, but was now obsolete. The RxNorm data set contains the obsolete record, but obsolete records are not used by the API or in RxNav. Similarly, the other 19 brands names also had obsolete branded drug names.
In set 2 (retrieving clinical drug forms starting with branded drug forms) it was expected there would be one target concept for each starting concept. While this was true in the direct path, 34 target concepts were missing in the indirect path. For example, the branded drug form Ketorolac Injectable Solution [Toradol IM] does not map to a clinical drug form in the indirect path. Further analysis showed that these branded drug forms had no current relationships to any branded drugs. The reason for this is that the branded drug names are obsolete, similar to those in set 1.
In set 3 (retrieving brand names starting with ingredients) the indirect path target concepts are not a subset of the direct path target concepts. There are 26 indirect path brand name instances identified that do not exist in the direct path. This would seem to indicate missing direct relationships between the ingredient and the brand name. Conversely, there are 174 instances of brand names in the direct path that are missing from the indirect path. All of these appear to be errors – for example, the ingredient Bisacodyl is related to the brand name Colax through the has_tradename relationship. However, the branded drug name, branded drug component and branded drug form related to Colax do not contain Bisacodyl as an ingredient. The direct path is this set appears not to be a better choice currently.
In set 4 (retrieving ingredients from clinical drug forms) the indirect path 1 used in RxNav produces many fewer target concepts than the direct path. This is because the path goes through the brand drug names even though both the start and end term types are associated with clinical (generic) drug. Many clinical drug forms do not have related brand names, so going through the branded drug names is in error. For example, hydrogen peroxide mouthwash has no branded drug names, so the indirect path 1 returns no ingredients.
Indirect path 2 uses only paths through clinical drug data, and as expected the results are much better. However, 60 clinical drug form concepts yielded no ingredients in this path because these drug forms contained no current relationship to a clinical drug name. An example of this is the clinical drug form magnesium citrate oral tablet. Once again, obsolete forms of clinical name drugs exist in the RxNorm data set for this concept, but there are no current clinical name drugs.
Practical implications
The implications of these quality assurance tests are two-fold. This experiment made it clear that the indirect paths originally implemented in RxNav are currently suboptimal and have the potential to misrepresent the RxNorm dataset. As a consequence, we decided to modify the implementation of RxNav in order to benefit from accurate direct paths whenever possible. (We traced the original design of RxNav and the use of indirect paths to issues with early versions of RxNorm data that have long been corrected.)
The discrepancies identified in the traversal of the RxNorm graph between direct and indirect paths and between alternative indirect paths may be indicative of errors in the RxNorm dataset. These potential problems have been reported to the curators of RxNorm and several have been fixed in releases since our testing with other changes scheduled for a future release. The relatively small number of discrepancies identified in the systematic examination of alternate paths in our study is a testimony to the high quality and careful curation of the RxNorm database overall. However, these potential errors also show how difficult it is to ensure the quality of data in a large, highly redundant and rapidly evolving database such as RxNorm.
This investigation was also an opportunity to apply the recently released RxNorm API in a relatively intensive application. The web services implementation provided support for easy integration of the RxNav functions in the program developed for checking the consistency of the RxNorm graph. The web services provided both convenience and speed.
Limitations.
The study only evaluated a subset of all the possible paths through the relationships in RxNorm. In the future, we plan to pursue the systematic investigation of the RxNorm dataset, using the knowledge of the obsolete clinical and branded drugs to filter out false positives and restricting the paths to stay within the clinical or branded relations when possible. In particular, we would like to use graph-based systems (e.g., Semantic Web technologies such as RDF, the Resource Description Framework) to develop a thorough and routine analysis of the RxNorm graph, therefore contributing to the quality assurance process of RxNorm.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM).
Annex 1.
List of functions of the RxNorm Web Services API
| •findRxcuiByString(searchString) |
| Search for a name in the RXNORM data set and return the RXCUIs of any concepts which have that name as an RxNorm term or as a synonym of an RxNorm term. |
| • findRxcuiById(idType, id) |
| Search for an identifier from another vocabulary and return the RXCUIs of any concepts which have an RxNorm term as a synonym or have that identifier as an attribute. |
| • getSpellingSuggestions(searchString) |
| Get spelling suggestions for a given term. The suggestions are RxNorm terms contained in the current version. |
| • getRxConceptProperties(rxcui) |
| Get the RxNorm Concept properties |
| • getRelatedByRelationship(rxcui, relationship-list) |
| Get the related RxNorm identifiers of an RxNorm concept specified by a relational attribute list. |
| • getRelatedByType(rxcui, type-list) |
| Get the related RxNorm identifiers of an RxNorm concept specified by one or more term types. |
| • getAllRelatedInfo(rxcui) |
| Get all the related RxNorm concepts for a given RxNorm identifier. |
| • getDrugs(name) |
| Get the drug products associated with a specified name. The name can be an ingredient, brand name, clinical drug form, branded drug form, clinical drug component, or branded drug component. |
| • getNDCs(rxcui) |
| Get the National Drug Codes (NDCs) for the RxNorm concept. |
| • getRxNormVersion() |
| Get the version of the RxNorm data set. |
| • getIdTypes() |
| Get the valid identifier types of the RxNorm data set. See findRxCuiById for use of these types. |
| • getRelaTypes() |
| Get the relationship names in the RxNorm data set. |
| • getTermTypes() |
| Get the valid term types in the RxNorm data set. |
Footnotes
References
- 1.Cimino JJ. Auditing the Unified Medical Language System with semantic methods. J Am Med Inform Assoc. 1998;5(1):41–51. doi: 10.1136/jamia.1998.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halper M, Wang Y, Min H, Chen Y, Hripc-sak G, Perl Y, et al. Analysis of error concentrations in SNOMED. AMIA Annu Symp Proc. 2007 [PMC free article] [PubMed] [Google Scholar]
- 3.Bangalore A, Thorn KE, Tilley C, Peters L. The UMLS knowledge source server: an object model for delivering UMLS data. AMIA Annu Symp Proc. 2003:51–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Mirhaji P, Kunapareddy N, Michea Y, Srinivasan A. A Web Services architecture for UMLS Knowledge Sources. AMIA Annu Symp Proc. 2005:1055. [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng K, Bodenreider O, Kilbourne J, Nelson S. RxNav: a web service for standard drug information. AMIA Annu Symp Proc. 2006:1156. [PMC free article] [PubMed] [Google Scholar]
- 6.HL7 Common Terminology Services: http://informatics.mayo.edu/LexGrid/downloads/CTS/specification/ctsspec/cts.htm
- 7.LexGrid Common Terminology Services: http://informatics.mayo.edu/LexGrid/index.php?page=ctsimpl
- 8.Anzbock R, Dustdar S. Modeling and implementing medical Web services. Data & Knowledge Engineering. 2005;55(2):203–236. [Google Scholar]
- 9.Liu S, Ma W, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange. IT Professional. 2005;7(5):17–23. [Google Scholar]
- 10.RxNorm: http://www.nlm.nih.gov/research/umls/rxnorm/