Abstract
Although it has long been recognized that the enteric community of bacteria that inhabit the human distal intestinal track broadly impacts human health, the biochemical details that underlie these effects remain largely undefined. Here, we report a broad MS-based metabolomics study that demonstrates a surprisingly large effect of the gut “microbiome” on mammalian blood metabolites. Plasma extracts from germ-free mice were compared with samples from conventional (conv) animals by using various MS-based methods. Hundreds of features were detected in only 1 sample set, with the majority of these being unique to the conv animals, whereas ≈10% of all features observed in both sample sets showed significant changes in their relative signal intensity. Amino acid metabolites were particularly affected. For example, the bacterial-mediated production of bioactive indole-containing metabolites derived from tryptophan such as indoxyl sulfate and the antioxidant indole-3-propionic acid (IPA) was impacted. Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. Multiple organic acids containing phenyl groups were also greatly increased in the presence of gut microbes. A broad, drug-like phase II metabolic response of the host to metabolites generated by the microbiome was observed, suggesting that the gut microflora has a direct impact on the drug metabolism capacity of the host. Together, these results suggest a significant interplay between bacterial and mammalian metabolism.
The human body is colonized by hundreds of trillions of microbes, which collectively possess hundreds of times as many genes as coded in the human genome. The combined genetic potential of the endogenous flora is referred to as the “microbiome” (1), and typically results in a mutualistic relationship between microbe and host. Normal host activities, including the processing of nutrients and the regulation of the immune system, are affected by the intestinal microbiome (2, 3), and the microbiome has been implicated in the pathogenesis of diseases such as nonalcoholic steatohepatitis (4), allergy (5), the formation of gallstones (6), and inflammatory bowel disease (7, 8). The composition of the gut microbiome is highly variable (9), and its diversity can be significantly affected by alterations in diet (2, 10) or antibiotic use (11). Alternatively, probiotic therapy is the attempt to alter the extant gut microbial environment through the ingestion of live consumable cultures of beneficial bacteria (12). Differences in commensal microflora are likely to impact human health and disease through any number of ways reflective of the complex nature of the microbiome itself.
Historically, classical microbiology methods including the isolation and culture of individual bacterial species associated with the gut were used to study microbial colonization of higher organisms. More recently, metagenomic techniques have been used to characterize both the composition and the potential physiological effects of entire microbial communities without having to culture individual community members. For example, genes for specific metabolic pathways such as amino acid and glycan metabolism were found to be overrepresented in the microbiome of the distal gut, reinforcing the notion that human metabolism is an amalgamation of microbial and human attributes (13). Also, obese mice were found to extract energy from their food more efficiently compared with lean counterparts due to alterations in the composition of their gut microflora that resulted in an increased complement of genes for polysaccharide metabolism (10). It has also been observed that bile salt hydrolase encoding genes are enriched in the gut microbiome, and that enteric bacteria carry out a wide range of bile acid modifications (6, 14). These metagenomic studies suggest that the metabolites derived from this diverse microbial community can have a direct role in human health and disease. To date, metabolomics-based investigations of aspects of the impact of the microbiome on mammalian biochemistry have detailed changes in the levels of well-documented metabolites based primarily on NMR-based analysis and subsequent multivariate statistics of unfractionated samples, such as urine, gut tissue, or cecum extracts (15–17). Recently, this same group reported the multicompartmental effects of the microbiome on murine metabolism by using NMR-based analysis of urine and tissue extracts from both conventional (conv) and germ-free (GF) mice (18). Although extremely powerful, these studies provide only limited opportunity for the discovery of differences in unexpected or lower level metabolites.
Here, we demonstrate the large effect of the microbiome on mammalian plasma biochemistry. Specifically, a broad, untargeted, mass spectrometry-based profiling of serum from GF and conv mice demonstrates that a significantly large number of chemical species found in systemic circulation arise because of the presence of the microbiome, whereas at least 10% of all detectable endogenous circulating serum metabolites vary in concentration by at least 50% between the 2 mouse lines. Several microbiome-affected molecules identified in the serum of conv mice are either potentially harmful (uremic toxins) or beneficial (antioxidant) to the host. Also, we observed a broad, drug-like phase II metabolic response of the host to species generated by the microbiome, as evidenced by the exclusive presence of numerous sulfated, glycine-conjugated, and glucuronide adducts in the serum of conv mice.
Results and Discussion
Mass Spectrometry Reveals That the Microbiome Has a Broad Effect on Mouse Plasma Biochemistry.
Mass spectrometry has become an increasing powerful tool for metabolomics studies due to its wide dynamic range, reproducible quantitative capabilities, and its ability to analyze samples of significant molecular complexity (19). However, the vast chemical diversity of the known metabolome creates numerous challenges for any MS-based “global” profiling effort, including analytical biases introduced by sample preparation, separation, and ionization techniques. Given these challenges, we sought to obtain an initial assessment of the extent of the effect of the microbiome on mammalian metabolism by analyzing uniformly processed samples employing various ionization methods, because studies have shown that such multiple ionization mode strategies increase both the number and types of analytes observed (20). We also chose to analyze plasma samples, because this biofluid provides an excellent opportunity to assess the extent of interplay between bacterial metabolic and systemic human pathways.
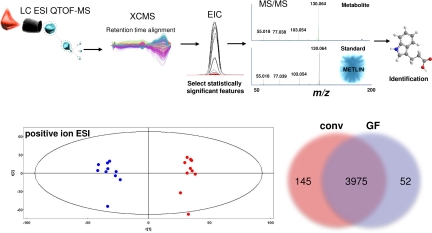
In Fig. 1 we show the experimental workflow for the metabolomics studies performed. Ten plasma samples each from both individual GF and conv mice were subjected to methanol protein precipitation and subsequent centrifugation, because this technique was previously reported to be the most effective, straightforward, and reproducible extraction method for such samples (21). The resulting supernatants were each analyzed by 4 different methods; reversed-phase liquid chromatography coupled to (i) electrospray ionization (ESI) in the positive mode (+), (ii) ESI in the negative mode (−), (iii) atmospheric pressure chemical ionization (APCI) in the + mode, and (iv) gas chromatography after derivatization with trimethylsilyl chloride. The raw data from the individual analyses were subjected to nonlinear data alignment (22), and the results were analyzed for both global changes by using multivariate statistics to determine group separation, as well as univariate statistics to evaluate the number and percentage of features that are either unique or vary significantly between the 2 sample sets. As seen in Fig. 1B, principle component analysis (plotting principle component 1 vs. principle component 2) shows an excellent separation of the GF and conv sample groups analyzed under + ESI conditions. Similar separations were also seen for the − ESI, + APCI, and GC-based analyses (Fig. S1). The majority of individual features detected were present in both the GF and conv samples. However, hundreds of features were detected in only 1 sample set, with the majority of these being unique to the conv samples. Also, as summarized in Table 1, ≈10% of all features detected in both sample sets under any of the analysis conditions showed significant changes in their relative signal intensity (defined as a ≥1.5-fold change with a P ≤ 0.0001). It should be noted that a given molecule may be represented by several different features, such as the naturally occurring components of its isotopic cluster or nonspecific adduct ions. Nevertheless, these comparative results show that the microbiome exerts a dramatic effect on mammalian plasma biochemistry.
Fig. 1.
The microbione exerts a broad effect on mammalian biochemistry. (Upper) The experimental workflow for the metabolomics studies performed. (Lower) Major differences are observed in the profiles of GF and conv plasma obtained employing + ESI. PCA analysis of the profiling data from XCMS (Left) shows an excellent separation between sample groups. As shown in the Venn diagram (Right), most species are observed in both GF and conv samples, but a significant number are unique to a given sample set, with ≈3-fold more ions unique to the conv sample.
Table 1.
Summary of comparative metabolomics profiles using different MS-based methodologies
| Total peaks | Total significant changed peaks* | Significant change conv > GF | Significant change GF > conv | Unique to conv | Unique to GF | |
|---|---|---|---|---|---|---|
| LC ESI + | 4,172 | 466, 11.2% | 318, 68% | 148, 32% | 145 | 52 |
| LC ESI − | 4,132 | 271, 6.6% | 168, 62% | 103, 38% | 100 | 61 |
| LC APCI + | 1,584 | 152, 9.5% | 96, 63% | 56, 37% | 27 | 5 |
| GCMS + | 1,803 | 253, 14.0% | 225, 88.9% | 28, 11.1% | 28 | 12 |
*Defined as having a fold-change ≥ 1.5 and P ≤ 0.0001.
We next turned our focus toward the identification of those features that showed significant differences in level between the 2 sample sets. This task is by far the most laborious part of any untargeted MS-based metabolomics profiling study, involving the integrated analysis of accurate mass measurements, tandem MS fragmentation patterns, and literature/database searches to produce candidate structures that still require experimental verification. For the purposes of this article, an observed feature was not designated as unequivocally identified unless an authentic sample of the proposed species was independently analyzed in our laboratories and found to exhibit the same chromatographic and mass spectrometric properties as the feature in question. By comparison, highly probable identification status was assigned when the experimentally determined properties of a given feature matched those reported in the literature for a particular molecule, or were fully consistent with those reported for a close analog. Although this process remains a work in progress, the identifications produced to date as listed in Table 2(and further detailed in Table S1) already exhibit several striking features. For example, despite the use of a single sample preparation method, we identified various chemically distinct compounds ranging from fatty acids to highly polar charged molecules, along with several groups of functionally related molecules.
Table 2.
Summary of metabolites identified that showed significant differences in concentration between the two sample sets
| Metabolite | Fold change | P value | Compound and metabolism class |
|---|---|---|---|
| Indole derivatives | |||
| Tryptophan | 1.7, GF | 8.42 × 10−12 | — |
| N-acetyltryptophan | 2.4, GF | 3.56 × 10−4 | — |
| Indoxyl sulfate | conv* | 1.34 × 10−7 | Phase I: hydroxyl; phase II: sulfate |
| Serotonin | 2.8, conv | 1.27 × 10−10 | — |
| IPA | conv* | 7.69 × 10−7 | Bacterial conjugation |
| Phenyl derivatives | |||
| Phenylalanine | 1.05, GF | 0.3 | — |
| Tyrosine | 1.44, GF | 1.14 × 10−4 | — |
| Hippuric acid | 17.4, conv | 1.98 × 10−9 | Phase II: glycine |
| Phenylacetylglycine | 3.8, conv | 4.70 × 10−8 | Phase II: glycine |
| Phenyl sulfate | conv* | 9.85 × 10−7 | Phase II: sulfate |
| p-Cresol sulfate | conv* | 0.002 | Phase II: sulfate |
| Phenylpropionylglycine | conv* | 3.07 × 10−7 | Phase II: glycine |
| Cinnamoylglycine | conv* | 2.93 × 10−7 | Phase II: glycine |
| Flavones | |||
| Equol sulfate | conv* | 1.44 × 10−5 | Phase II: sulfate |
| Methyl equol sulfate | conv* | 2.18 × 10−6 | Phase II: sulfate |
| Others | |||
| Urate | 1.99, conv | 1.51 × 10−6 | — |
| Creatinine | 1.08, conv | 0.071 | — |
| Dihydroxyquinoline glucuronide | conv* | 7.64 × 10−6 | Phase II: glucuronide |
| 12-Hydroxy-5Z,8Z,10E,14Z,17Z-eicosapentaenoic acid | 4.0, conv | 8.20 × 10−5 | Fatty acid |
| 3-Carboxy-4-methyl-5-pentyl-2-furanpropionic acid glucuronide | 3.4, conv | 1.37 × 10−6 | Phase II: glucuronide |
Fold change equals the fold difference in concentration observed between conv and GF samples, with the group indicating which is higher. Names in italics refer to compound identifications that are highly probable, as defined in the text.
*Compound observed only in a single group (conv or GF).
The Microbiome Affects the Diversity of Indole-Containing Compounds in Serum.
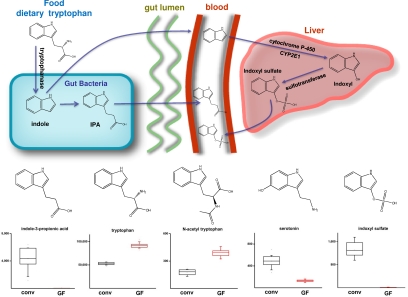
Multiple features that differed significantly between conv and GF mice correspond to compounds bearing indole moieties. For example, the plasma concentrations of tryptophan and N-acetyltryptophan in conv mice were 40 and 60% lower than in their respective GF counterparts. A subset of enteric bacteria express tryptophanase, which converts tryptophan to indole, pyruvate, and ammonia. Given that tryptophanase activity in conv mice has been shown to increase nearly 2-fold after dosing with tryptophan (23), it is highly likely that the decreased concentrations of tryptophan and N-acetyltryptophan observed in conv mouse serum resulted from the metabolism of dietary tryptophan by the direct action of enteric bacteria.
Interestingly, serotonin plasma levels were 2.8-fold higher in conv animals. However, this increase in serotonin is difficult to attribute to a direct metabolic transformation mediated by gut bacteria, because the production of serotonin by characterized enteric bacterial species has not been described. Despite the well-documented role of serotonin as a neurotransmitter, Enterochromaffin cells in the gut are the largest source of serotonin production in the body, and this molecule has been implicated in gastrointestinal pathologies such as irritable bowel syndrome and Crohn's disease (24). These findings suggest that the increased plasma serotonin levels observed could indirectly result from an as yet undefined host microbe interaction.
As seen in Fig. 2, other indole-containing molecules were also affected by the microflora. For example, indoxyl sulfate (indican) was identified only in the serum of conv mice. This molecule, a known nephrotoxin that accumulates in the blood of patients suffering from chronic kidney failure (25), arises from hepatic transformation of the bacterial metabolite indole. Because tryptophanase activity derives from only a subset of enteric bacteria, non-indole-producing bacteria, such as various Bifidobacterium species, have been administered as a test probiotic to dialysis patients to decrease their plasma levels of indoxyl sulfate (26). Conversely, a different set of enteric bacteria has been implicated in the metabolic transformation of indole to indole-3-propionic acid (IPA) (27). IPA, also identified only in the plasma of conv mice, has been shown to be a powerful antioxidant (28), and is currently being investigated as a possible treatment for Alzheimer's disease (29, 30).
Fig. 2.
The diversity of indole-containing compounds in serum is affected by the microbiome. (Upper) Various indole-containing molecules arise only in the presence of the microbiome and ultimately enter the plasma by means of different routes. (Lower) Differences in the plasma levels of indole-containing compounds attributed to the action of the microbiome. The integrated signal intensities plotted on the y axes are reduced by a factor of 1,000.
Although the presence of IPA in mammals has long been ascribed in the literature to bacterial metabolic processes, this conclusion was based on either the production of IPA in ex vivo cultures of individual bacterial species (31) or observed decreases in IPA levels in animals after administration of antibiotics (32). In our own survey of IPA production by representative members of the intestinal flora, only Clostridium sporogenes was found to produce IPA in culture (Table S2). Based on these results, individual GF mice were intentionally colonized with C. sporogenes strain ATCC 15579, and blood samples were taken at several intervals after colonization. IPA was undetectable in the samples taken shortly after introduction of the microbes, and was first observed in the serum 5 days after colonization, reaching plateau values comparable with conv mice by day 10. These colonization studies demonstrate that the introduction of enteric bacteria capable of IPA production in vivo into the gastrointestinal tract is sufficient to introduce IPA into the bloodstream of the host. Also, other GF animals were injected i.p. with either IPA (at 10, 20, or 40 mg/kg) or sterile PBS vehicle, and their serum concentrations of IPA were measured over time. As seen in Table S3, the high serum levels of IPA observed 1 h after injection decreased more than 90% within 5 h, showing that IPA is rapidly cleared from the blood, and that its presence in the serum of conv animals must result from continuous production from 1 or more bacterial species associated with the mammalian gut. The ability to measure the metabolic profiles of animals selectively colonized with individual species or even complex communities of defined bacterial populations promises to be a powerful new tool for deconvoluting the contributions of the various species comprising the microbiome and for dissecting complex microbial-mammalian metabolic interactions
Sulfation Is Observed as a Prevalent Host Response to Bacterial Metabolites.
Serum metabolites from GF and conv mice differed significantly in their level of sulfation. For example, both phenyl and p-cresol sulfate were present only in conv animals (Fig. S2). These molecules likely arose from the sulfation of direct bacterial metabolites of tyrosine (33, 34), the plasma concentration of which was 1.4-fold higher in GF animals.
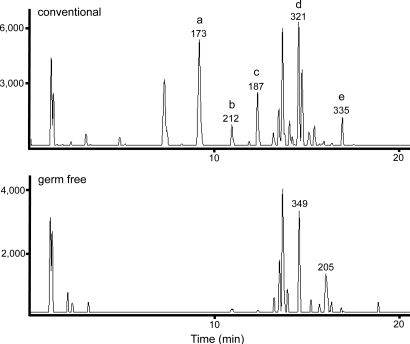
Sulfation is a well-studied phase II drug metabolism mechanism used by the body to facilitate the removal of hydrophobic endogenous species and xenobiotics. Given the identification of several sulfated molecules found only in conv animals, we decided to investigate the extent to which this human chemical detoxification mechanism targets microbiome affected vs. endogenous metabolites. One MS-based methodology for the detection of putative sulfated molecules employs a triple quadrupole mass spectrometer to scan for species that exhibit a neutral loss of 80 m/z when analyzed under negative ionization conditions (35). Thus, pooled serum samples from GF and conv mice were subjected to this more targeted analysis. As seen in Fig. 3, the conv sample resulted in nearly double the number of peaks (36) compared with its GF counterpart (23). Importantly, several of the peaks unique to the conv sample were later confirmed to be the already identified phenyl (peak a), indoxyl (peak b), and p-cresol (peak c) sulfates; thus, demonstrating the potential of this targeted scanning technique to quickly and effectively detect potential sulfated bacterial metabolites.
Fig. 3.
Sulfate profiling based on constant neutral loss scanning of 80 m/z in − ESI mode for conv (Upper) and GF (Lower) pooled plasma samples. The m/z values for several species are listed above their respective peak. Identified sulfates include: phenyl sulfate (a), indoxyl sulfate (b), p-cresol sulfate (c), equol sulfate (d), and methyl equol sulfate (e).
Next, we sought to confirm whether other unique features resulting from the targeted scanning technique were also sulfated molecules. Thus, equol sulfate was identified as being present only in the conv sample, whereas a second conv-specific feature was identified with high confidence as methyl equol sulfate (Fig. S3). Equol is itself a reported metabolite of the dietary isoflavone dihydrodaidzein. Purported to have the potential to lower the incidence of prostate cancer (37), equol has been shown to arise exclusively as a result of bacterial metabolic processes (38), although only a small percentage of humans harbor the gut bacteria that affect this biotransformation (39). It is important to note that neither equol sulfate nor its methyl ester was listed in any metabolite database, again emphasizing the degree of complexity involving the microbiome that remains to be explored.
Other Phase II Metabolic Products Are also Greatly Elevated in Response to the Microbiome.
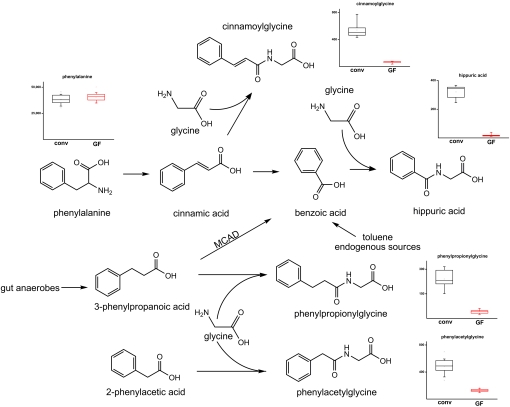
We next sought to determine whether other bacterial metabolites were modified by mammalian chemical detoxification processes. Indeed, numerous products of another prevalent phase II drug metabolism mechanism, glycine conjugation, were found to be either exclusively present or present at much higher concentrations in conv animals. As shown in Fig. 4, the plasma concentration of hippuric acid was 17-fold higher in conv samples compared with their GF counterparts. Hippuric acid is produced by the conjugation of benzoic acid with glycine, a reaction that occurs not only in the liver (40) but also directly in the intestine (41) and kidney (42). Also, it has been reported that hippuric acid levels increase when animals are fed increased levels of phenylalanine (43). However, the concentration of circulating phenylalanine was not observed to differ significantly between the 2 sample sets, suggesting that unlike the presence of indican or IPA in conv mice, the observed increased levels of hippuric acid may not simply result from the removal of some of the dietary supply of an essential amino acid by the direct action of enteric bacteria. These findings again illustrate the broad interplay of mammalian and enteric bacterial metabolism.
Fig. 4.
Differences in the plasma levels of various glycine-conjugated compounds attributed to the action of the microbiome. Potential metabolic pathways leading to the formation of hippuric acid as well as 3 other glycine conjugates arising from the presence of the microbiome are provided. The integrated signal intensities plotted on the y axes are reduced by a factor of 1,000.
In Fig. 4 we indicate several additional glycine conjugates identified exclusively in the serum of conv animals. For example, cinnamoylglycine was identified as being present only in conv samples despite not being listed in any metabolite database, illustrating the fact that our knowledge of the metabolome even in biofluids as well-studied as plasma is far from complete. Also, phenylpropionylglycine was also detected exclusively in conv samples. This molecule almost certainly arose from the direct conjugation of glycine to phenylpropionic acid, a known metabolic product of anaerobic bacteria (36). However, in humans, phenylpropionic acid is instead normally converted to benzoic acid by the action of the β-oxidizing enzyme medium chain acyl-CoA dehydrogenase (MCAD) (44). Thus, phenylpropionylglycine is detected only in the urine of MCAD-deficient patients, and has been used as a biomarker for the diagnosis of this condition (45). These results illustrate the specificity of host–bacteria mutualism, and suggest the likelihood of coevolution between symbionts and their hosts.
Several products of yet another mammalian chemical detoxification mechanism, glucuronidation, were also elevated in the presence of bacterial metabolism. For example, a glucuronide of 3-carboxy-4-methyl-5-pentyl-2-furanpropionic acid, a known uremic retention solute shown to inhibit the binding of salicylate and 5,5-diphenylhydantoin to albumin (46), was identified exclusively in the serum of conv animals. The greatly increased concentrations of this furan dicarboxylic acid, as well as other classic uremic retention solutes such as indoxyl sulfate, p-cresol sulfate, urate, and hippuric acid in the blood of these animals, point to the large burden that the metabolic processes of enteric bacteria impose on the chemical detoxification mechanism of their hosts.
Conclusion
In summary, we have demonstrated a significant effect of the microbiome on mammalian blood metabolites by applying an untargeted MS-based metabolomics approach to a germ-free mouse model system. Numerous circulating molecules were determined to arise exclusively in the presence of gut microflora, whereas ≈10% of commonly observed features differed significantly in concentration between GF and conv animals. Several pathways including the metabolic processing of indole-containing molecules were seen to particularly interact with the microbiome. Numerous molecules apparently resulting from phase II drug-like chemical processing of microbial metabolites were also significantly elevated. Such broad metabolomics profiling studies combined with the selective colonization of germ-free animals with defined bacterial population promise to be a powerful tool for the deconvolution of the complex interplay between mammalian and bacterial metabolic processes.
Materials and Methods
Serum Sample Collection and Preperation.
Blood from 10 conv and 10 GF male Swiss Webster mice age 8–10 weeks was ordered from Taconic Farms. Both sets of animals were maintained on autoclaved NIH-31 chow. Blood was collected by retro-orbital bleed into heparinized tubes, spun down at 3,000 g for 10 min, and the entire serum content of each animal was frozen immediately at −80 °C and shipped on dry ice. Metabolites were extracted from plasma with methanol. We added 4 volumes of cold methanol to 50 μL of plasma, vortexed, and incubated at −20 °C for 1 h. Samples were centrifuged 10 min at 14,000 × g, the supernatant was collected, and the centrifugation was repeated. The supernatant was dried in a SpeedVac and resuspended in 50 μL of 95:5 water:acetonitrile, and clarified for 5 min at 14,000 × g.
Metabolomics Profiling with ESI.
The HPLC system consisted of a degasser, capillary pump, binary pump, and autosampler (1100 series, Agilent); 3 μL of extracted plasma maintained at 4 °C in a thermostated autosampler was applied to a capillary reversed-phase column (Zorbax C18 SB-300, Agilent) with dimensions 300-μm ID × 150-mm length. The flow rate was 4 μL/min with solvent A composed of water + 0.1% formic acid and solvent B composed of acetonitrile + 0.1% formic acid in positive ion mode. For negative ion mode data, 5 mM ammonium acetate was substituted for the formic acid in solvents A and B. The gradient consisted of 5% B for 5 min, followed by a gradient to 95% B over 45 min, hold at 95% B for 5 min, and a reequilibration at 5% B for 10 min.
To reduce systematic error associated with instrumental drift, samples were run in an order that alternated between wild type and knockout. Data were collected in + and − ESI mode in separate runs on a TOF (Agilent 6210) operated in full scan mode from 100 to 1,000 m/z. The capillary voltage was 3,500 (+) or 2,000 V (−) with a scan rate of 0.5 scan per second; the nebulizer flow rate was 12 L/hr.
Data Analysis and Statistics.
Data in instrument specific format (.wiff) were converted to common data format (.cdf) files. The program XCMS (22) was used for nonlinear alignment of the data in the time domain and automatic integration and extraction of the peak intensities. Accurate masses of features representing significant differences were searched against the METLIN, KEGG, HMDB, and LIPIDMAPS databases. SIMCA-P (Umetrics) was used for multivariate statistical calculations and plotting. Differences between wild type and GF plasma were evaluated for individual metabolites by using a 2-tailed t test, assuming unequal variance (Welch's t test), calculated by using Excel. Statistical plots were calculated by using Origin version 6.1.
Compound Identification.
The identity of compounds was confirmed by LC/MS/MS by using a QTOF (model 6510, Agilent). Pooled plasma extracts were made from the same set of conv or GF samples, and the experiment was repeated with the identical chromatography conditions on the TOF, with the exceptions that the column was 2.1-mm ID (SB-C18, Zorbax), the flow rate was 200 μL/min, and the nebulizer flow rate was 30 L/hr. Ions were targeted for collision-induced dissociation (CID) fragmentation on the fly based on the previously determined accurate mass and retention time. The exact retention time was determined at the higher flow rate by manually comparing the pooled extracted ion chromatograms from the 2 groups. The fragmentation patterns for plasma unknowns and authentic model compounds were compared after an initial optimization of the collision energy.
Other.
Supplementary Material
Acknowledgments.
This work was supported by the U.S. Department of Energy Office of Science under contracts DE-FG02-07ER64325 and DE-AC02-05CH11231 through the Genomics: GTL Research Program (G.S.), National Institutes of Health Grant P30 MH062261 (G.S.), and the Novartis Research Foundation. This is manuscript 19923 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812874106/DCSupplemental.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Alverdy JC, Chang EB. The re-emerging role of the intestinal microflora in critical illness and inflammation: Why the gut hypothesis of sepsis syndrome will not go away. J Leukoc Biol. 2008;83:461–466. doi: 10.1189/jlb.0607372. [DOI] [PubMed] [Google Scholar]
- 8.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. Isme J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 12.Hord NG. Eukaryotic-microbiota crosstalk: Potential mechanisms for health benefits of prebiotics and probiotics. Annu Rev Nutr. 2008;28:215–231. doi: 10.1146/annurev.nutr.28.061807.155402. [DOI] [PubMed] [Google Scholar]
- 13.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Metabonomic investigations in mice infected with Schistosoma mansoni: An approach for biomarker identification. Proc Natl Acad Sci USA. 2004;101:12676–12681. doi: 10.1073/pnas.0404878101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin FP, et al. Effects of probiotic Lactobacillus paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res. 2007;6:1471–1481. doi: 10.1021/pr060596a. [DOI] [PubMed] [Google Scholar]
- 17.Yap IK, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 18.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: The inevitable imprint of mass spectrometry in metabolomics. J Proteome Res. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- 20.Nordstrom A, Want E, Northen T, Lehtio J, Siuzdak G. Multiple ionization mass spectrometry strategy used to reveal the complexity of metabolomics. Anal Chem. 2008;80:421–429. doi: 10.1021/ac701982e. [DOI] [PubMed] [Google Scholar]
- 21.Want EJ, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 22.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 23.Botsford JL, Demoss RD. Escherichia coli tryptophanase in the enteric environment. J Bacteriol. 1972;109:74–80. doi: 10.1128/jb.109.1.74-80.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costedio MM, Hyman N, Mawe GM. Serotonin and its role in colonic function and in gastrointestinal disorders. Dis Colon Rectum. 2007;50:376–388. doi: 10.1007/s10350-006-0763-3. [DOI] [PubMed] [Google Scholar]
- 25.Deguchi T, et al. Major role of organic anion transporter 3 in the transport of indoxyl sulfate in the kidney. Kidney Int. 2002;61:1760–1768. doi: 10.1046/j.1523-1755.2002.00318.x. [DOI] [PubMed] [Google Scholar]
- 26.Takayama F, Taki K, Niwa T. Bifidobacterium in gastro-resistant seamless capsule reduces serum levels of indoxyl sulfate in patients on hemodialysis. Am J Kidney Dis. 2003;41(Suppl 1):S142–S145. doi: 10.1053/ajkd.2003.50104. [DOI] [PubMed] [Google Scholar]
- 27.Smith EA, Macfarlane GT. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol. 1997;33:180–188. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- 28.Karbownik M, et al. Indole-3-propionic acid, a melatonin-related molecule, protects hepatic microsomal membranes from iron-induced oxidative damage: Relevance to cancer reduction. J Cell Biochem. 2001;81:507–513. [PubMed] [Google Scholar]
- 29.Chyan YJ, et al. Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J Biol Chem. 1999;274:21937–21942. doi: 10.1074/jbc.274.31.21937. [DOI] [PubMed] [Google Scholar]
- 30.Bendheim PE, et al. Development of indole-3-propionic acid (OXIGON) for Alzheimer's disease. J Mol Neurosci. 2002;19:213–217. doi: 10.1007/s12031-002-0036-0. [DOI] [PubMed] [Google Scholar]
- 31.Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by clostridia. Arch Microbiol. 1976;107:283–288. doi: 10.1007/BF00425340. [DOI] [PubMed] [Google Scholar]
- 32.Young SN, Anderson GM, Gauthier S, Purdy WC. The origin of indoleacetic acid and indolepropionic acid in rat and human cerebrospinal fluid. J Neurochem. 1980;34:1087–1092. doi: 10.1111/j.1471-4159.1980.tb09944.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- 34.Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr. 1976;29:1448–1454. doi: 10.1093/ajcn/29.12.1448. [DOI] [PubMed] [Google Scholar]
- 35.Yi L, Dratter J, Wang C, Tunge JA, Desaire H. Identification of sulfation sites of metabolites and prediction of the compounds' biological effects. Anal Bioanal Chem. 2006;386:666–674. doi: 10.1007/s00216-006-0495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert PW, Roos BA. Paired-ion reversed-phase high-performance liquid chromatography of human and rat calcitonin. J Chromatogr. 1980;198:293–299. doi: 10.1016/s0021-9673(00)84769-x. [DOI] [PubMed] [Google Scholar]
- 37.Akaza H, et al. Comparisons of percent equol producers between prostate cancer patients and controls: Case-controlled studies of isoflavones in Japanese, Korean and American residents. Jpn J Clin Oncol. 2004;34:86–89. doi: 10.1093/jjco/hyh015. [DOI] [PubMed] [Google Scholar]
- 38.Wang XL, Hur HG, Lee JH, Kim KT, Kim SI. Enantioselective synthesis of S-equol from dihydrodaidzein by a newly isolated anaerobic human intestinal bacterium. Appl Environ Microbiol. 2005;71:214–219. doi: 10.1128/AEM.71.1.214-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankenfeld CL, et al. High concordance of daidzein-metabolizing phenotypes in individuals measured 1 to 3 years apart. Br J Nutr. 2005;94:873–876. doi: 10.1079/bjn20051565. [DOI] [PubMed] [Google Scholar]
- 40.Chiba M, Poon K, Hollands J, Pang KS. Glycine conjugation activity of benzoic acid and its acinar localization in the perfused rat liver. J Pharmacol Exp Ther. 1994;268:409–416. [PubMed] [Google Scholar]
- 41.Strahl NR, Barr WH. Intestinal drug absorption and metabolism. 3. Glycine conjugation and accumulation of benzoic acid in rat intestinal tissue. J Pharm Sci. 1971;60:278–281. doi: 10.1002/jps.2600600227. [DOI] [PubMed] [Google Scholar]
- 42.Poon K, Pang KS. Benzoic acid glycine conjugation in the isolated perfused rat kidney. Drug Metab Dispos. 1995;23:255–260. [PubMed] [Google Scholar]
- 43.Armstrong MD, Chao FC, Parker VJ, Wall PE. Endogenous formation of hippuric acid. Proc Soc Exp Biol Med. 1955;90:675–679. doi: 10.3181/00379727-90-22134. [DOI] [PubMed] [Google Scholar]
- 44.Rinaldo P, O'Shea JJ, Welch RD, Tanaka K. The enzymatic basis for the dehydrogenation of 3-phenylpropionic acid: In vitro reaction of 3-phenylpropionyl-CoA with various acyl-CoA dehydrogenases. Pediatr Res. 1990;27:501–507. doi: 10.1203/00006450-199005000-00017. [DOI] [PubMed] [Google Scholar]
- 45.Rinaldo P, O'Shea JJ, Welch RD, Tanaka K. Diagnosis of medium chain acyl-CoA dehydrogenase deficiency by stable isotope dilution analysis of urinary acylglycines: Retrospective and prospective studies, and comparison of its accuracy to acylcarnitine identification by FAB/mass spectrometry. Prog Clin Biol Res. 1990;321:411–418. [PubMed] [Google Scholar]
- 46.Niwa T, Takeda N, Maeda K, Shibata M, Tatematsu A. Accumulation of furancarboxylic acids in uremic serum as inhibitors of drug binding. Clin Chim Acta. 1988;173:127–138. doi: 10.1016/0009-8981(88)90250-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.