Abstract
Since the demonstration that the sequence of a protein encodes its structure, the prediction of structure from sequence remains an outstanding problem that impacts numerous scientific disciplines, including many genome projects. By iteratively fixing secondary structure assignments of residues during Monte Carlo simulations of folding, our coarse-grained model without information concerning homology or explicit side chains can outperform current homology-based secondary structure prediction methods for many proteins. The computationally rapid algorithm using only single (φ,ψ) dihedral angle moves also generates tertiary structures of accuracy comparable with existing all-atom methods for many small proteins, particularly those with low homology. Hence, given appropriate search strategies and scoring functions, reduced representations can be used for accurately predicting secondary structure and providing 3D structures, thereby increasing the size of proteins approachable by homology-free methods and the accuracy of template methods that depend on a high-quality input secondary structure.
Keywords: protein folding, secondary structure prediction, tertiary structure prediction, iterative fixing, statistical potential
The protein folding process is integral to multiple cellular processes, and errors can result in amyloidgenic diseases. The structure of a protein affords a window on its function, and the huge growth in the number of sequenced genomes provides codes for an enormous number of new proteins with unknown functions (1), a number far exceeding experimental capabilities and requiring fast throughput theoretical methods for deducing protein structure from sequence. To this end, great progress in predicting structure has emerged by using homology-based methods (2).
However, the goal of predicting structure and pathways beginning only from the sequence remains an elusive goal. Furthermore, methods for secondary and tertiary (2° and 3°) structure prediction, although often quite accurate, can fail for lack of sufficient sequences that are homologous to the target sequence. Even if a multiple sequence alignment (MSA) exists, e.g., as generated by using PSI-BLAST (3), the alignment may diminish any structural propensity that is specific to the target sequence (in its 3° context) in favor of the consensus of the alignment. This disadvantage can adversely affect 3° structure prediction because the homology-based 2° structure prediction and MSA generally serve as crucial inputs.
The reliance on homology also precludes identifying the underlying physiochemical principles that govern protein folding, including determining the minimal information and model of protein structure that are required for accurate structure prediction. This inadequacy arises from the failure of many 2° structure prediction methods (4, 5) to incorporate 3° context explicitly. Context dependence can overrule local biases (6–8), and its neglect has limited 2° structure accuracy to ≈80% for decades (9). Previous attempts to improve 2° structure predictions by including 3° structure predictions achieve limited success (10), perhaps because of a reliance on sequence homology.
We present a homology-free strategy using a Cβ-level representation in which 2° and 3° structure predictions emerge as an integral component of the folding process. Consequently, our strategy may share some benefits that authentic proteins gain by folding along a robust and efficient pathway. Although others have integrated 2° and 3° structure determination (11, 12) with an iterative fixing (ItFix) of 2° structure (13–15), our approach differs by (i) not using any exogenous 2° structure prediction or homology; (ii) removing side-chain degrees of freedom from the model, which greatly reduces computation time; and (iii) allowing the whole chain to interact throughout the entire folding process. Furthermore, our moves involve changes only in a single pair of dihedral angles (φ,ψ) that is obtained from the Protein Data Base (PDB) and that includes the influence of the identity and 2° structure of the neighboring residues. These results demonstrate that although our model lacks explicit side chains or information from homology, our predictions are often as accurate while requiring orders of magnitude less computing time. In addition, information about folding pathways can be extracted from the simulations.
Integration of 2° and 3° Structure.
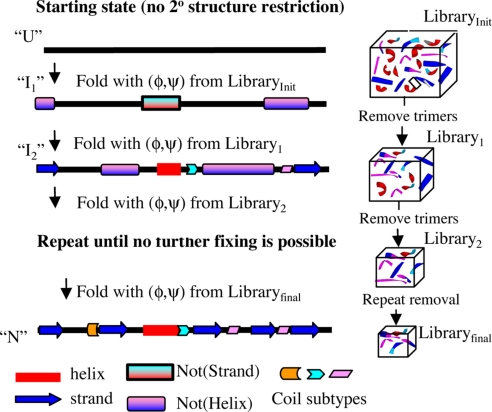
Our ItFix algorithm focuses on three fundamental protein properties: the sequence-dependent backbone torsional angle preferences, the backbone hydrogen-bonding requirements, and the different chemical properties and packing preferences of the 20 amino acid side chains (Fig. 1). Because each factor strongly influences the other two, a major challenge lies in simultaneously including all three factors with appropriate weights into a folding algorithm. Because the model retains the backbone heavy atoms and the side-chain Cβ atoms (16–19), the 2N backbone dihedral φ,ψ angles are the major degrees of freedom for a chain of N residues in our treatment (cis-ω conformers are occasionally allowed, see Methods).
Fig. 1.
Interrelated themes of protein folding. Protein backbone motions, 2° structure and hydrogen bonding, and side-chain packing are the necessary components of any folding model. Secondary and 3° structure formation are coupled processes whereby the formation of each type of structure influences the formation of the other type.
The neglect of side groups raises questions of how the model describes packing and individual residue preferences. As demonstrated below, our Monte Carlo simulated annealing (MCSA) algorithm, using a statistical potential (StatPot) (16–19) and an increasingly restrictive PDB-based move set (Fig. 1), recaptures the requisite side-chain information and performs remarkably well in the prediction of 2° and 3° structure without invoking homology information (Figs. 2 and 3 and Tables 1 and 2).
Fig. 2.
ItFix 2° and 3°structure prediction protocol. At the end of each round, the 2° structure frequencies are used to eliminate H, E, or C when a frequency falls below a specified threshold.
Fig. 3.
Secondary structure prediction. ItFix predicts 2° structure at the Q8 level (H, E, CG, CN, CI, CS, CB, or CT). Results are displayed for the targets in Table 2, which have a large number of sequence homologs, making them ideal targets for the PSIPRED and SSPro homology-based prediction methods.
Table 1.
Structure prediction for low-homology set
| Protein |
2° structure and percent accuracy |
3° structure, Å |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PDB ID code | Description | Length | Fold | R0* Q3 | ItFix Q3 (Q8†) | SSPro‡ Q3 (Q8) | PSIPRED§ Q3 | Meiler and Baker¶ Q3 | ItFix (best‖) | Meiler and Baker (best**) |
| 1ail | Protein fragment | 70 | αβ | 46 | 76 (73) | 70 (74) | 73 | 64 | 5.4 | 6.0 |
| 1aoy | Single domain repressor | 78 | αβ | 54 | 82 (72) | 81 (65) | 87 | 89 | 5.7 | 5.7 |
| 1c8cA | DNA-binding | 64 | αβ | 56 | 86 (70) | 72 (59) | 59 | 67 | 3.7 | 5.0 |
| 1cc5 | Heme-binding | 76 | α | 70 | 92 (68) | 74 (75) | 88 | 86 | 6.5 | 6.2 |
| 1dtdB | Disulfide bonds | 61 | αβ | 57 | 71 (57) | 64 (57) | 75 | 69 | 6.5 | 5.7 |
| 1hz6A | Protein L | 67 | αβ | 57 | 80 (72) | 80 (75) | 83 | 87 | 3.8 | 3.4 |
| 1fwp | CheY-binding domain | 69 | αβ | 45 | 70 (55) | 48 (30) | 61 | 68 | 8.1 | 7.3 |
| 1isuA | Iron-binding | 62 | αβ | 65 | 82 (44) | 66 (39) | 81 | 89 | 6.5 | 6.9 |
| 1sap | Hyper-thermophile | 66 | αβ | 65 | 85 (67) | 76 (67) | 65 | 65 | 4.6 | 6.6 |
| 1wapA | Oligomer in crystal structure | 68 | β | 43 | 80 (68) | 73 (64) | 81 | 68 | 8.0 | 7.7 |
| 2ezk | DNA-binding | 93 | α | 58 | 80 (75) | 71 (64) | 91 | 85 | 5.5 | 6.6 |
Target sequences were taken from previous study by Meiler and Baker (10).
*Round 0 accuracy of the initial, sequence-dependent trimer library before any 2° structure restrictions are made. This reflects local 2° propensity.
†ItFix predicts 2° structure at the Q8 level (H, E, and the 6 types of coil, including turn (CT), bend (CS), 3–10 helix (CG), π helix (CI), β bridge (CB), and other (CN).
‡SSPro predictions were taken from the SSPro online server (31).
§PSIPRED predictions were taken from the PSIPRED online server (32).
‖Lowest rmsd obtained.
Table 2.
Structure prediction for high-homology set
| Protein |
2° structure % accuracy |
3° structure, Å |
||||||
|---|---|---|---|---|---|---|---|---|
| PDB ID code | Length | Fold | R0* Q3 | ItFix Q3 (Q8) | SSPro† Q3 (Q8) | PSIPRED‡ Q3 | ItFix§ lowest energy (best) | Bradley et al.¶ |
| 1af7 | 69 | α | 70 | 97 (86) | 86 (81) | 90 | 2.9 (2.5) | 10.4 |
| 1b72A | 50 | α | 62 | 88 (84) | 68 (72) | 84 | 3.5 (1.6) | 1.1 |
| 1csp | 67 | β | 49 | 79 (67) | 75 (67) | 88 | 10.5 (6.0) | 4.7 |
| 1di2 | 68 | αβ | 68 | 88 (79) | 74 (75) | 97 | 6.1 (4.6) | 2.6 |
| 1dcj | 72 | αβ | 38 | 45 (29) | 65 (56) | 89 | 13.3 (7.6) | 2.5 |
| 1mky | 77 | αβ | 66 | 86 (70) | 87 (71) | 90 | 6.9 (6.1) | 6.3 |
| 1o2Fb | 77 | αβ | 65 | 78 (69) | 79 (66) | 75 | 11.2 (5.8) | 10.1 |
| 1r69 | 61 | α | 79 | 93 (89) | 84 (72) | 92 | 4.2 (2.4) | 1.2 |
| 1shfA | 59 | β | 53 | 76 (56) | 85 (69) | 80 | 12.2 (6.7) | 10.8 |
| 1tif | 57 | αβ | 47 | 89 (79) | 86 (70) | 93 | 11.3 (4.2) | 4.1 |
| 1tig | 86 | αβ | 53 | 83 (70) | 69 (67) | 83 | 6.4 (5.3) | 3.5 |
| 1ubq | 73 | αβ | 60 | 92 (69) | 88 (67) | 90 | 5.3 (3.1) | 1.0 |
Target sequences are the same as a previous study (27).
*Round 0 accuracy of the initial, sequence-dependent trimer library before any 2° structure restrictions are made. This reflects local 2° propensity.
†SSPro predictions were taken from the SSPro online server (31).
‡PSIPRED predictions were taken from the PSIPRED online server (32).
§ItFix lowest energy and lowest observed Cα rmsd (in parentheses) structures.
Iterative Fixing and Trimer Selection.
A critical aspect of the algorithm is the selection of a single dihedral angle pair from an increasingly refined library of amino acid trimers, similar in spirit to earlier studies (13–15). During the initial round of the simulations, trimer selection is conditional only on the amino acid identity of the three residues (Fig. 2). Trimer selection in subsequent rounds depends on the 2° structure type at each position that is identified from the previous round by the prescriptions described in Methods. The specification of 2° structure is enabled because each trimer in the trimer library is labeled by the 2° structure assignments for each of the three residues in the original PDB structure in which they originate using the Dictionary of Protein Secondary Structure (DSSP) definition (20). The frequencies of occurrence for each originating 2° structure type, H(elix), E(xtended), or C(oil), are calculated from the last inserted trimer at each position in the 200–300 final structures emerging from each round of folding. Following Sherlock Holmes' deductive strategy “Eliminate all other factors, and the one which remains must be the truth” (21), if the frequency of occurrence for a particular 2° structure type falls below a ≈1–10% threshold at a given position or across a contiguous stretch of sequence (see Methods), any trimer inconsistent with that 2° structure is removed from the trimer library used in subsequent folding rounds. The process continues until no additional positions can be further restricted. After the last round of 2,000 trajectories, the lowest energy and best 3° structures are identified, whereas the 2° structure is predicted from the frequencies of appearance of H, E, and C in all final structures (see Methods).
Our MCSA algorithm is designed to resemble a true folding pathway. Each round in the ItFix process begins from a configuration devoid of any 3° structure rather than a collapsed structure generated from a previous round. Consequently, the chains execute a new global search each round. The backbone geometry is simulated by replacing only one of the three pairs of φ,ψ dihedral angles at a randomly chosen position with those from the equivalent position in a φ,ψ trimer selected from the trimer library. In principle, all-atom simulations for tripeptides could be used, but the accuracy of current methods makes this approach less reliable (22). The starting chain is built by using angles from trimers specified solely by the amino acid sequence. The trimer library becomes increasingly conditional on 2° structure type as the rounds proceed. Each round of ItFix consists of 200–300 individual folding trajectories. Each trajectory involves a global search guided by φ,ψ insertion moves, a Metropolis acceptance criterion, and a StatPot for a scoring function. The trajectory ends when the collapsed structure cannot undergo additional moves. The end result of the iterative rounds is a folding-enhanced 2° structure prediction that emerges simultaneously with an ensemble of 3° structures.
Retaining Lost Side-Chain Information.
The retention of the side-chain information lost by the use of the Cβ-level representation poses a serious challenge. Central to this goal is our φ,ψ dihedral angle sampling procedure that is conditional on both the chemical identity and the increasingly refined 2° structure specificity for each position and its neighboring residues. The backbone dihedral angles are strongly correlated with the side-chain rotamer angles and both the neighboring residues' side-chain identities and conformations (18, 23–25). Hence, even without explicitly depicting the side-chain atoms, much of their influence is retained by choosing φ,ψ values using our conditional trimer selection strategy.
Given this retention of the interplay of side chain–backbone interactions, the other elements of our algorithm focus on optimizing 3° interactions. The 3° interaction energies are obtained from the StatPot discrete optimized protein energy (DOPE)-Cβ (18, 26) derived from an all-atom pairwise additive StatPot (16, 17) that uses a novel reference state and distinguishes the backbone atoms according to amino acid type. Our version removes all contributions involving hydrogen and side-chain atoms beyond the Cβ atom. To eliminate bias toward specific 2° structure types, the attractive potential is removed between atoms in continuous stretches of 2° structure, whereas the repulsive portion is retained to prevent steric overlap. In addition, interactions are conditional on backbone geometry and the relative orientation of the Cα–Cβ bonds of the two interacting side chains [supporting information (SI) Figs. S1 and S2], a feature particularly helpful in setting up the overall chain topology so that collapse generates native-like structures. Beyond the prescription used to eliminate a 2° structure option in the trimer library, the only adjustable parameters are the four linear weight factors in the StatPot (Table S1).
Results
Improvement in 2° Structure Prediction Arising from Folding.
The first set of targets (Table 1 and Figs. S3–S5) originates from a previous study that integrates 2° and 3° structure prediction (10). The set contains proteins with 11 diverse folds and relatively low sequence homology. The second set of targets (Table 2, Figs. 3 and 4, and Fig. S3) originates from a study focusing on improving 3° structure prediction by using high sequence homology and extensive side-chain refinement.
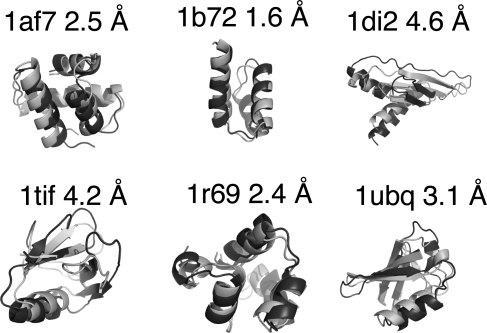
Fig. 4.
Tertiary structure determination. Alignments of the ItFix lowest observed rmsd 3° structure (dark) with the native structure (light) were made by using PyMol visualization software. Cα rmsd between ItFix model and native structure from column 8 of Table 2 is listed above each target.
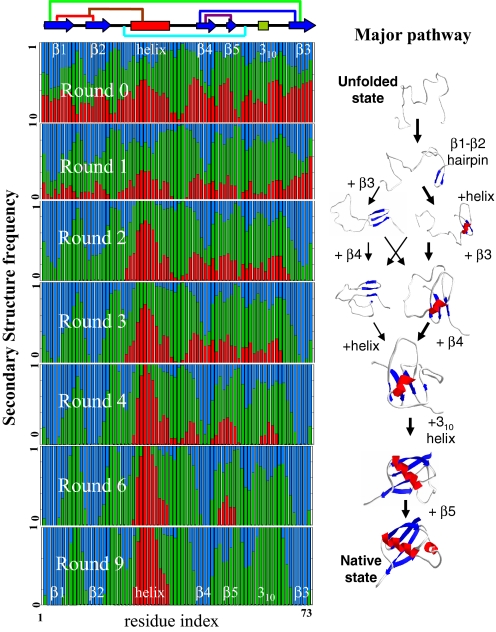
The ItFix folding algorithm significantly improves the accuracy of predicting the three major 2° structure types, H, E, and C (termed “Q3 level”) compared with the intrinsic, locally determined biases. This improvement is apparent by comparing the final 2° structure accuracy with that from the initial trimer library that is contingent only on the sequence. This Round 0, or “R0,” accuracy of 58 ± 10% improves to 82 ± 11% over the 6–9 rounds of the ItFix process for the various proteins (Fig. 5 and Fig. S6). The process of specifying 2° structure by eliminating options is well illustrated by the evolution of 2° structure frequencies at each position in 1ubq (PDB ID code). The R0 frequencies display some bias to the native 2° structure but provide only 60% accuracy. Only as 2° structure options are eliminated does the native 2° structure pattern emerge over the course of the nine rounds to an accuracy of 92%. A notable example is the carboxyl-terminal region where the high intrinsic helicity is overridden by 3° context and the region becomes a native-like strand.
Fig. 5.
ItFix algorithm mimics the experimentally determined ubiquitin folding pathway. The position dependence of the 2° structure frequencies at the end of each round, E (blue), H (red), and C (green), is shown. A single color bar represents a residue assigned to a single 2° structure type (native 2° structure shown at the top, along with long-range contacts). As the rounds progress, uncertainties in 2° structure diminish. The major steps in the proposed folding pathway (30, 31) are similar to the order of structure fixing over the multiple rounds: the hairpin forms, followed by the helix and β3 strand, and then β4. The final two events are the folding of the 3–10 helix and β5. Their formation appears in some trajectories but not at a high enough frequency to be fixed for the next round.
The importance of 3° context in determining 2° structure within ItFix is demonstrated for the five best performing targets (PDB ID codes 1af7, 1b72A, 1r69, 1di2, 1ubq) by comparison with ItFix simulations that eliminate the distant attractive terms between amino acids farther than six residues (|i–j| > 6). The ItFix process is repeated but without attractive terms between amino acids farther than six residues (|i–j| > 6). When all of the repulsive terms are retained, the chain adopts extended geometries to avoid steric overlap, and the accuracy of the 2° structure prediction decreases sharply, even compared with the initial R0 2° structure accuracy. When long-range chain overlap is permitted, the quality of the 2° structure prediction also degrades relative to R0 because the only remaining favorable interaction term is between two residues on strands with |i–j| > 4. By itself, this term is insufficient to drive stable, native-like sheet formation. A slight improvement over R0 occurs simply from the 2° structure-fixing protocol without any simulated annealing. However, the improvement is marginal, 0–2%, compared with 13–30% obtained when the long-range interactions are included.
Hence, accurate 2° structure prediction requires 3° context, which serves to stabilize or buttress weak local biases and 2° structural elements. For example, the amino β hairpin in 1ubq emerges when the formation of a weak turn brings two potential strands together. Similarly, an unstable amphipathic helix can be mutually stabilized by a β hairpin with a hydrophobic face. Such 3° contacts may not always be completely native-like because significant increases in 2° structure accuracy can arise even when the global 3° fold is inaccurate (e.g., rmsd >6 Å).
Comparison with Existing 2° and 3° Structure Prediction Methods.
The ItFix accuracy can surpass the accuracy of the 2° structure prediction servers SSPRO (4) and PSIPRED (5) and the previous study (10). The high homology of these sequences is responsible for the prediction accuracy to meet or exceed 80% for both the SSPRO (4) and PSIPRED (5) servers. Nevertheless, our ItFix protocol achieves comparable accuracy without invoking any homology information (Table 2). The average ItFix accuracy is only slightly smaller for the low-homology targets, 80% vs. 83%. However, the lack of homology significantly degrades PSIPRED and SSPro performances, 77% vs. 88% and 70% vs. 79%, respectively. Furthermore, the ItFix method is able to predict all eight types of 2° structure where coil is subdivided into the six of the DSSP-defined subtypes (CG, CN, CI, CS, CB, CT), termed “Q8 level.” This ability also is available by using SSPro, but it is slightly less accurate for most targets. As illustrated below, ItFix provides much better predictions for the location of turns and the ends of helices and strands, features that are crucial in 3° structure prediction.
The ItFix algorithm describes α, α/β, and β proteins within each set with comparable accuracy, although 3° predictions for the more challenging low-homology set are generally poorer (Tables 1 and 2 and Fig. 3) because we have difficulty predicting metal- and heme-binding proteins and disulfide-bonded proteins. The high-homology set lacks these challenging protein types. The accuracy of the ItFix 3° structure predictions are comparable with those of the highly successful Rosetta fragment-based insertion algorithm, as implemented in the papers from which the test sets are obtained (10, 27). Our structures are more similar in quality for the low-homology set than the high-homology set. The high-homology targets have been chosen by Baker and coworkers (27) because improved predictions are obtained for them by using data from the folding of an extensive number of homologs. In addition, the Rosetta algorithm requires extensive side-chain refinement and thus orders of magnitude more computation time (27) than our algorithm that omits side-chain degrees of freedom. Hence, it is clear why this implementation of Rosetta performs better for 9/12 of this target set.
Feedback Between 2° and 3° Structure Prediction.
Although the average accuracy of a 2° structure prediction is a useful metric, it underreports the importance of the feedback between 2° and 3° structure as illustrated for two of the many examples. The ItFix 2° structure accuracy for 1c8c is only modestly superior to those of PSIPRED and SSPro; but crucially, ItFix correctly predicts as β strand a region that the other methods incorrectly assign as helix (Fig. S4). Similarly, the SSPro Q8 level prediction incorrectly assigns positions 9 and 10 as turn in 1ubq, whereas ItFix correctly assigns the turn residues to positions 8 and 9 (Fig. 3). Only through successive rounds of folding does the proper 3° context override the local propensities to correctly determine the location of the turn. Although seemingly insignificant, this difference is crucial because the alignment of the hairpin, and therefore the quality of the overall structure, depends on properly identifying the turn location. Thus, extensive sequence homology information and intrinsic propensities can be insufficient for 2° structures that depend strongly on 3° context.
Our main limitation in predicting 2° structure is the occasional deficiency of our starting trimer library. For example, when we predict 2° structure for target 1dcj from the initial trimer library contingent only on the sequence (R0), the accuracy is <40%, implying very poor local 2° structure context exists for this target (Table 2). Our 46% accuracy for this target suggests that the 3° context of folding is insufficient to compensate for poor local propensity. In fact, we assign the second helix of 1dcj as coil because a proline–glycine pair in the center of that helix has a very high preference for coil. PSIPRED performs well on this target, presumably because of the influence of sequence homology. SSPro underperforms for this protein and some others, perhaps because local preferences are weighted more heavily than the contribution of the sequence alignment compared with PSIPRED.
Tertiary Structure Predictions.
Even though our StatPot can routinely distinguish a native structure from a set of folding decoys, the folding simulations cannot always generate native-like models. This limitation is often caused by the vast size of the conformational search space for some sequences. We reduce the search space by specifying the sequence and then iteratively identifying the 2° structure. Local propensities, however, often are so strong that even enormous amounts of sampling and 3° context cannot overcome the bias. For example, the turn of the second β-hairpin of 1di2 contains residues whose turn propensities are very low. Even through many rounds of ItFix folding, the turn probability of that region never becomes high enough to fix, which severely limits the quality of the 3D models generated. Other prediction methods circumvent this problem and accurately predict this structure by using the degeneracy of sequence homology to properly predict the turns and by sampling larger structure fragments that may contain long range information that specifies the turn (27), suggesting that employing sequence homology can smooth over any incorrect local biases. Although ItFix uses no homology information and samples one position at a time, it still correctly predicts the structure of 1di2 by including the crucial 3° structure context.
The lack of homology-based information is actually beneficial to predictions for some sequences, specifically when the MSA incorrectly biases the 2° structure. ItFix fares exceptionally well for the 2° and 3° structure of 1sap (Table 1) because the 3° context drives the central region of the protein to be β sheet rather than the helix preferred by the sequence homology-based methods (Fig. S4). The very high confidence of PSIPRED in this region suggests that the MSA strongly biases the 2° structure toward helix, resulting in less accurate 3° structure.
Folding Pathway.
Many aspects of ItFix replicate the folding behavior of authentic proteins. During the ItFix process, subunits of structure, or “foldons,” are fixed cooperatively, just as observed by hydrogen exchange experiments (28). The foldons add to existing structure in a process of sequential stabilization (29) that may resemble the pathway taken by authentic proteins. In contrast to methods using preformed fragments or exogenous 2° structure predictions where the connection to the authentic pathway is murky at best, the ItFix protocol begins with an initial unstructured chain, and the buildup of structure evolves out of the folding process. Hence, the order of fixing of structural elements may recapitulate major features of the authentic pathway followed as the real chain progresses along the free energy surface (Fig. S6).
For the α/β protein ubiquitin, the order of fixing structure (Fig. 5) and their interactions are in remarkable accord with the experimental pathway (30). A notable feature is the formation of the parallel β strand interaction between the amino and the carboxyl termini. This long-range contact occurs before the 2° structure assignment of 30 intervening residues and is possible with our method because the simulation includes the entire chain at all times. Further, this parallel interaction overrides the initial R0 trimer propensities that favored helix for the carboxyl-terminal strand, as noted. Irrespective of whether the ItFix algorithm replicates experiment, the pathway nature of the algorithm and the interplay of 2° and 3° structure formation contribute to the success, just as a pathway helps real proteins fold reproducibly and expediently.
Conclusions
The ItFix algorithm predicts 2° structure without resorting to homology and yet delivers an accuracy and specificity that can often match or exceed current methods that rely heavily on homology. The success is because of the integration of 3° structure context during the folding simulations and the recursive refinement of the 2° structure assignments. Concurrently, accurate 3° structures are often generated. Although the model lacks explicit side chains, our PDB-based backbone sampling protocol and scoring functions largely recapture the lost information. Hence, we avoid the computationally expensive search along the rugged side-chain rotamer energy surface that is frequently involved in other successful prediction methods. In addition to highlighting the basic principles required for ab initio structure prediction, our work extends the size of proteins that can be predicted by using homology-free methods. Furthermore, the ItFix 2° structure predictions provide improved prediction of turns and ends of helices and strands, features that are important in describing 3° structure. Thus, the ItFix predictions can be used as inputs to increase the accuracy of template-based predictions that have inherent restrictions imposed by requiring sequence homology. Moreover, now that the basic principles have been established, the performance of ItFix can be improved further by using homology.
Methods
Fixing Protocol.
The protocol for eliminating a 2° structure option at a position is determined by using the 2° structure frequencies in the trimer library at the beginning of the round, PXInit (X = E, H, or C), the frequencies calculated by using DSSP for the 200–300 final structures, PXFin_1, and the frequencies of the trimers' original 2° structure, PXFin_0, according to the following main criteria (see SI Methods). For i consecutive positions (in order of precedence):
(i >6): [HEC] → [EC] if PHFin_1 < 0.03.
(i >10): [HEC] → [EC] if PHFin_1 < 0.05.
(i >2): [HEC] → [EC] if PEFin_0 > 0.50 and PEFin_0 > PEInit and PEFin_1 > 0.
(All positions in protein) [HEC] → [HC] if PEFin_1 < 0.01.
(i >3): [HEC] → [HC] if PHFin_0 > 0.50 and PHFin_0 > PHInit.
(i >4): [HEC] → [HC] if PHFin_1 > 0.40.
(i >0): [H or C] → [H only] if PCFin_0 < 0.10, or (PHFin_0 > 0.50 for i−1, i−2, i+1, i+2).
(i >0): [H or C] → [C only] if PHFin_1 < 0.10.
(i >0): [E or C] → [C only] if PEFin_0 < 0.05.
(i >0): [E or C] → E only] if PCFin_0 < 0.10.
(i >0): [E or C] → [E only] if PEFin_0 > 0.50 and (PEFin_0 > 0.50 for i−1, i+1).
(i >0): [E or C] → [E only] if PEFin_0 > 0.50 and PEFin_1 > 0.00 and total positions fixed >80% of sequence length.
Energy Function.
The reduced Cβ model includes only the backbone heavy atoms and the side-chain Cβ. The energy function is a pairwise additive statistical potential based on the DOPE function (16). We further divide interaction types as contingent on 2° structure type and continuity, sequence separation, and orientation (SI Methods, Figs. S1 and S2, and Table S1). The 2° structure types are defined as per DSSP. An atom pair is defined as in a continuous segment of 2° structure if each residue in the pair and all intervening residues in sequence have the same 2° structure classification. The orientation dependence is determined by the angle between the side-chain Cα–Cβ vector and the Cα–Cα vector connecting the interacting pair.
MCSA Simulations.
Our MCSA energy minimization and sampling methods have been described in detail (18). The ϕ,ψ are sampled from a PDB-derived library (resolution <2.5 Å, homology <90%). To test whether 90% homology provides a native-like bias, five of the best performing targets (1af7, 1b72A, 1r69, 1di2, 1ubq) are refolded by using a library with only a 25% homology threshold. The average accuracy of the 2° structure prediction changes from 91.6 to 90.2%, whereas the 3D structures change on average from 2.84 to 3.16 Å rmsd. These slight differences probably arise from a 1.5–2-fold decrease in trimer diversity rather than the 90% homology, which is at most a minimal factor in the success of the algorithm.
The annealing simulations only consider the heavy atoms of the main chain and the β carbons (Cβ) of the side chains. The backbone planar angles and bond lengths are fixed at their ideal values, except ω, which is chosen as cis at a frequency of 5% and 0.1% for prolines and all other residues, respectively. The cis-proline predictions in Table 2 yield 2 true positives, 4 false positives, 41 true negatives, and 2 false negatives based on an increase above the 5% baseline. All nonproline residues are correctly predicted to be trans.
Supplementary Material
Acknowledgments.
We thank G. Rose, D. Shortle, J. Xu, G. Hocky, H. Gong, R.S. Berry, and members of the Sosnick and Freed laboratories for helpful discussions. This work was supported by National Institutes of Health research (to T.R.S. and K.F.F.) and training grants and by the National Science Foundation through TeraGrid resources provided by National Center for Supercomputing Applications.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811363106/DCSupplemental.
References
- 1.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Service RF. Problem solved* (*sort of) Science. 2008;321:784–786. doi: 10.1126/science.321.5890.784. [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 5.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 6.Kihara D. The effect of long-range interactions on the secondary structure formation of proteins. Protein Sci. 2005;14:1955–1963. doi: 10.1110/ps.051479505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minor DL, Jr, Kim PS. Context-dependent secondary structure formation of a designed protein sequence. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- 8.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dor O, Zhou Y. Achieving 80% 10-fold cross-validated accuracy for secondary structure prediction by large-scale training. Proteins. 2007;66:838–845. doi: 10.1002/prot.21298. [DOI] [PubMed] [Google Scholar]
- 10.Meiler J, Baker D. Coupled prediction of protein secondary and tertiary structure. Proc Natl Acad Sci USA. 2003;100:12105–12110. doi: 10.1073/pnas.1831973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JS, Chen WW, Skolnick J, Shakhnovich EI. All-atom ab initio folding of a diverse set of proteins. Structure. 2007;15:53–63. doi: 10.1016/j.str.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Liwo A, Khalili M, Scheraga HA. Ab initio simulations of protein-folding pathways by molecular dynamics with the united-residue model of polypeptide chains. Proc Natl Acad Sci USA. 2005;102:2362–2367. doi: 10.1073/pnas.0408885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan R, Rose GD. LINUS: A hierarchic procedure to predict the fold of a protein. Proteins. 1995;22:81–99. doi: 10.1002/prot.340220202. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan R, Fleming PJ, Rose GD. Ab initio protein folding using LINUS. Methods Enzymol. 2004;383:48–66. doi: 10.1016/S0076-6879(04)83003-9. [DOI] [PubMed] [Google Scholar]
- 15.Ozkan SB, Wu GA, Chodera JD, Dill KA. Protein folding by zipping and assembly. Proc Natl Acad Sci USA. 2007;104:11987–11992. doi: 10.1073/pnas.0703700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eramian D, et al. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colubri A, et al. Minimalist representations and the importance of nearest neighbor effects in protein folding simulations. J Mol Biol. 2006;363:835–857. doi: 10.1016/j.jmb.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Fitzgerald JE, Jha AK, Colubri A, Sosnick TR, Freed KF. Reduced Cβ statistical potentials can outperform all-atom potentials in decoy identification. Protein Sci. 2007;16:2123–2139. doi: 10.1110/ps.072939707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 21.Doyle AC. The Sign of the Four. London: Penguin Books; 1989. [Google Scholar]
- 22.Zaman MH, Shen MY, Berry RS, Freed KF, Sosnick TR. Investigations into sequence and conformational dependence of backbone entropy, interbasin dynamics and the Flory isolated-pair hypothesis for peptides. J Mol Biol. 2003;331:693–711. doi: 10.1016/s0022-2836(03)00765-4. [DOI] [PubMed] [Google Scholar]
- 23.Jha AK, et al. Helix, sheet, and polyproline II frequencies and strong nearest neighbor effects in a restricted coil library. Biochemistry. 2005;44:9691–9702. doi: 10.1021/bi0474822. [DOI] [PubMed] [Google Scholar]
- 24.Jha AK, Colubri A, Freed KF, Sosnick TR. Statistical coil model of the unfolded state: Resolving the reconciliation problem. Proc Natl Acad Sci USA. 2005;102:13099–13104. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu TT, Kabat EA. An attempt to evaluate the influence of neighboring amino acids (n−1) and (n+1) on the backbone conformation of amino acid (n) in proteins: Use in predicting the three-dimensional structure of the polypeptide backbone of other proteins. J Mol Biol. 1973;75:13–31. doi: 10.1016/0022-2836(73)90526-3. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald JE, Jha AK, Colubri A, Sosnick TR, Freed KF. Reduced Cβ statistical potentials can outperform all-atom potentials in decoy identification. Protein Sci. 2007;16:2123–2139. doi: 10.1110/ps.072939707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 28.Bai Y, Englander SW. Future directions in folding: The multi-state nature of protein structure. Proteins. 1996;24:145–151. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 29.Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Protein folding: The stepwise assembly of foldon units. Proc Natl Acad Sci USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krantz BA, Dothager RS, Sosnick TR. Discerning the structure and energy of multiple transition states in protein folding using ψ analysis. J Mol Biol. 2004;337:463–475. doi: 10.1016/j.jmb.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Sosnick TR, Krantz BA, Dothager RS, Baxa M. Characterizing the protein folding transition state using ψ analysis. Chem Rev. 2006;106:1862–1876. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.