Abstract
The Bicoid (Bcd) transcription factor is distributed as a long-range concentration gradient along the anterior posterior (AP) axis of the Drosophila embryo. Bcd is required for the activation of a series of target genes, which are expressed at specific positions within the gradient. Here we directly tested whether different concentration thresholds within the Bcd gradient establish the relative positions of its target genes by flattening the gradient and systematically varying expression levels. Genome-wide expression profiles were used to estimate the total number of Bcd target genes, and a general correlation was found between the Bcd concentration required for activation and the positions where target genes are expressed in wild-type embryos. However, concentrations required for target gene activation in embryos with flattened Bcd were consistently lower than those present at each target gene's position in the wild-type gradient, suggesting that Bcd is in excess at every position along the AP axis. Also, several Bcd target genes were positioned in correctly ordered stripes in embryos with flattened Bcd, and we suggest that these stripes are normally regulated by interactions between Bcd and the terminal patterning system. Our findings argue strongly against the strict interpretation of the Bcd morphogen hypothesis, and support the idea that target gene positioning involves combinatorial interactions that are mediated by the binding site architecture of each gene's cis-regulatory elements.
Keywords: morphogen, patterning, transcription
According to the gradient morphogen hypothesis, diffusible signaling molecules or transcription factors may provide positional information that organizes metazoan body plans (1). In Drosophila, the maternal transcription factor Bicoid (Bcd) is distributed as a long-range nuclear gradient along the anterior posterior (AP) axis (2). Embryos lacking functional Bcd protein fail to form any anterior structures (3), and microinjection of bcd mRNA causes anterior structures to be formed near the point of injection (4). These findings suggest that Bcd functions as a true morphogen, which establishes a precise order of cell fate decisions by the threshold-dependent positioning of target gene expression patterns.
Intense efforts have been expended to determine whether the Bcd gradient is the primary determinant that positions and sharpens the posterior boundary of the zygotic expression domain of the target gene hunchback (hb) (5–8). However, a complete understanding of anterior patterning will require the identification of all target genes activated by the Bcd morphogen, and the elucidation of the mechanisms that position each one. Previous studies identified 18 direct target genes (including hb), each of which is associated with one or more cis-regulatory modules (CRMs) containing 5–15 clustered Bcd-binding sites (9–11). Recent ChIP-chip experiments estimate the total number of Bcd bound regions in the genome to be in the range of 500–600 (12), but the actual number of direct target genes is unknown.
The known Bcd target genes are expressed in bands or stripes, with posterior boundaries at specific positions along the AP axis of the early embryo. It has been hypothesized that differences in aggregate Bcd-binding strength among CRMs could control the differential placement of these boundaries (13). In this hypothesis, CRMs containing stronger binding clusters would establish posterior boundaries in regions that contain lower Bcd concentrations, and those with weaker clusters would set boundaries at positions with higher Bcd concentrations.
The differential sensitivity hypothesis makes two major predictions. First, if posterior boundaries of target genes are established by specific Bcd concentrations, then there should be little or no change in the amount of Bcd associated with each boundary when the gradient is genetically manipulated. This is not what is observed. For example, in embryos produced by bcd/+ heterozygotes, there are anterior shifts of the posterior boundaries of hb and orthodenticle (otd), but these shifts are significantly less severe than those expected if the boundaries were set by specific Bcd concentrations (14, 15). Second, if differential sensitivity to Bcd binding controls target gene positioning, then there should be a correlation between the posterior boundary position (PBP) and the relative Bcd-binding strength of each target gene's CRM. This hypothesis was examined in two recent studies, but no significant correlations were found (9, 16). However, most CRMs were shown to contain binding sites for other transcriptional regulators, including Hunchback (Hb), which functions synergistically in Bcd-mediated activation, and Kruppel (Kr), which can act as a repressor to set the posterior boundaries of some expression patterns. These studies argue that Bcd functions in combination with other transcription factors, but did not directly test the role of different Bcd concentrations.
Here we used genetic manipulations to produce embryos with nearly uniform levels of Bcd along the AP axis. We further changed bcd copy number to produce pure populations of embryos with different levels of flattened Bcd. We then performed microarray experiments on these embryos to monitor target gene activation genome-wide and tested whether individual target genes respond to specific levels of Bcd by in situ hybridization. Our findings argue against the hypothesis that Bcd target genes are differentially positioned by specific thresholds within the wild-type gradient.
Results
Manipulating Bcd Concentrations in Vivo.
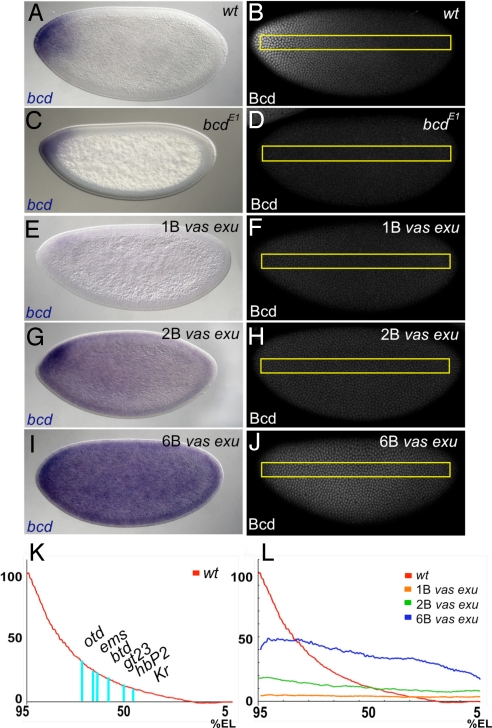
A double mutant in the maternal genes vasa (vas) and exuperantia (exu) was used to generate embryos with flattened Bcd protein gradients (Fig. 1). exu mutants display a partial delocalization of bcd mRNA from the anterior tip (ref. 17; Fig. 1 E, G, and I). Posterior translational repression of bcd through the Nanos (Nos) response element (NRE) in its 3′ UTR (18) was alleviated by the vas mutation, which prevents posterior localization and translation of nos RNA (19).
Fig. 1.
Flattening the Bcd gradient. (A–J) bcd mRNA and protein gradients in wild-type (A and B), bcdE1 (C and D), 1B vas exu (E and F), 2B vas exu (G and H), and 6B vas exu (I and J) embryos. Yellow rectangles in the right-hand panels represent regions of interest (ROIs) used for quantification of fluorescence intensities in (K) and (L). The bcdE1 mutant (C and D) is a protein null, but still produces and localizes mRNA. (K and L) Bcd concentration profiles in wild-type embryos (K and L), and in embryos containing different bcd gene copy numbers in the vas exu background (L). X axes represent percent EL, Y axes represent the percentage of maximum mRNA concentrations. Each line represents an average profile from 3–18 individual embryos of the same genotype at early nuclear cycle 14. The blue lines in (K) represent PBPs for otd, ems, btd, gt23, hb P2, and Kr, which correspond to regions along the AP axis that contain 35%, 27%, 24%, 20%, 11%, and 9% of [Bcd]max in wild type. All embryos in this article are oriented with anterior to the left and dorsal up.
We varied Bcd concentrations in the vas exu background by changing bcd gene copy number, using vas exu; bcd/+ females to generate embryos with one copy (1B vas exu), vas exu; +/+ females to generate embryos with two copies (2B vas exu), and vas exu; +/+ females that were also homozygous for two insertions of a bcd rescue transgene (17) to generate embryos with six copies (6B vas exu). We further quantified Bcd levels in these embryos and normalized them to the maximum Bcd concentration ([Bcd]max) in wild-type embryos [supporting information (SI) Fig. S1; see Methods]. As expected, increasing bcd copy number raises the levels of Bcd protein throughout the embryo (Fig. 1 F, H, J, L). 1B vas exu embryos expressed Bcd levels that represent ≈4 ± 1% [Bcd]max of wild type. 2B vas exu embryos contained 11 ± 4% [Bcd]max, and 6B vas exu embryos contained 40 ± 7% [Bcd]max. Both 2B and 6B vas exu embryos showed consistent differences in Bcd levels along the AP axis, with very flat distributions in anterior regions, and gradual AP gradients in middle to posterior regions. (Fig. 1 H, J, and L).
Genome-Wide Expression Patterns in Embryos with Different Bcd Concentrations.
If Bcd target genes are activated by specific concentration thresholds, they should show dramatic changes in expression levels (on/off responses) in embryos containing different levels of flattened Bcd. We tested this hypothesis by performing microarray expression experiments, with the additional idea that by looking on a genome-wide scale, we would be able to identify new Bcd targets (see Methods and Tables S1–S3).
Here we focus on genes that showed at least a 1.5-fold increase in gene expression in comparisons between 0B and 1B (87 genes), 0B and 2B (152 genes), and 0B to 6B (155 genes). Some genes showed significant changes in more than one comparison, so that the three lists contain a total of 242 unique genes (Fig. 2A and Table S1). Of these, 97 genes have been assayed for RNA expression in embryos, based on the literature, the BDGP expression database (20), and our in situ hybridization experiments (data not shown). Of these, 48 genes showed AP expression patterns in stage 5 embryos, 7 showed dorsal ventral (DV) expression patterns, 30 showed ubiquitous expression, and 11 showed no expression (Table S1).
Fig. 2.
Summary of microarray experiments. (A) Overlaps between lists of genes showing at least a 1.5-fold increase in gene expression in 1B, 2B, or 6B vas exu embryos compared with those lacking Bcd (0B→1B, 0B→2B, and 0B→6B). (B) The correlation between PBPs of putative Bcd target genes and the concentration of Bcd required for activation in the microarray experiments (Table S2). Genes activated by one copy of Bcd (teal squares) include those present only in the 0B→1B group, the intersection between the 0B→1B and 0B→2B groups, the intersection between 0B→1B and 0B→6B, and the intersection between all three groups. Similarly, genes activated by two copies of Bcd (purple circles) include those present only in the 0B→2B, and the intersection between the 0B→2B and 0B→6B groups. Finally, genes activated by six copies of “flat” Bcd include those present only in the 0B→6B group. Bold symbols represent previously known Bcd target genes (Table S1).
Thirteen of the 48 AP-expressed genes are known Bcd targets with defined enhancers: hb, giant (gt), Kruppel (Kr), knirps (kni), orthodenticle (otd), empty spiracles (ems), buttonhead (btd), CG9571, spalt major (salm), bowl, hairy (h), sloppy-paired 1 (slp1), and sloppy-paired 2 (slp2). Five known Bcd targets did not appear as significantly upregulated: tailless (tll), even-skipped (eve), paired (prd), Dichaete (D), and bancal/miR7. Of the 35 remaining genes with AP expression patterns, 17 are putative direct Bcd targets, based on their proximity to a cluster of Bcd-binding sites (data not shown) or a Bcd-bound region detected in ChIP-chip experiments (ref. 12; Table S1). The most significant gene ontology (GO) term for this group of genes is transcription factor activity (P = 5.744375e-04). The other 18 genes are putative indirect targets (Table S1), and their most significant GO term is transcription regulator activity (P = 6.996488e-07).
We then asked whether the AP-expressed genes that vary significantly in embryos with different levels of flattened Bcd are expressed at specific positions in wild-type embryos. We focused on 25 of the 48 genes based on their relatively simple expression patterns, which made it possible to unambiguously assign expression domains that may be activated by Bcd. All 25 genes contain known Bcd-dependent CRMs nearby (within 10 kb of the transcriptional start site), and/or CRMs predicted by the presence of a nearby cluster of Bcd-binding sites (data not shown), and/or a region bound by Bcd in ChIP-chip experiments (12). We then measured the PBPs (50% of max mRNA) of these domains along the AP axis and grouped them according to their behavior in the microarray experiments (Fig. 2B; Table S1). PBPs of genes strongly activated in 1B vas exu compared with bcd mutants (0B–1B group) ranged from 44% to 58% EL, PBPs of genes in the 0B–2B group ranged from 60% to 82% EL, and PBPs of genes in the 0B–6B group ranged from 65% to 93% EL. Thus, there is a reasonable correlation between PBPs of target genes along the AP axis and their expression level changes in the microarray experiments.
All or None: Target Gene Responses to Flattened Bcd Gradients.
We next performed in situ hybridization experiments to test whether individual Bcd target genes are activated in an all-or-none fashion in embryos with different levels of flattened Bcd. Of six tested target genes, only three, hb, gt, and Kr, responded in this way (Fig. 3). Zygotic hb is normally activated only in the anterior half of early wild-type embryos (ref. 21; Fig. 3A), but was expanded throughout most of the length of the embryo in 1B vas exu embryos (Fig. 3G). This expression pattern did not change significantly in 2B and 6B vas exu embryos (Fig. 3 J and M). A lacZ reporter gene driven by the Bcd-dependent hb P2 promoter (HB0.8-lacZ; ref. 22) was also weakly activated in 1B vas exu embryos (Fig. S2), confirming that the observed expression is zygotic.
Fig. 3.
All-or-nothing responses to Bcd-dependent activation. hb, gt, and Kr mRNA expression patterns are shown for wild-type embryos (A–C); embryos produced by bcd mutant females (D–F); and embryos produced by 1B vas exu (G–I), 2B vas exu (J–L), and 6B vas exu females (M–O).
The target gene gt is expressed in anterior and posterior regions in wild-type embryos (ref. 23; Fig. 3B), but was expressed only in weak stripes near the anterior and posterior poles in 1B vas exu embryos (Fig. 3H). However, gt was activated throughout the embryo (but not at the poles) in 2B and 6B vas exu (Fig. 3 K and N). These patterns were precisely recapitulated by a lacZ transgene containing only the Bcd-dependent gt23 CRM (ref. 9; Fig. S2), which suggests that the near ubiquitous expression in 2B and 6B vas exu embryos represents an expansion of the anterior gt expression domain.
Finally, the target gene Kr, which is expressed in a central domain in wild-type embryos (ref. 24; Fig. 3C), was expressed throughout middle-body regions in 1B vas exu embryos (Fig. 3I). This suggests that Kr can respond to levels of Bcd similar to those required for hb activation or that Bcd activates Kr indirectly by activating Hb. The latter hypothesis is consistent with the activation of Kr in embryos lacking bcd altogether (25, 26). Strikingly, however, Kr was completely repressed in 2B and 6B vas exu embryos (Fig. 3 L and O). This repression is probably caused by the nearly ubiquitous expression of gt in 2B and 6B vas exu embryos (Fig. 3K), as Gt has been previously shown to be an effective repressor of Kr (27–29).
The posterior boundaries of the hb P2 and Kr domains are positioned more posteriorly than the posterior boundary of the gt23 domain (Fig. 1K), and activated by lower levels of Bcd (1B vas exu for hb and Kr compared with 2B vas exu for gt). These findings are consistent with the differential sensitivity hypothesis, which proposes that more-posteriorly expressed target genes should be more sensitive to Bcd-dependent activation. However, we noticed that the Bcd levels required for activation of all three genes in vas exu backgrounds were much lower than those present at the PBPs of hb, gt, and Kr in wild-type embryos (Fig. 1K). 1B vas exu embryos contain on average ≈4% [Bcd]max, but the posterior boundaries of hb P2 and Kr in wild-type embryos are positioned in regions that contain ≈11% and ≈9% [Bcd]max respectively. Similarly, 2B vas exu embryos contain ≈11% [Bcd]max, but the posterior boundary of the anterior gt domain in wild type corresponds to a position with 20% [Bcd]max.
Stripes of Target Gene Expression in the Absence of a Bcd Gradient.
The head gap genes otd, ems, and btd are Bcd-dependent genes that are expressed in overlapping domains in anterior regions of wild-type embryos (refs. 30–32; Fig. 4 A–C). The otd expression domain is normally positioned most anteriorly, followed by ems and btd, in that order (Fig. 5A, B, E, and F). In the microarray experiments described previously, these genes showed increased expression levels with increased bcd copy number, but they did not show all-or-none responses by in situ hybridization. In 1B vas exu embryos, the posterior-most gene (btd) was activated in a thin anterior stripe (Fig. 4I). In 2B vas exu embryos, all three genes were activated in strong anterior stripes, and also in weak stripes near the posterior pole (Fig. 4 J–L). The Bcd concentrations in 1B and 2B vas exu embryos (≈4% and ≈11% [Bcd]max) are much lower than those present in the wild-type gradient at the positions of the posterior boundaries of otd, ems, and btd (35%, 27%, and 24% [Bcd]max respectively). This is inconsistent with the hypothesis that these boundaries are formed by specific thresholds of Bcd concentration in vivo. In 6B vas exu embryos, the anterior stripes of these genes expanded throughout much of the anterior half of the embryo, with relatively sharp posterior boundaries, and the posterior stripes became very strong (Fig. 4 M–O).
Fig. 4.
Striped expression patterns in embryos with flattened Bcd. otd, ems, and btd mRNA expression patterns are shown for wild-type embryos (A–C); embryos produced by bcd mutant females (D–F); and embryos produced by 1B vas exu (G–I), 2B vas exu (J–L), and 6B vas exu females (M–O).
Fig. 5.
Mirror-image patterning at the poles of embryos with flattened Bcd. Embryos were double-stained for otd (red) and ems (green) mRNAs (A and C) or ems (green) and btd (red) mRNAs (E and G). White rectangles represent regions of interest used for quantification of fluorescence intensities, shown in (B), (D), (F), and (H). X axes represent percent EL; Y axes represent the percentage of maximum mRNA concentrations.
The expression of otd, ems, and btd in stripes in 1B, 2B, and 6B vas exu embryos shows that sharp expression boundaries can be formed in the absence of a Bcd concentration gradient. Because these stripes appeared in both anterior and posterior regions of the embryo, we postulated that they may be under the control of the Torso (Tor)-dependent patterning system that patterns the termini of the embryo. This is consistent with previous experiments. For example, there are anterior shifts of the PBPs of otd and btd in loss-of-function tor mutants, and posterior shifts in gain-of-function tor mutants (15, 33).
To test whether the correct AP registry of the anterior and posterior stripes is maintained in the vas exu background, we performed double in situ hybridization experiments to simultaneously detect otd and ems, or ems and btd (Fig. 5). These experiments showed that the PBPs of the anterior expression domains in the 2B (data not shown) and 6B vas exu embryos (Fig. 5 C, D, G, and H) are registered as in wild type, with the btd PBP more posterior than the ems and otd PBPs, in that order. In contrast, the ectopic stripes in posterior regions were positioned in a mirror-image order compared with the anterior stripes, with the anterior boundary of otd most posterior, followed by the anterior boundaries of ems and btd, in that order.
The mirror-image order of otd, ems, and btd boundaries in posterior regions of 6B vas exu embryos argues strongly against the model that specific Bcd concentration thresholds set the positions of these stripes. Clearly there is no posterior-to-anterior Bcd gradient in these embryos (Fig. 1L). A more likely hypothesis is that Tor terminal signaling pathway synergizes with Bcd for the activation of these stripes. Supporting this hypothesis is a previous study that showed that the posterior otd stripe in 2B vas exu embryos is abolished in vas exu tor triple mutants, which also remove the terminal patterning system (15).
Discussion
Toward a Complete List of Bcd Target Genes.
In this study, we have used genetic and transgenic manipulations to create pure populations of embryos with flattened Bcd gradients. These manipulations expanded specific subregions of the body plan, which reduced the complexity of cell fates in the embryo compared with wild type, and increased signal-to-noise ratios in the microarray experiments. The three levels of Bcd generated in our experiments, ≈4%, 11%, and ≈40%, cover the lower half of the full range of the Bcd gradient, and these experiments identified 13 of the 18 known Bcd target genes.
The 13 known Bcd target genes are included in a set of 242 genes that are differentially activated by increasing levels of Bcd. Ninety-seven of these genes have been tested for expression in the early embryo, and 48 are expressed differentially along the AP axis. Of these, 30 are likely to be direct targets based on known or predicted Bcd-dependent CRMs. If we use a linear extrapolation of this number to the full set of 242 genes, our genome-wide estimate is ≈74 genes, and if we take into account the fact that our experiments did not identify five previously known Bcd target genes (27%), the estimate increases to ≈103 genes.
Six other genes were identified as Bcd targets based on the microarray experiments and the presence of nearby clusters of Bcd sites, but these genes are either expressed ubiquitously or in dorsal-ventral patterns, with no apparent modulation along the AP axis (Table S2). It is possible that Bcd-dependent activation may partially contribute to these patterns by activating expression in anterior regions, which is consistent with recent studies that showed ChIP-chip binding of DV transcription factors to AP-expressed genes and vice versa (12, 34). If these are real target genes, they would slightly increase the estimate of the total number of Bcd target genes.
Role of the Bcd Gradient in Embryonic Patterning.
Bicoid has been considered as one of the best examples of a gradient morphogen in all of biology. Several lines of evidence suggest that Bcd does indeed function as a morphogen, including the coordinated shifts of morphological features and target gene expression patterns in embryos with different copy numbers of the bcd gene (22, 35), and the ability of bcd mRNA to establish anterior cell fates when microinjected into ectopic positions (4). Furthermore, manipulations of the Bcd-binding sites in the hb P2 promoter and synthetic constructs with defined Bcd sites showed that cis-regulatory elements can be designed to be more or less sensitive to Bcd-mediated transcription (13, 36, 37). These studies led to the hypothesis that differential sensitivity to Bcd binding may control the relative positioning of different target genes.
Our findings suggest that differential sensitivity to Bcd binding is not the primary mechanism that controls the relative positioning of its target genes. Though some target genes respond in an all-or-none fashion to different levels of flattened Bcd, the levels required for activation are much lower than those present in the wild-type gradient in the regions where those genes are activated. These findings suggest that Bcd concentrations are in excess of those required for activation at every position along the length of the wild-type gradient.
We also show that the head gap genes otd, ems, and btd are expressed in correctly ordered stripes in embryos containing flattened Bcd gradients. This is most dramatically demonstrated by the mirror-image duplication of otd, ems, and btd stripes in the posterior region of 6B vas exu embryos (Fig. 5), where the Bcd gradient slopes in the opposite direction to the order of striped expression. We propose that these genes are patterned by the terminal system in the absence of a Bcd gradient, and though Bcd function is required for their activation, the Bcd gradient does not play a major role in establishing their relative positions along the AP axis.
Bcd seems capable of bypassing the terminal system if expressed at high levels. For example, the anterior defects in terminal-system mutants can be partially rescued by increasing bcd copy number (38). Also, in 6B vas exu embryos, higher levels of Bcd are present throughout the embryo, with a relatively weak gradient along the AP axis (Fig. 1L). This causes expansions of the anterior otd, ems, and btd expression patterns into central regions of the embryo. The posterior boundaries of these patterns are positioned correctly (Fig. 5), suggesting that the Bcd protein gradient is sufficient to position these target genes in regions where the terminal system does not reach. This is consistent with the observation that microinjected bcd mRNA can autonomously specify anterior structures (4).
An Integrated System for AP Patterning.
Our data are consistent with previous studies that failed to find a strong correlation between the relative positioning of target genes and the Bcd-binding “strength” of their associated cis-regulatory elements (9, 16). They further support a model in which Bcd functions as only one component of an integrated patterning system that establishes gene expression patterns along the AP axis (Fig. 6). A second major component is maternal Hb, which is expressed in an AP protein gradient (39). Hb synergizes with Bcd in the activation of several specific target genes (37). In vas exu embryos, the loss of vas causes ectopic translation of maternal hb in posterior regions, so Hb protein is ubiquitously expressed and available for combinatorial activation with Bcd. This combination is likely sufficient to lead to the near ubiquitous expression of zygotic hb and Kr in 1B vas exu embryos, and gt in 2B vas exu embryos.
Fig. 6.
An integrated anterior patterning system. (A) Model for interactions between the Bcd gradient, maternal Hb, and the Tor-dependent terminal system (TS). Interactions between Bcd and Hb provide the potential for target gene activation throughout the anterior half of the embryo. Activation of the terminal system at the poles directly modifies Bcd and/or represses the activity of ubiquitously expressed repressors, resulting in enhanced morphogenetic activity close to the anterior pole. (B) Anterior expression patterns of seven Bcd target genes (black lines) are colinear with the morphogenetic functions illustrated in (A). The terminal system synergizes with Bcd and Hb for the correct positioning of otd, ems, btd, slp1, and gt23. In more central regions, out of the range of the terminal system, Bcd functions only with Hb to establish the posterior boundaries of zygotic hbP2 and Kr (adapted from ref. 47).
A third major component is the terminal system, which seems to affect the expression patterns of Bcd target genes in two ways. First, it causes a repression of all known Bcd target genes at the anterior pole by a mechanism that is not clearly understood (40). Second, our data and other studies suggest that the terminal system functions with Bcd for the establishment of the posterior boundaries of the head gap genes. This interaction appears to be important for regulating at least two other target genes, gt and slp1, which are expressed in anterior domains that shift toward the anterior pole in terminal system mutants (41, 42). Both gt and slp1 are also activated in anterior and posterior stripes in embryonic regions containing low levels of flattened Bcd (ref. 41; Fig. 4H). These findings suggest that interactions with the terminal system may be required for positioning most Bcd target genes. The only known target genes that may not be directly influenced by the terminal system are zygotic hb and Kr, which are expressed in middle embryonic regions, far from the source of the terminal system activity.
How synergy between Bcd and the terminal system is achieved for each target gene is not clear. One possibility is that the Torso phosphorylation cascade directly modifies the Bcd protein, increasing its potency as a transcriptional activator. Mutations in Bcd's MAP-kinase phosphorylation sites partially reduce the ability of Bcd to activate otd, consistent with this hypothesis (43). Alternatively, the terminal system has been shown to repress the activities of ubiquitously expressed repressor proteins (reviewed in ref. 44). Perhaps repression by the terminal system creates posterior to anterior gradients of these proteins, which then compete with Bcd-dependent activation mechanisms to establish posterior boundaries of target gene expression.
Interactions between Bcd, maternal Hb, and the terminal system may be critical for the initial positioning of target gene expression patterns, but it is clear that other layers of regulation are required for creating the correct order of gene expression boundaries in the anterior part of the early embryo. Almost all known Bcd target genes are transcription factors, and there is evidence that they regulate each other by feed-forward activation and repression mechanisms. Each target gene contains one or more CRMs, each of which is composed of a specific combination and arrangement (code) of transcription factor binding sites (e.g., refs. 45 and 46). Unraveling the mechanisms that differentially position Bcd target will require the detailed dissections of CRMs that direct spatially distinct expression patterns.
Materials and Methods Summary.
The maternal-effect alleles used in this study were vasPD, exuPJ, and bcdE1. mRNA and protein staining patterns were assayed using standard techniques. Fluorescent expression data were collected by confocal microscopy and analyzed using MATLAB (Mathworks). For microarray analysis, we isolated total RNA from 2–4 h embryos lacking bcd (0B; from bcdE1 females), and 1B, 2B, and 6B vas exu embryos, and hybridized cDNA made from each genetic background to whole-genome Drosophila tiling arrays from Affymetrix. We then performed statistical analyses to identify a set of 3,600 genes that varied significantly when Bcd levels were changed. Genes were ranked in terms of the fold change of their expression for all pairs of mutants: 0B–1B vas exu, 0B–2B vas exu, and 0B–6B vas exu. Complete methods are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Jerry Huang and Hongtao Chen for providing Bcd cluster data, Alison Mello for assistance with microarray data analysis, Huey Ling Kao for assistance in MATLAB programming, Gary Struhl for the HB0.8 LacZ strain, Claude Desplan for helpful comments on the manuscript, Yuri Goltsev and Uli Loehr for sharing data before publication, and Gozde Yucel for stimulating discussions. This work was supported by National Institutes of Health Grant GM 51946 in a facility constructed with support from Research Facilities Improvement Grant C06 RR-15518–01 from the National Center for Research Resources, National Institutes of Health.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Networks in Animal Development and Evolution,” held February 15–16, 2008, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at: http://www.nasonline.org/SACKLER_Gene_Networks.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807878105/DCSupplemental.
References
- 1.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theoret Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 2.Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54(1):83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 3.Frohnhofer HG, Nusslein-Volhard C. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature. 1986;324:120–124. [Google Scholar]
- 4.Driever W, Siegel V, Nusslein-Volhard C. Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development. 1990;109(4):811–820. doi: 10.1242/dev.109.4.811. [DOI] [PubMed] [Google Scholar]
- 5.Crauk O, Dostatni N. Bicoid determines sharp and precise target gene expression in the Drosophila embryo. Curr Biol. 2005;15(21):1888–1898. doi: 10.1016/j.cub.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 6.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130(1):153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann S, et al. Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 2007;5(2):e46. doi: 10.1371/journal.pbio.0050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He F, et al. Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev Cell. 2008;15(4):558–567. doi: 10.1016/j.devcel.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochoa-Espinosa A, et al. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2005;102(14):4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann B, Reichert H, Walldorf U. Interaction of gap genes in the Drosophila head: Tailless regulates expression of empty spiracles in early embryonic patterning and brain development. Mech Dev. 2001;109(2):161–172. doi: 10.1016/s0925-4773(01)00519-6. [DOI] [PubMed] [Google Scholar]
- 11.Liaw GJ, Lengyel JA. Control of tailless expression by bicoid, dorsal and synergistically interacting terminal system regulatory elements. Mech Dev. 1993;40(1–2):47–61. doi: 10.1016/0925-4773(93)90087-e. [DOI] [PubMed] [Google Scholar]
- 12.Li XY, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6(2):e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340(6232):363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 14.Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415(6873):798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Wang Y, Finkelstein R. Orthodenticle regulation during embryonic head development in Drosophila. Mech Dev. 1996;56(1–2):3–15. doi: 10.1016/0925-4773(96)00504-7. [DOI] [PubMed] [Google Scholar]
- 16.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451(7178):535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 17.Berleth T, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7(6):1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamberi C, Peterson DS, He L, Gottlieb E. An anterior function for the Drosophila posterior determinant Pumilio. Development. 2002;129(11):2699–2710. doi: 10.1242/dev.129.11.2699. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112(3):679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 20.Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3(12):RESEARCH0088. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 22.Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57(7):1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 23.Mohler J, Eldon ED, Pirrotta V. A novel spatial transcription pattern associated with the segmentation gene, giant, of Drosophila. EMBO J. 1989;8(5):1539–1548. doi: 10.1002/j.1460-2075.1989.tb03538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackle H, et al. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–473. doi: 10.1101/sqb.1985.050.01.058. [DOI] [PubMed] [Google Scholar]
- 25.Gaul U, Jackle H. Pole region-dependent repression of the Drosophila gap gene Kruppel by maternal gene products. Cell. 1987;51(4):549–555. doi: 10.1016/0092-8674(87)90124-3. [DOI] [PubMed] [Google Scholar]
- 26.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69(2):237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 27.Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Kruppel define middle body regions of the Drosophila embryo. Development. 1991;111(2):611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, Vakani R, Small S. Two distinct mechanisms for differential positioning of gene expression borders involving the Drosophila gap protein giant. Development. 1998;125(Pt 19):3765–3774. doi: 10.1242/dev.125.19.3765. [DOI] [PubMed] [Google Scholar]
- 29.Yu D, Small S. Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr Biol. 2008;18(12):868–876. doi: 10.1016/j.cub.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkelstein R, Perrimon N. The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature. 1990;346(6283):485–488. doi: 10.1038/346485a0. [DOI] [PubMed] [Google Scholar]
- 31.Walldorf U, Gehring WJ. Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J. 1992;11(6):2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wimmer EA, Jackle H, Pfeifle C, Cohen SM. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature. 1993;366(6456):690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 33.Wimmer EA, Simpson-Brose M, Cohen SM, Desplan C, Jackle H. Trans- and cis-acting requirements for blastodermal expression of the head gap gene buttonhead. Mech Dev. 1995;53(2):235–245. doi: 10.1016/0925-4773(95)00439-8. [DOI] [PubMed] [Google Scholar]
- 34.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21(4):385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54(1):95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 36.Hanes SD, Riddihough G, Ish-Horowicz D, Brent R. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol. 1994;14(5):3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson-Brose M, Treisman J, Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78(5):855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffer V, Killian D, Desplan C, Wimmer EA. High bicoid levels render the terminal system dispensable for Drosophila head development. Development. 2000;127(18):3993–3999. doi: 10.1242/dev.127.18.3993. [DOI] [PubMed] [Google Scholar]
- 39.Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332(6161):281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- 40.Janody F, Sturny R, Schaeffer V, Azou Y, Dostatni N. Two distinct domains of Bicoid mediate its transcriptional downregulation by the Torso pathway. Development. 2001;128(12):2281–2290. doi: 10.1242/dev.128.12.2281. [DOI] [PubMed] [Google Scholar]
- 41.Grossniklaus U, Cadigan KM, Gehring WJ. Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development. 1994;120(11):3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- 42.Kraut R, Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991;111(2):601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- 43.Janody F, Sturny R, Catala F, Desplan C, Dostatni N. Phosphorylation of bicoid on MAP-kinase sites: Contribution to its interaction with the torso pathway. Development. 2000;127(2):279–289. doi: 10.1242/dev.127.2.279. [DOI] [PubMed] [Google Scholar]
- 44.Li WX. Functions and mechanisms of receptor tyrosine kinase Torso signaling: Lessons from Drosophila embryonic terminal development. Dev Dyn. 2005;232(3):656–672. doi: 10.1002/dvdy.20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrioli LP, Vasisht V, Theodosopoulou E, Oberstein A, Small S. Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development. 2002;129(21):4931–4940. doi: 10.1242/dev.129.21.4931. [DOI] [PubMed] [Google Scholar]
- 46.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122(1):205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 47.Paroush Z, Wainwright SM, Ish-Horowicz D. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development. 1997;124(19):3827–3834. doi: 10.1242/dev.124.19.3827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.