Abstract
GPR109B (HM74) is a putative G protein-coupled receptor (GPCR) whose cognate ligands have yet to be characterized. GPR109B shows a high degree of sequence similarity to GPR109A, another GPCR that was identified as a high-affinity nicotinic acid (niacin) receptor. However, the affinity of nicotinic acid to GPR109B is very low. In this study, we found that certain aromatic D-amino acids, including D-phenylalanine, D-tryptophan, and the metabolite of the latter, D-kynurenine, decreased the activity of adenylate cyclase in cells transfected with GPR109B cDNA through activation of pertussis toxin (PTX)-sensitive G proteins. These D-amino acids also elicited a transient rise of intracellular Ca2+ level in cells expressing GPR109B in a PTX-sensitive manner. In contrast, these D-amino acids did not show any effects on cells expressing GPR109A. We found that the GPR109B mRNA is abundantly expressed in human neutrophils. D-phenylalanine and D-tryptophan induced a transient increase of intracellular Ca2+ level and a reduction of cAMP levels in human neutrophils. Furthermore, knockdown of GPR109B by RNA interference inhibited the D-amino acids-induced decrease of cellular cAMP levels in human neutrophils. These D-amino acids induced chemotactic activity of freshly prepared human neutrophils. We also found that D-phenylalanine and D-tryptophan induced chemotactic responses in Jurkat cells transfected with the GPR109B cDNA but not in mock-transfected Jurkat cells. These results suggest that these aromatic D-amino acids elicit a chemotactic response in human neutrophils via activation of GPR109B.
Keywords: D-kynurenine, D-phenylalanine, D-tryptophan, nicotinic acid (niacin), orphan receptor
GPR109B (also known as HM74) is an orphan G protein-coupled receptor (GPCR), which was cloned during a search for novel leukocyte chemoattractant receptors relating to receptors for IL-8, N-formyl peptide, and C5a (1). A related receptor, GPR109A (HM74A), has been identified as a receptor for nicotinic acid (niacin). GPR109A is highly expressed in adipose tissue and the spleen (2). Nicotinic acid inhibits adipocyte lipolysis by inhibiting hormone-sensitive triglyceride lipase (3). This antilipolytic effect of nicotinic acid involves the inhibition of cAMP accumulation in adipocytes through Gi protein-mediated inhibition of adenylyl cyclase (4–6). Independently, Tunaru et al. (7) reported that the mouse orthologue of GPR109A, named “protein upregulated in macrophages by IFN-γ” (mouse PUMA-G), is highly expressed in adipose tissue and is a nicotinic acid receptor. They reported that the nicotinic acid-mediated decrease in plasma levels of fatty acids and triglyceride was abrogated in mice lacking PUMA-G, indicating that PUMA-G mediates the antilipolytic and lipid-lowering effects of nicotinic acid in vivo. However, the mouse does not have an orthologue of GPR109B, which seems to be specific in primates.
Although GPR109A and GPR109B are highly similar, displaying 96% sequence identity at the protein level (differing by only 15 aa), the 2 receptors are not simply polymorphic variants or splice variants. They are encoded by separate genes colocated with another orphan GPCR, GPR81, at chromosome 12q24.31 (8). Comparison of the cDNA sequences of GPR109A and GPR109B reveal 15 base changes as well as a 5-nucleotide insertion at the 3′ end, resulting in GPR109A possessing a shorter C-terminal tail. Although GPR109B was once reported to be a low-affinity receptor for nicotinic acid, the affinity of nicotinic acid to GPR109B is quite low at millimolar levels (2). Furthermore, an independent study reported that GPR109B does not appreciably bind nicotinic acid (7), suggesting the possibility that GPR109B is a receptor for another biologically active substance or substances. The mRNA expression profiles of GPR109A and GPR109B partially overlap and are partially distinct. GPR109A is detected in adipose tissue, lung, trachea, and spleen, whereas GPR109B is detected in adipose tissue, lung, spleen, and peripheral blood leukocytes (2). Here we have identified a novel mechanism by which some aromatic D-amino acids may regulate cellular responses through activation of GPR109B. We identified D-phenylalanine (D-Phe), D-tryptophan (D-Trp), and D-kynurenine (D-Kyn, a metabolite of D-Trp) as agonists for GPR109B. These D-amino acids elicit chemotactic responses in human neutrophils through GPR109B.
Results
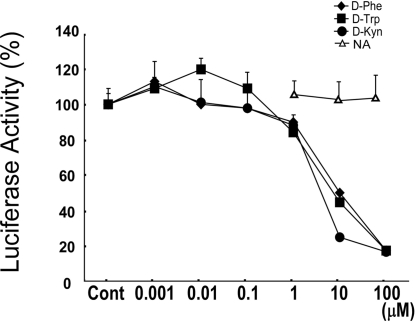
In the framework of a general strategy for characterizing ligands for orphan GPCRs, we established CHO-K1 cell lines coexpressing orphan receptor cDNAs and a luciferase reporter driven by a cAMP response element (CRE). We screened a library of ≈600 natural bioactive compounds and their derivatives, including lipids, amino acids, and others [supporting information (SI) Table S1]. During this procedure, we found that certain D-amino acids, namely D-Phe, D-Trp, and D-Kyn, inhibited the CRE-luciferase activity (up to ≈80%) induced by forskolin in CHO-K1 cells stably expressing GPR109B (CHO-GPR109B cells) in a dose-dependent manner (Fig. 1). The EC50 values of the responses to these ligands were 9.02 μM, 3.72 μM, and 2.61 μM, respectively (Fig. 1). The inhibitory effects of these D-amino acids on luciferase activity were confirmed in 3 independent lines of stable clones expressing GPR109B (data not shown). Despite the high degree of sequence similarity between GPR109A and GPR109B, D-Phe, D-Trp, and D-Kyn showed no effect on the CRE-luciferase activity induced by forskolin in CHO-GPR109A cells (data not shown). Other amino acids, including L-Phe, L-Trp, L-Kyn, and their related compounds showed no effect on luciferase activity induced by forskolin in CHO-GPR109B cells (Table S1). In cells stably expressing another orphan GPCR, GPR81, which shows high similarity to GPR109B (52% amino acid identity), D-amino acids, including D-Phe, D-Trp, and D-Kyn, did not inhibit the CRE-luciferase activity induced by forskolin (data not shown).
Fig. 1.
Identification of D-amino acids as agonists for GPR109B. CHO-K1 cell lines expressing GPR109B along with the CRE-luciferase reporter plasmid were incubated with various concentrations of designated D-amino acids, together with 0.3 μM forskolin. All values are expressed as a percentage of the activity induced by 0.3 μM forskolin only. Data are mean ± SEM of 3 independent experiments, each performed in triplicate. NA, nicotinic acid (niacin).
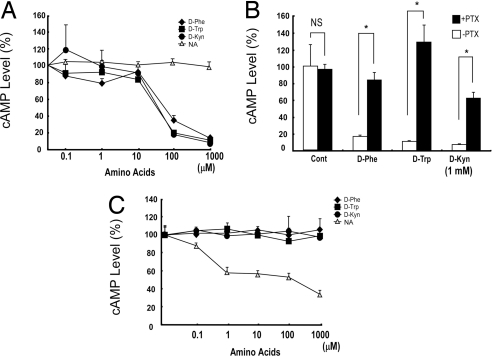
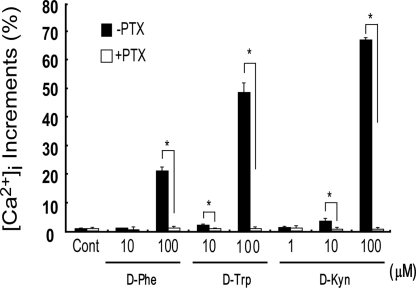
The results from these CRE-luciferase assays suggested that GPR109B couples to the Gi class of G proteins. In fact, the forskolin-stimulated production of cAMP was also inhibited by these D-amino acids in CHO-GPR109B cells (Fig. 2A). This inhibition was abolished by pretreatment of the cells with pertussis toxin (PTX) (Fig. 2B). In contrast, D-Phe, D-Trp, and D-Kyn showed no effect on forskolin-stimulated cAMP accumulation in CHO-GPR109A cells, whereas nicotinic acid significantly lowered cAMP levels, as expected (2) (Fig. 2C). We also examined the effect of D-amino acids, including D-Phe, D-Trp, and D-Kyn, on intracellular calcium ion mobilization in CHO-GPR109B cells. These amino acids elicited a transient rise of intracellular Ca2+ levels in CHO-GPR109B cells in a dose-dependent manner (Fig. 3). This response was also abolished by PTX pretreatment, suggesting that this calcium response is mediated via activation of PTX-sensitive G proteins in CHO cells (Fig. 3). These observations suggested that GPR109B predominantly couples to the Gi/o class of G proteins. These amino acids also elicited a transient rise of intracellular Ca2+ levels in COS7 cells transiently transfected with GPR109B (data not shown). D-Phe, D-Trp, and D-Kyn, did not induce a rise of intracellular Ca2+ level in CHO cells stably expressing GPR81 or GPR109A (data not shown).
Fig. 2.
D-amino acids act on GPR109B and activate PTX-sensitive classes of G proteins in CHO cells. (A) D-amino acids inhibit cAMP accumulation through GPR109B via activation of PTX-sensitive G proteins. A CHO-K1 cell line expressing GPR109B was incubated with various concentrations of the D-amino acids D-Phe, D-Trp, and D-Kyn together with 10 μM forskolin. Content of cAMP is expressed as a percentage of control (value in absence of D-amino acids). Data are mean ± SEM of 3 independent experiments, each performed in triplicate. (B) A CHO-K1 cell line expressing GPR109B was incubated with D-amino acids (1 mM) together with forskolin (10 μM) after incubation with 100 ng/mL PTX or vehicle for 24 h. Content of cAMP is expressed as a percentage of control (value in absence of D-amino acids). Data are mean ± SEM of 3 independent experiments, each performed in triplicate. (C) D-Phe, D-Trp, and D-Kyn do not act on GPR109A. D-Phe, D-Trp, and D-Kyn did not decrease level of intracellular cAMP in CHO cells expressing GPR109A, whereas nicotinic acid (NA) potently inhibited it. Data are mean ± SEM of 3 independent experiments, each performed in triplicate.
Fig. 3.
Effect of D-amino acids on cytoplasmic Ca2+ concentration. Increase in intracellular calcium concentration after exposure to various amino acids in a CHO-K1 cell line expressing GPR109B and effects of PTX treatment on D-amino acid-induced increases in intracellular calcium concentration in CHO-GPR109B cells are shown. Data are expressed as percentage of the peak response induced by UTP (100 μM). *, P < 0.05 vs. PTX (-).
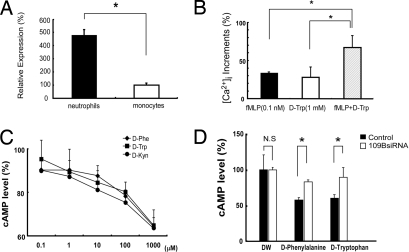
GPR109B mRNA has been reported to be expressed in adipose tissue, lung, spleen, and peripheral blood leukocytes in humans (2, 9). We also found that GPR109B mRNA was abundantly expressed in human neutrophils. By quantitative real-time RT-PCR analysis, we found that the expression of GPR109B mRNA was 4.8-fold higher in neutrophils than in monocyte fractions (Fig. 4A). Given the identification of GPR109B as a receptor for specific aromatic D-amino acids, we investigated the effects of these D-amino acids on freshly prepared human neutrophils. Although we found that GPR109B couples to the Gi/o class of G proteins in transfected CHO cells, it is well established that many GPCRs couple to phospholipase C through the promiscuous G15/16 proteins in blood cells. Therefore, we first examined the effect of these D-amino acids on intracellular Ca2+ mobilization in neutrophils. D-Trp significantly elicited a transient rise of intracellular Ca2+ levels in human neutrophils (Fig. 4B) but did not induce degranulation, in contrast to some other chemotaxic factors, such as formyl-methionyl-leucyl-phenylalanine (fMLP) (data not shown). We also found that D-Trp (1 mM) augmented the intracellular Ca2+ mobilization induced by fMLP (0.1 nM) (Fig. 4B), suggesting that D-Trp and fMLP can act synergistically on neutrophils. In contrast, L-Phe, L-Trp, or L-Kyn did not elicit a transient rise of intracellular Ca2+ levels in human neutrophils (data not shown). We also measured intracellular cAMP levels after stimulation of human neutrophils with these D-amino acids and found that D-Phe, D-Trp, and D-Kyn decreased the forskolin-stimulated production of cAMP with similar potencies (Fig. 4C).
Fig. 4.
(A) Expression of GPR109B in human neutrophils. Expression of GPR109B mRNA in human neutrophils is determined by real-time RT-PCR analysis, and the relative expression level is expressed as percentage of that in human monocytes. Data represent mean value ± SEM (n = 4). *, P < 0.05 vs. monocytes. (B) D-Trp enhanced intracellular calcium mobilization induced by fMLP at a low dose in human neutrophils. Neutrophils were loaded with 10 μM Fura-2AM for 2 h at room temperature, and intracellular calcium mobilization was measured. All values are expressed as a percentage of control (fMLP, 100 nM). Data represent mean value ± SEM (n = 4). *, P < 0.05 vs. fMLP (0.1 nM) or D-Trp (1 mM). (C) D-Phe, D-Trp, and D-Kyn inhibit cAMP production in neutrophils through GPR109B. Neutrophils were incubated with D-amino acids together with 50 μM forskolin. After 15 min, cAMP accumulation was measured. cAMP content is expressed as a percentage of control in the absence of D-amino acids. Data are mean ± SEM of 3 independent experiments, each performed in triplicate. (D) Effects of GPR109B knockdown on D-amino acids-induced decrease in cAMP level in human neutrophils. Transfection with GPR109B siRNA significantly inhibited the decrease of cAMP level by D-Phe and D-Trp in neutrophils as compared with control siRNA.
To examine whether these effects are mediated by GPR109B expressed in human neutrophils, we performed an RNA interference experiment by transfecting human neutrophils with siRNA sequences targeting GPR109B mRNA. Quantitative real-time RT-PCR analysis of GPR109B mRNA levels in neutrophils after transfection with GPR109B siRNA demonstrated a marked reduction of GPR109B mRNA levels to 49.5% ± 2.0% compared with control (neutrophils treated with a mixture of unrelated siRNA sequences). Although the inhibitory response to D-Trp of cAMP production induced by forskolin remained intact in neutrophils transfected with scrambled siRNA, transfection with GPR109B siRNA significantly inhibited the decrease of cAMP levels by D-Phe and D-Trp (Fig. 4D). These observations confirmed that the effect of D-Trp on neutrophils is mediated by GPR109B.
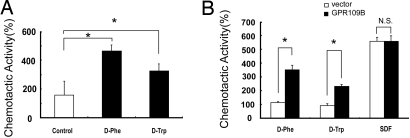
The observation that GPR109B seems to share a similar signaling pathway with some chemotactic factors led us to test for chemoattractant activity of D-amino acids in neutrophils using an optical assay device (the EZ-TAXIScan system; Effector Cell Institute). D-Phe at an endpoint concentration of 100 mM induced marked chemotactic activity in human neutrophils. D-Trp at 10 mM at the endpoint also induced significant chemotactic activity (Fig. 5A) (we could not investigate the chemotactic activity of D-Trp at 100 mM because D-Trp is not soluble in water at 100 mM). It is worth noting here that, under our experimental conditions, the ratio of the attractant concentrations at the endpoint (where the attractants are applied) and the start point (where the cells initially reside) is ≈100:1. Therefore, under the conditions depicted in Fig. 5A, cells were exposed to ≈1 mM of D-Phe and 0.1 mM of D-Trp (see Materials and Methods).
Fig. 5.
D-amino acids elicit chemotactic activity through activation of GPR109B. (A) D-amino acids induced chemotaxis in human neutrophils. Chemotactic activities to D-Phe, D-Trp, or control (distilled water) recorded for 60 min are shown. All values are expressed as a percentage of control. Data are the mean of 2 independent preparations of cells. (B) D-amino acids induced chemotaxis in Jurkat cells expressing GPR109B. Chemotactic activities of Jurkat cells expressing GPR109B to D-Phe, D-Trp, SDF-α, or control (distilled water) recorded for 240 min are shown. All values are expressed as a percentage of control. Data are mean ± SEM of 2 independent experiments, each performed in duplicate.
The fact that the chemotaxis assay requires very fresh preparations of neutrophils rendered siRNA transfection experiments highly difficult. Therefore, to examine whether the chemotactic responses of neutrophils to D-Phe and D-Trp are mediated by GPR109B in heterologous cells, we transfected GPR109B cDNA into Jurkat cells, which do not detectably express endogenous GPR109B. D-Phe and D-Trp induced significant chemotactic responses in Jurkat cells expressing GPR109B, whereas these D-amino acids exerted no effects on Jurkat cells transfected with the empty vector (Fig. 5B). These results suggest that D-Phe and D-Trp can act as chemoattractants for blood cells through activation of GPR109B.
Discussion
Our present study suggests that GPR109B is a functional receptor responsible for the action of D-Phe and D-Trp on neutrophils. The identification of signal-transducing receptor for D-amino acids in neutrophils opens a new perspective on the modulatory mechanism of immune system functions in various pathophysiologic situations.
Recently, Jilek et al. (10) reported that D-amino acids are present in some peptides from amphibian skin and that these residues are derived from the corresponding L-amino acids present in the respective precursors. They isolated an enzyme from frog skin that catalyzes the isomerization of an L-lle in position 2 of a model peptide to D-allo-lle. They also reported that polypeptides related to the frog skin enzyme are present in several vertebrate species, including humans (10). Their findings raise the intriguing possibility that peptides containing a D-amino acid or acids are also present in humans. The presence of D-isomers may have been overlooked in hormonal, antibacterial, or modulatory peptides of the immune system, or neuropeptides that have been characterized earlier. Neutrophils play a key role in the early steps of the inflammatory process. D-amino acids, as peptide metabolites, could induce activation of the immune system, leading to inflammatory responses. The development of therapeutic strategies to block neutrophil recruitment and activation might be beneficial in a number of diseases characterized by an inflammatory component. Therefore, the identification of GPR109B as a signaling receptor for D-Phe and D-Trp could potentially lead to new therapeutic strategies.
hGPCR48 (TG1019), which has high sequence similarity to GPR109B (41% amino acid identity), was revealed to be a receptor for 5-oxo-eicosatetraenoic acid (11). This eicosanoid is also known to be a potent chemotactic factor for eosinophils and neutrophils (12, 13). In addition to several GPCRs for leukocyte chemoattractants [such as IL-8, C5a, fMLP, Platelet activating factor (PAF), and Leukotriene B4 (LTB4)], human GPR43, which displayed dual coupling through the Gi/o and PTX-insensitive Gq protein families, was uncovered as a neutrophil-specific receptor for short chain fatty acids (SCFAs) (14). SCFAs, such as sodium acetate and sodium propionate, are produced as metabolic byproducts of anaerobic bacteria and induce neutrophil chemotaxis at an optimal concentration of 1 mM (14). Because some D-amino acids are also contained in bacterial components, such as D-Phe in peptidoglycans, they might also act as bacterially derived chemoattractants through GPR109B. However, the possibility that these D-amino acids are surrogate rather than natural ligands for GPR109B is not excluded, because there is no proof to date that D-amino acids exist in the organism at the local concentrations required to activate GPR109B.
We and others have not been able to identify ESTs or genomic fragments encoding a GPR109B orthologue in mammalian species other than humans and chimpanzees. This suggests that GPR109B may be the result of a very recent gene duplication event, and GPR109A and GPR109B might serve as a model to study the evolution of the GPCR gene family. Considering that GPR109B might play a role in the defensive mechanism by leukocytes, distinct environmental conditions and evolutionary niches for different species could lead to different selective pressures to the GPR109 family of genes. Because rodents do not have a GPR109B gene, there is a possibility that other receptor(s) would be activated by D-amino acids. This point should be evaluated by future screening studies that use a rodent GPCR library.
Both GPR109A and GPR109B are expressed in adipose tissue. Nicotinic acid (niacin), via GPR109A, is reported to exert lipid-lowering effects by decreasing the fatty acid output from adipose tissue. These observations suggest the intriguing possibility that D-Phe and D-Trp may also have a lipid-lowering effect in humans. Because niacin has well-documented adverse effects, such as severe persistent hypotension and cutaneous flushing, a GPR109B-selective agonist might potentially be a better therapeutic agent.
Materials and Methods
Cloning of Human GPR109B and GPR109A.
Transcriptional ORFs of GPR109A or GPR109B were amplified by PCR from human genomic DNA as a template using the following primers: 5′-CCCTTAAGGCCACCATGAATCGGCACCATCTGCA and 3′-CCGGAATTCCTCGATGCAACAGCCCAACTG for GPR109B; and 5′-CCCTTAAGGCCACCATGAATCGGCACCATCTGCA and 3′-CCGGAATTCAGGAGAGGTTGGGCCCAGATA for GPR109A. The amplified products were tagged with GFP at their 3′ termini and subcloned into a mammalian expression vector (pIRESneo2; Clontech). Both strands of the cloned DNA were sequenced with a DNA sequencer.
Cell Culture and Transfection.
CHO-K1 cells were cultured in DMEM/HamF12 supplemented with 10% FCS (Sigma-Aldrich), 100 IU/mL penicillin, and 100 μg/mL streptomycin. For transfection, 4 μg DNA was mixed with 6 μL FuGENE6 in 0.3 mL Opti-MEM (Invitrogen) and incubated at room temperature for 30 min. The cells were exposed to the FuGENE6/DNA mixture in 2 mL of 10% FCS DMEM/HamF12.
For the generation of stable cell lines for the CRE-luciferase assay, CRE-luc reporter (15) and pcDNA3.1/Zeo (Invitrogen) were used to transfect CHO-K1 cells seeded in 6-well dishes and grown to 50% confluence as described above. These cells were maintained in DMEM/HamF12 containing 10% FCS. At 24 h after transfection, the medium was supplemented with 500 μg/mL Zeocin (Invitrogen) for selection of antibiotic-resistant clones. Several resistant clones were isolated by limiting dilution and examined for the expression of the reporter protein by stimulation with forskolin (Sigma-Aldrich). The clone that showed highest luciferase activity (CHO-CRE6) was used for the experiment.
For the generation of stable cell lines for GPR109A and GPR109B, CHO-CRE6 cells were again transfected with pIRESneo-GPR109A::GFP or pIRESneo-GPR109B::GFP. At 24 h after transfection, 500 μg/mL G418 was added to the medium for selection of neomycin-resistant cells. The expression levels of each receptor were validated by fluorescent microscopy. Three lines with high expression for each receptor were selected, maintained in DMEM/HamF12 containing 10% FCS, 500 μg/mL Zeocin, and G418, and used for further analysis.
For the generation of stable cell lines of Jurkat cells expressing GPR109B, the GPR109B gene was subcloned into the pEB vector (16). pEBGPR109B::GFP was used to transfect Jurkat cells by electroporation (voltage: 260 V). These cells (107 cells) were maintained in RPMI 1640 (Invitrogen) containing 10% FCS. At 24 h after transfection, the medium was supplemented with 1 mg/mL G418 for selection of antibiotic-resistant cells.
CRE-Luciferase Activity.
Cells were seeded in a 96-well-plate and grown to confluence. Cells were treated with 0.3 μM forskolin and the indicated compounds. Six hours later, luciferase activity was determined as described previously (15).
Measurement of Intracellular Calcium Concentration.
Cells were loaded with 10 μM Fura-2AM (Dojindo Laboratories) in Hepes-Tyrode's buffer at room temperature for 2 h. The cells were washed twice and resuspended in Hepes-Tyrode's buffer at a density of 106 cells/mL and seeded in a 96-well plate. Intracellular Ca2+ concentration was measured from the ratio of emission fluorescence of 500 nm by excitation at 340 or 380 nm using a Functional Drug Screening System 3000 (Hamamatsu Photonics).
Measurement of cAMP.
The cells were seeded on 96-well plates (2 × 104 cells per well) and cultured for another 24 h. The medium was replaced with 75 μL serum-free DMEM containing 1 mM isobutyl methylxanthine and incubated at 37 °C for 20 min. The reaction was initiated by adding 25 μL of ligand solution containing designated ligands and forskolin (final 10 or 50 μM). After 15 min of incubation, cAMP level was assayed by a homogeneous time-resolved fluorescence cAMP femtomolar assay system (Schering).
Chemotaxis Assay.
Chemotaxis was measured using an EZ-TAXIScan (Effector Cell Institute). The granulocyte fraction prepared from human whole blood was used as neutrophils (17). Human neutrophils or GPR109B-Jurkat cells were examined for their chemotactic responses to D-amino acids as described previously (18). The neutrophils were suspended at 106 cells/mL in RPMI 1640 medium supplemented with 20 mM Hepes and 0.1% BSA. Medium containing 10−1 M D-Phe, 10−2 M D-Trp, 10−6 M stromal cell-derived factor-1α (SDF-1α), or none (distilled water) was injected into the opposite compartment after injection of the cells. The concentration gradient between the start and end points reached a maximum at approximately 5–15 min and the maximum concentration difference per 10 μm (almost 1:100). To observe and record the migration of cells in the channel, a CCD camera or CCD video camera connected to a monitor display was used. The migration of cells in the channel was recorded for 60–240 min.
RNA Interference.
RNA interference was performed with DeliverX transfection reagents (Panomics) according to the manufacturer's instructions (19). Each pair of 21-nt oligonucleotides was synthesized and annealed. Neutrophils were transfected with 1 of 3 different targeting regions of GPR109B. The target sequences used for GPR109B were 5′-CCGUUCGUGAUGGACUACU-3′, 5′-CAUCACUGUUGGCCUAACA-3′, and 5′-GCAUCUCUGGAGAAACAGU-3′ (target position: 241–259, 459–477, and 1126–1144). At 18–20 h after transfection of human neutrophils with the siRNA, cAMP levels following ligand administration were determined. To measure GPR109B RNA level, total RNA from the transfected neutrophils was extracted using an RNeasy kit (Invitrogen) and subjected to quantitative real-time RT-PCR analysis using primers for GPR109B or human GAPDH (TaqMan Gene expression assays; Applied Biosystems). Quantitative PCR was carried out using the ABI TaqMan Universal Master Mix kit with an ABI Prism 7900HT sequence detection system (Applied Biosystems).
Data Analysis.
Data were expressed as mean ± SEM. One-way ANOVA followed by Bonferroni test was used for statistical comparison among the various treatment groups. Student's t test was also used. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments.
We thank Satomi Takahashi for technical support and Wendy Gray for the critical reading of the manuscript. This study was supported in part by a grant from Exploratory Research for Advanced Technology from the Japan Science and Technology Agency and Program, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811844106/DCSupplemental.
References
- 1.Nomura H, Nielsen BW, Matsushima K. Molecular cloning of cDNAs encoding a LD78 receptor and putative leukocyte chemotactic peptide receptors. Int Immunol. 1993;5:1239–1249. doi: 10.1093/intimm/5.10.1239. [DOI] [PubMed] [Google Scholar]
- 2.Wise A, et al. Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem. 2003;278:9869–9874. doi: 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- 3.Carlson LA. Studies on the effect of nicotinic acid on catecholamine stimulated lipolysis in adipose tissue in vitro. Acta Med Scand. 1963;173:719–722. doi: 10.1111/j.0954-6820.1963.tb17457.x. [DOI] [PubMed] [Google Scholar]
- 4.Aktories K, Jakobs KH, Schultz G. Nicotinic acid inhibits adipocyte adenylate cyclase in a hormone-like manner. FEBS Lett. 1980;115:11–14. doi: 10.1016/0014-5793(80)80715-0. [DOI] [PubMed] [Google Scholar]
- 5.Aktories K, Schultz G, Jakobs KH. Regulation of adenylate cyclase activity in hamster adipocytes. Inhibition by prostaglandins, alpha-adrenergic agonists and nicotinic acid. Naunyn Schmiedebergs Arch Pharmacol. 1980;312:167–173. doi: 10.1007/BF00569726. [DOI] [PubMed] [Google Scholar]
- 6.Aktories K, Schultz G, Jakobs KH. Inactivation of the guanine nucleotide regulatory site mediating inhibition of the adenylate cyclase in hamster adipocytes. Naunyn Schmiedebergs Arch Pharmacol. 1982;321:247–252. doi: 10.1007/BF00498508. [DOI] [PubMed] [Google Scholar]
- 7.Tunaru S, et al. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- 8.Lee DK, et al. Discovery and mapping of ten novel G protein-coupled receptor genes. Gene. 2001;275:83–91. doi: 10.1016/s0378-1119(01)00651-5. [DOI] [PubMed] [Google Scholar]
- 9.Soga T, et al. Molecular identification of nicotinic acid receptor. Biochem Biophys Res Commun. 2003;303:364–369. doi: 10.1016/s0006-291x(03)00342-5. [DOI] [PubMed] [Google Scholar]
- 10.Jilek A, et al. Biosynthesis of a D-amino acid in peptide linkage by an enzyme from frog skin secretions. Proc Natl Acad Sci USA. 2005;102:4235–4239. doi: 10.1073/pnas.0500789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosoi T, et al. Identification of a novel human eicosanoid receptor coupled to G(i/o) J Biol Chem. 2002;277:31459–31465. doi: 10.1074/jbc.M203194200. [DOI] [PubMed] [Google Scholar]
- 12.O'Flaherty JT, Taylor JS, Thomas MJ. Receptors for the 5-oxo class of eicosanoids in neutrophils. J Biol Chem. 1998;273:32535–32541. doi: 10.1074/jbc.273.49.32535. [DOI] [PubMed] [Google Scholar]
- 13.Schwenk U, Schroder JM. 5-Oxo-eicosanoids are potent eosinophil chemotactic factors. Functional characterization and structural requirements. J Biol Chem. 1995;270:15029–15036. doi: 10.1074/jbc.270.25.15029. [DOI] [PubMed] [Google Scholar]
- 14.Le Poul E, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Y, et al. Endothelin-1 stimulates leptin production in adipocytes. J Biol Chem. 2001;276:28471–28477. doi: 10.1074/jbc.M103478200. [DOI] [PubMed] [Google Scholar]
- 16.Saeki Y, Wataya-Kaneda M, Tanaka K, Kaneda Y. Sustained transgene expression in vitro and in vivo using an Epstein-Barr virus replicon vector system combined with HVJ liposomes. Gene Ther. 1998;5:1031–1037. doi: 10.1038/sj.gt.3300711. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, et al. Characterization of the superoxide-generating system in human peripheral lymphocytes and lymphoid cell lines. J Biochem. 1995;117:758–765. doi: 10.1093/oxfordjournals.jbchem.a124773. [DOI] [PubMed] [Google Scholar]
- 18.Kanegasaki S, et al. A novel optical assay system for the quantitative measurement of chemotaxis. J Immunol Methods. 2003;282:1–11. doi: 10.1016/j.jim.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Deshayes S, et al. On the mechanism of non-endosomal peptide-mediated cellular delivery of nucleic acids. Biochim Biophys Acta. 2004;1667:141–147. doi: 10.1016/j.bbamem.2004.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.