Abstract
NLRP1 (NLR family, pyrin domain-containing 1) is a contributor to innate immunity involved in intracellular sensing of pathogens, as well as danger signals related to cell injury. NLRP1 is one of the core components of caspase-1-activating platforms termed “inflammasomes,” which are involved in proteolytic processing of interleukin-1β (IL-1β) and in cell death. We previously discovered that anti-apoptotic proteins Bcl-2 and Bcl-XL bind to and inhibit NLRP1 in cells. Using an in vitro reconstituted system employing purified recombinant proteins, we studied the mechanism by which Bcl-2 and Bcl-XL inhibit NLRP1. Bcl-2 and Bcl-XL inhibited caspase-1 activation induced by NLRP1 in a concentration-dependent manner, with Ki ≈ 10 nM. Bcl-2 and Bcl-XL were also determined to inhibit ATP binding to NLRP1, which is required for oligomerization of NLRP1, and Bcl-XL was demonstrated to interfere with NLRP1 oligomerization. Deletion of the flexible loop regions of Bcl-2 and Bcl-XL, which are located between the first and second α-helices of these anti-apoptotic proteins and which were previously shown to be required for binding NLRP1, abrogated ability to inhibit caspase-1 activation, ATP binding and oligomerization of NLRP1. Conversely, synthetic peptides corresponding to the loop region of Bcl-2 were sufficient to potently inhibit NLRP1. These findings thus demonstrate that the loop domain is necessary and sufficient to inhibit NLRP1, providing insights into the mechanism by which anti-apoptotic proteins Bcl-2 and Bcl-XL inhibit NLRP1.
Keywords: apoptosis, innate immunity
NLR-family proteins [NOD-like receptors (NLRs)] are recently recognized components of innate immunity, constituting large families of related proteins that contain a nucleotide-binding oligomerization domain called NACHT and several leucine-rich repeat (LRR) domains involved in pathogen sensing (1, 2). Binding of pathogen-derived molecules to the LRRs is thought to induce conformational changes that allow NACHT-domain mediated oligomerization, thus initiating downstream signaling events, including activation of proteases involved in cytokine processing and activation. In this regard, the NACHT and LRRs are often associated with other domains that allow many of the NLRs to bind directly to pro-caspase-1 (via CARD domains) or indirectly through adaptor proteins (via PYRIN domains). Caspase-1 belongs to the inflammatory group of caspases, which cleave pro-interleukin-1β (IL-1β), pro-IL-18, and pro-IL-33 (3) in the cytosol, thus preparing them for secretion. Excessive activation of caspase-1 can also induce cell death, either by apoptosis (4) or by a variant, recently termed “pyroptosis,” especially during host responses to pathogens (for review, see ref. 5).
The human genome contains at least 22 NLR-encoding genes (1). NLRP1 is among the best characterized, with the NLRP1 protein representing the central component of a multiprotein, caspase-1-activating complex termed the “inflammasome” (6). Microbial ligands capable of activating NLRP1 include muramyl dipeptide (MDP), a component of peptidoglycans produced by Gram-positive and Gram-negative bacteria (7, 8). The murine NLRP1b isoform was reported to be crucial for anthrax lethal toxin-mediated macrophage cell killing (9), whereas the human ortholog is involved in tissue injury in the context of UV-irradiated keratinocytes (10, 11). In addition, hereditary polymorphisms in NLR-encoding genes are involved in several autoimmune and inflammatory diseases (12), including NLRP1, which is associated with vitiligo and related autoimmune diseases (13).
Recently, we reported that anti-apoptotic proteins Bcl-2 and Bcl-XL are capable of binding and inhibiting NLRP1, thus suppressing caspase-1 activation and IL-1β production in cultured cells and in mice (8). Structure–function studies demonstrated that an ≈50-aa flexible loop between the first and second α-helices of Bcl-2 and Bcl-XL is critical for their association with NLRP1 in cells. However, the biochemical mechanism by which Bcl-2 and Bcl-XL accomplish their inhibition of NLRP1 is unknown. Using an in vitro reconstituted system employing purified recombinant proteins, we investigated the mechanism of NLRP1 suppression by Bcl-2 and Bcl-XL.
Results
Bcl-2 and Bcl-XL Inhibit NLRP1-Mediated Caspase-1 Activation in Vitro.
Previously, we used the baculovirus expression system to produce and purify NLRP1 and pro-caspase-1 proteins and showed that the combination of MDP and ATP induces NLRP1 oligomerization and caspase-1 activation (7). Using this in vitro reconstituted system, we then tested the effects of recombinant Bcl-2 and Bcl-XL proteins produced in bacteria. In addition, we contrasted these proteins with a mutant of Bcl-XL lacking a “loop” between the first and second α-helices, which we previously determined to be important for binding of Bcl-XL to NLRP1 in cells (8) [see Fig. S1 for depiction of proteins used for these studies]. For these initial experiments, Bcl-2-family proteins and various control proteins were added at 2× molar excess relative to NLRP1, then MDP and ATP were added, and caspase-1 activity was subsequently measured using a fluorigenic peptide substrate. Both Bcl-2 and Bcl-XL inhibited NLRP1-driven caspase-1 activity, whereas Bcl-XLΔLoop, Bid, and GST control proteins did not (Fig. 1A). Consistent with the inhibition by Bcl-2 and Bcl-XL of NLRP1-mediated caspase-1 activity, we observed that MDP/ATP-induced proteolytic processing of pro-caspase-1 to produce the cleaved p10 catalytic subunit (detected by immunoblot analysis of reactions) was also inhibited by Bcl-2 and Bcl-XL (Fig. 1B Upper). Moreover, in vitro proteolytic processing of pro-IL-1β was also inhibited by Bcl-2 and Bcl-XL (Fig. 1B Lower), thus confirming the results for a natural caspase-1 substrate. In contrast, Bcl-XLΔLoop failed to inhibit caspase-1 cleavage and IL-1β processing, consistent with the data above. In accordance with these functional data, recombinant His-6-NLRP1 was found to bind GST-Bcl-2 and GST-Bcl-XL in vitro, as shown by GST-pull down assays, but not GST-Bfl1 (Fig. 1C), a Bcl-2-family member that we previously reported lacks NLRP1 inhibitory activity in cells (8). In addition, deletion of the loop domain from Bcl-XL abolished binding to NLRP1 in vitro (Fig. 1D), as shown by GST pull down assays where GST-NLRP1 was mixed with His-6-Bcl-XL versus His-6-Bcl-XLΔLoop. Taken together, these results show that Bcl-XL mediates inhibition of NLRP1-driven caspase-1 activation through Loop-dependent binding to NLRP1.
Fig. 1.
Bcl-2 and Bcl-XL suppress in vitro reconstituted NLRP1 inflammasome. (A) Reaction mixtures contained His-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP (Alexis) and GST-Bcl-2, or GST-Bcl-XL, or His-Bcl-XLΔLoop, or GST, or Bid (17 nM). Caspase-1 activity was measured after 60 min by hydrolysis of Ac-WEHD-7-amino-4-methylcoumarin (AMC) substrate (20 μM) (Calbiochem), expressing data as mean ± SD, n = 3. RFU, relative fluorescence units. Asterisks indicate P < 0.05. (B) His-6-NLRP1 with or without GST-Bcl-2, GST-Bcl-XL, or His-Bcl-XLΔLoop (molar ratio: 1/5) was incubated 15 min in ice, then with pro-caspase-1 in the presence of 1 mM ATP, 1 mM Mg2+, 20 μg/mL MDP for 30 min at 37 °C with or without recombinant GST-pro-IL-1β (Novus Biologicals). Proteins were separated by SDS/PAGE and then immunoblotted using anti-p10 caspase-1 antibodies (Santa Cruz Biotechnology) (Upper), or anti-IL-1β (cell signaling) (Lower). (C and D) Analysis of NLRP1 binding to proteins by GST pull-down assay. His-6-NLRP1 was incubated with GST-Bcl-2, GST-Bcl-XL, or GST-Bfl-1 fusion proteins (C), or GST-NLRP1 was incubated with His-6-Bcl-XL versus His-6-Bcl-XLΔloop (D). Proteins associated with glutathione-Sepharose were analyzed by protein staining using Sypro Ruby. Note that gel mobility of GST-NLRP1 was reproducibly altered in reactions containing Bcl-XL, suggesting perhaps induction of a SDS-resistant conformational change in the protein.
Next, we determined the concentration dependence of NLRP1 inhibition by Bcl-2 and Bcl-XL, observing that suppression of NRLP1-induced caspase-1 activity follows a hyperbolic curve to reach essentially complete inhibition at concentrations of Bcl-2 or Bcl-XL that are 2× the concentration of NLRP1 (Fig. 2A). To exclude a direct effect of Bcl-2 and Bcl-XL on caspase-1, as opposed to NLRP1, we tested the effects of these anti-apoptotic proteins on fully active, proteolytically processed caspase-1 comprised of p20 and p10 catalytic subunits purified from bacteria (Fig. S1). When added at 2-fold molar excess relative to caspase-1, neither Bcl-2 nor Bcl-XL suppressed caspase-1 activity (Fig. 2B). In contrast, caspase-1 was completely inhibited by a peptide inhibitor, Ac-Trp-Glu-His-Asp-aldehyde (Ac-WEHD-CHO) (Fig. 2B).
Fig. 2.
Bcl-2 and Bcl-XL suppress caspase-1 activation induced by NLRP1 but not caspase-1. (A) Reactions contained His-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP and various concentrations of recombinant GST-Bcl-2 (black symbols) or GST-Bcl-XL (white symbols). Caspase-1 activity was measured after 60 min by hydrolysis of Ac-WEHD-AMC substrate (20 μM), expressing data as mean ± SD, n = 3. (B) Purified active p10/p20 caspase-1 [5 nM (black bars) or 0.625 nM (white bars) (Gift from G. Salvesen/Burnham Institute for Medical Research)] in presence or absence of GST-Bcl-2, or GST-Bcl-XL, or Ac-WEHD-CHO was incubated with 10 mM DTT 10 min at 37 °C. Caspase-1 activity was measured after 60 min by hydrolysis of Ac-WEHD-AMC substrate (20 μM), expressing data as mean ± SD, n = 3. Note that the lower concentration (0.625 nM) of active caspase-1 was used to achieve enzyme activity levels comparable to those obtained in reactions where NLPR1 was used to activate pro-caspase-1, thus achieving similar levels of protease activity.
Bcl-2 and Bcl-XL Inhibit ATP Binding to NLRP1 and Suppress Oligomerization.
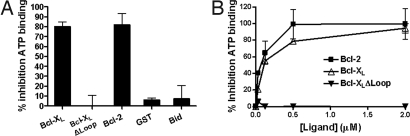
Previously, using the reconstituted NLRP1 inflammasome (7), we described that caspase-1 is activated after a 2-step process, in which MDP binding to NLRP1 first induces a conformational change of the protein, which is secondarily followed by ATP binding to mediate NLRP1 oligomerization (7). Upon oligomerization of NLRP1, caspase-1 monomers associate with NLRP1 oligomers, which results in protease activation, presumably via an induced dimerization mechanism (14). Among these molecular events, the binding of ATP to MDP-primed NLRP1 is readily measured by fluorescence polarization assay (FPA), using fluorescein isothiocyanate (FITC)-conjugated ATP (7). We therefore measured FITC-ATP binding to recombinant NLPR1 by FPA in presence and absence of Bcl-2-family proteins. The portion of specific binding of FITC-ATP to NLRP1 was determined by competition with unlabeled ATP, and data were then normalized to exclude the non-specific binding component (Fig. S2). Addition of Bcl-2 or Bcl-XL at 8-fold molar excess relative to NLRP1 reduced ATP binding by ≈80%, whereas control proteins GST, Bid, and Bcl-XLΔLoop had negligible effects (Fig. 3A). Moreover, titrating various amounts of Bcl-2 or Bcl-XL proteins into reactions showed that suppression of FITC-ATP binding to NLRP1 by Bcl-2 and Bcl-XL was concentration-dependent and saturable (Fig. 3B), with half maximal inhibition obtained at a molar ratio of ≈1 for both Bcl-2: NLRP1 and Bcl-XL:NLRP1. In contrast, Bcl-XLΔLoop failed to inhibit ATP binding at all concentrations tested.
Fig. 3.
Bcl-2 and Bcl-XL inhibit ATP binding to NLRP1. (A) His-6-NLRP1 (0.125 μM) was incubated for 15 min in ice in the presence of GST-Bcl-XL, His-Bcl-XLΔLoop, GST-Bcl-2, GST, or Bid (1 μM). The mixture was then incubated for additional 15 min in ice with 1 μM MDP-LD, FITC-conjugated ATP analog (10 nM) and Mg2+ (0.5 mM). ATP binding was analyzed by FPA (n = 3), measuring milliPolars (mP), and the percentage of inhibition was determined vs. NLRP1 incubated only with MDP-LD (mean ± SD). (B) His-6-NLRP1 (0.1 μM) was incubated for 15 min in ice with various amounts of GST-Bcl-2 (black squares), GST-Bcl-XL (white triangles), or GST-Bcl-XLΔLoop (black triangles) and then incubated 15 min on ice with 1 μM MDP-LD, FITC-conjugated ATP analog (10 nM) and Mg2+ (0.5 mM). ATP binding to NLRP1 was analyzed by FPA (n = 3), measuring milliPolars (mP), and the percentage of inhibition calculated (mean ± SD). Percentage inhibition values were derived by subtracting non-specific FITC-ATP binding, based on experiments where an excess of unlabeled ATP (100 nM) was applied to determine the competitive component of FITC-ATP binding (Fig. S2).
Since Bcl-2 and Bcl-XL inhibit ATP binding to NLRP1, we predicted they would inhibit NLRP1 oligomerization. Oligomerization of NLRP1 induced by MDP and ATP can be monitored using 2D gel-electrophoresis analysis, in which the first dimension is non-denaturing and thus preserves macromolecular complexes during electrophoresis and the second dimension contains SDS for subsequent identification of proteins based on their known molecular mass (7). For these experiments, NLRP1 monomers were incubated with various proteins (GST, GST-Bcl-XL, His-6-Bcl-XLΔLoop) at ≈10-fold molar excess, then MDP and ATP were added to induce oligomerization (Fig. 4). In the absence of other proteins, NLRP1 migrated in the ≈150- to 450-kDa range before addition of MDP/ATP. Addition of MDP/ATP caused much of the NLRP1 to shift to a larger complex (≥1 MDa) (Fig. 4), including material that failed to enter gels, and this oligomerization was not affected by addition of GST control protein (Fig. 4). Adding GST-Bcl-XL suppressed oligomerization of NLRP1. Moreover, GST-Bcl-XL protein was found comigrating with non-oligomerized NLRP1 (range ≈150–450 kDa), consistent with the ability of Bcl-XL to bind NLRP1. In contrast, Bcl-XLΔLoop did not inhibit formation of NLRP1 oligomers, and this protein did not comigrate with nonoligomerized NLRP1 (Fig. 4). Altogether, these data suggest that Bcl-XL interacts with unoligomerized NLRP1 to inhibit ATP binding and to suppress oligomerization.
Fig. 4.
Bcl-XL inhibits oligomerization of NLRP1. Purified His-6-NLRP1 monomers obtained from gel filtration were incubated 15 min in ice with or without GST-Bcl-XL, His-Bcl-XLΔLoop, or GST (1/10 molar ratio). The mixture was then incubated without [no treatment (NT)] or with Mg2+ (1 mM), ATP (1 mM) and MDP-LD (20 μg/mL) for 30 min at 37 °C. Proteins were separated by a first native-PAGE dimension, then by a second denaturing SDS/PAGE dimension, and stained using Sypro-Ruby (7). Arrows at bottom indicate positions of NLRP1 monomers and oligomers.
Loop Region of Bcl-2 Is Sufficient to Inhibit NLRP1.
We previously determined that the “loop” domains of Bcl-2 and Bcl-XL are required for binding to and suppression of NLRP1 in cells (8), and extended these observations here to purified proteins. While necessary, it was unknown whether the loop region is sufficient for suppressing NLRP1. To study the effect of the loop domain in isolation, we chemically synthesized peptides corresponding to the loop region and other segments of Bcl-2 (Fig. S3) and tested their effects in vitro on the reconstituted NLRP1 inflammasome when added at a 6-fold molar excess relative to NLRP1. Addition of Bcl-2 loop peptide (residues 35–83) inhibited NLRP1-mediated caspase-1 activation to nearly baseline levels (Fig. 5A). In contrast, this 35–83 Bcl-2 loop peptide did not directly inhibit caspase-1, whereas the peptidyl inhibitor Ac-WEHD-CHO potently suppressed the activity of this protease (Fig. 5B).
Fig. 5.
Bcl-2 Loop peptide inhibits NLRP1. (A) Reactions contained His-6-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP, and Bcl-2 Loop peptide 35–83 (50 nM). Asterisk indicates P < 0.05. (B) Purified active p10/p20 caspase-1 (5 nM or 0.625 nM) was incubated with or without Bcl-2 Loop peptide 35–83 or Ac-WEHD-CHO (10 nM). (C) Reactions contained His-6-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP, and various 20-mer Bcl-2 Loop peptides (50 nM), or Ac-WEHD-CHO (10 nM). Asterisks indicate P < 0.05. (D) Reactions contained His-6-NLRP1ΔLRR (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, with (black bars) or without (white bars) 0.1 μg/mL MDP, and with or without Bcl-2 Loop peptide 35–83, 71–90 peptide (50 nM), or Ac-WEHD-CHO (10 nM). (E) Reactions contained His-6-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP, and various amounts of Bcl-2 Loop peptide 35–83, 71–90 Loop peptide, or BH4 peptide 9–30. For all experiments, caspase-1 activity was measured after 60 min by hydrolysis of Ac-WEHD-AMC substrate (20 μM), expressing data as mean ± SD, n = 3.
To characterize the sequence of the Bcl-2 loop responsible for inhibiting NLRP1, we synthesized a series of overlapping 20-mer peptides corresponding to the 35–83 region (Fig. S3) and tested their effects on NLRP1-mediated caspase-1 activation. These experiments showed that a peptide corresponding to residues 71–90 of Bcl-2 is sufficient to potently inhibit NLRP1, whereas other 20-mer peptides corresponding to 31–50, 41–60, 51–70, and 61–80 are inactive (Fig. 5C). The effects of the full-loop peptide (35–83) and the active segment (71–90) were specific, as determined by experiments using a NLRP1 mutant lacking the LRRs. Unlike full-length NLRP1, the ΔLRR mutant is constitutively active (not requiring MDP) and does not bind Bcl-2 or Bcl-XL (8). As shown in Fig. 5D, neither the 35–83 nor the 71–90 loop peptides suppressed caspase-1 activation by NLRP1ΔLRR, whereas Ac-WEHD-CHO peptide completely neutralized caspase-1 activity. Finally, we compared the concentration dependence of NLRP1 inhibition by the 35–83 and 71–90 loop peptides. Both peptides demonstrated similar concentration-dependent inhibition, with sigmoidal curves, reducing caspase-1 activity to background levels (Fig. 5E). The deduced IC50 values (concentrations to inhibit NLRP1-induced caspase-1 activity by 50%) for the full-length 35–83 and 71–90 loop peptides were 11.38 ± 4.51 nM and 9.92 ± 0.86 nM, respectively. In contrast, a control peptide corresponding to residues 9–30 (BH4 domain) had negligible activity in this assay. These results indicate that residues 71–90 of Bcl-2 loop region are sufficient to inhibit NLRP1.
Next, we tested still shorter peptides within the region 71–90 peptide, examining the NLRP1 inhibitory activity of a series of 5- and 10-mers (Fig. 6A). We observed that 10-mer 71–80 effectively inhibited NLRP1, while the 10-mer 81–90 did not. The 5-mer corresponding to the 71–80 peptide (71–75) was less potent. Furthermore, the importance of the 71–75 segment of the loop was suggested by comparison of a 76–90 peptide lacking this region with peptides 71–90 and 71–80, which contained this sequence (Fig. 6A).
Fig. 6.
Enzymology of NLRP1 inflammasome inhibition by Bcl-2, Bcl-XL, and Bcl-2 Loop peptide 71–80. (A) Caspase-1 activity induced by stimulation of NLRP1 with MDP and ATP was measured in the presence or absence of various peptides. Reactions contained His-6-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP, and various 10-mer or 5-mer Bcl-2 Loop peptides (50 nM). Asterisk indicates P < 0.05. (B) ATP binding by NLRP1 was measured in presence or absence of peptides. Reactions contained His-6-NLRP1 (0.125 μM), GST-Bcl-2 protein or Bcl-2 loop peptides (2 or 20 μM), 1 μM MDP[LD], FITC-ATP analog (10 nM), and Mg2+ (0.5 mM). FITC-ATP binding was analyzed by FPA (n = 3), measuring milliPolars (mP), and values corrected for non-specific FITC-ATP binding as determined by competition with excess unlabeled ATP. The percentage inhibition was determined compared to NLRP1 incubated only with MDP-LD (mean ± SD). (C and D) Enzyme kinetics analysis of NLRP1 inhibition. Reactions contained His-6-NLRP1 (8.5 nM), pro-caspase-1 (8.5 nM), 0.25 mM ATP, 0.5 mM Mg2+, 0.1 μg/mL MDP, with or without 2 nM GST-Bcl-2, or GST-Bcl-XL, or (D) 2 nM Bcl-2 loop peptides 31–50 or 71–80. Caspase-1 activity was measured after 60 min by hydrolysis of various concentrations of Ac-WEHD-AMC substrate, expressing data as mean ± SD, n = 3. Experimental data were analyzed by linear regression to fit the Lineweaver–Burk equation.
Consistent with the ability of the 71–80 peptide to suppress NLRP1-driven caspase-1 activity, this peptide also inhibited NLRP1 binding to ATP, with results obtained for the Bcl-2 Loop peptides 71–90 and 71–80 comparable to recombinant Bcl-2 protein and full-length Bcl-2 Loop peptide 35–83 (Fig. 6B). In contrast, inactive Bcl-2 peptide 31–50 suppressed neither caspase-1 activation nor ATP binding to NLRP1.
Bcl-2, Bcl-XL, and Bcl-2 peptide 71–80 Are Competitive Inhibitors of NLRP1.
We explored the mechanism of NLRP1 inhibition by Bcl-2 and Bcl-XL using classical enzyme kinetics analysis. First, we determined the concentration dependence of NLRP1 inhibition by Bcl-2 and Bcl-XL, observing that suppression of NRLP1-induced caspase-1 activity follows a hyperbolic curve to reach essentially complete inhibition at concentrations of Bcl-2 or Bcl-XL that are 2× the concentration of NLRP1 (Fig. 2A). Second, we performed a Lineweaver–Burk double-reciprocal transformation of the data, observing that addition of Bcl-2 or Bcl-XL (Fig. 6C), or Bcl-2 Loop peptide 71–80 (Fig. 6D) changed the x axis intercept (−1/Km) but not the y axis intercept (1/Vmax), thus suggesting a competitive mechanism of inhibition. Of note, the affinity (Km) of caspase-1 for its substrate Ac-WEHD-AMC was reduced 2-fold in presence of Bcl-2, Bcl-XL, or 71–80 peptide compared to control (5.04 ± 0.5 μM for Bcl-2, 6.05 ± 0.24 μM for Bcl-XL, and 5.62 ± 1.58 μM for 71–80 Bcl-2 peptide versus 2.62 ± 0.35 μM without inhibitor, see Table 1). From these values, we also calculated the inhibitory constants (Ki) for Bcl-2, Bcl-XL, and 71–80 peptide suppression of NLRP1 as 13.5 ± 1.35 nM, 9.57 ± 0.38 nM, and 10.9 ± 3.07, respectively (Table 1). Thus, Bcl-2, Bcl-XL, and the 71–80 Bcl-2 peptide are potent (low nanomolar) inhibitors of NLRP1 with respect to its ability to activate caspase-1.
Table 1.
Kinetic parameters of NLRP1 inflammasome inhibition by Bcl-2, Bcl-XL, and Bcl-2 Loop peptide 71–80
| Parameter | No inhibitor | GST-Bcl-2 | GST-Bcl-XL | Peptide 71–80 |
|---|---|---|---|---|
| 1/Kmapp, μM−1 | 0.38 ± 0.05 | 0.19 ± 0.02 | 0.16 ± 0.01 | 0.18 ± 0.05 |
| Kmapp, μM | 2.62 ± 0.35 | 5.04 ± 0.50 | 6.05 ± 0.24 | 5.62 ± 1.58 |
| Ki, nM | — | 13.5 ± 1.35 | 9.57 ± 0.38 | 10.9 ± 3.07 |
Parameters are obtained from the data presented in Fig. 6 using the double-reciprocal Lineweaver-Burk equation (mean ± SD; n = 3).
Discussion
We demonstrate here that Bcl-2 and Bcl-XL directly and potently inhibit NLRP1. These studies were performed using purified recombinant proteins, thus eliminating the contributions of other factors, and unambiguously demonstrating a direct inhibitory effect of Bcl-2 and Bcl-XL on NLRP1. The Ki values for Bcl-2 and Bcl-XL were ≈10 nM, suggesting physiologically relevant suppression. By comparison, the Ki for inhibition of caspase-9 by XIAP is ≈30 nM (15), showing that Bcl-2 and Bcl-XL suppress their target (NLRP1) with similar potency as this well-known caspase inhibitor. Furthermore, the concentration-dependent suppression of NLRP1 by Bcl-2 and Bcl-XL suggests a 1:2 molar complex. The mechanism of Bcl-2- and Bcl-XL-mediated suppression of NLRP1 is competitive, as shown by Lineweaver–Burk analysis. Because competitive inhibitors reduce the affinity of enzymes for their substrates, we interpret this result to mean that in the presence of Bcl-2-family proteins, the active site of caspase-1 does not form, thus effectively impairing the affinity (Km) of caspase-1 for its substrate (Ac-WEHD-AMC). In this regard, we observed that Bcl-2 and Bcl-XL inhibit ATP binding by MDP-activated NLRP1, an event required for oligomerization of NLRP1, and we documented by nondenaturing gel electrophoresis experiments that Bcl-XL suppresses oligomerization of NLRP1. Thus, we surmise that the active site of caspase-1 does not form because Bcl-2 and Bcl-XL prevent NLRP1 oligomerization, and thus caspase-1 molecules are not brought into close apposition for activation. This interpretation is consistent with evidence that initiator caspases such as caspase-8 and -9 require assisted dimerization to induce conformational changes that establish the functionally active site of these enzymes (16).
Because Bcl-2 and Bcl-XL inhibit NLRP1 even when its activating ligand MDP is provided, it seems likely that 2 events may be required to achieve NLRP1 activation in those cell-types where Bcl-2 or Bcl-XL is playing an inhibitory role. First, repression by Bcl-2 and Bcl-XL must be relieved. Second, once freed of Bcl-2 and Bcl-XL, NLRP1 must be activated by MDP or functionally equivalent ligands. How Bcl-2 and Bcl-XL are dissociated from NLRP1 in cellular contexts remains to be clarified. Previous domain mapping experiments showed that the NACHT and LRRs of NLRP1 are required for Bcl-XL binding (8). Thus, posttranslation modifications, changes in protein conformation, and interaction of other proteins or of small non-protein ligands with the NACHT-LRR region of NRLP1 may free this NLR-family member from Bcl-2 and Bcl-XL. Alternatively, control of dissociation may occur at the level of Bcl-2 and Bcl-XL, affecting directly or indirectly the interacting loop domain (see below). Regardless, requirement for 2 steps—removal of repression (Bcl-2/Bcl-XL) and addition of activating ligand (MDP)—would help to ensure that NLRP1 activation occurs only in appropriate cellular contexts, thus reducing chances for dysregulated production of IL-1β and other pro-inflammatory cytokines activated by caspase-1.
We extended our prior studies, in which the loop regions of Bcl-2 and Bcl-XL were shown, to be required for association with and suppression of NLRP1 in cells (8), by showing here that deletion of the loop from purified recombinant Bcl-XL eliminates suppression of NLRP1 in vitro. Bcl-XLΔloop also failed to inhibit ATP binding and oligomerization of MDP-activated NLRP1, unlike intact Bcl-XL. Moreover, in this report, we also demonstrated that the loop of Bcl-2 is sufficient to inhibit NLRP1, based on experiments using synthetic peptides corresponding to this region of Bcl-2. Previously, it was unclear whether the loop domain directly inhibits NLRP1, versus the possibility that it affects the overall conformation of Bcl-2 and Bcl-XL in a manner such that the loop is required to generate conformational states that are competent to bind NLRP1. The data provided here argue that the loop directly inhibits NLRP1. Moreover, we observed that a synthetic 10-mer loop peptide corresponding to residues 71–80 is sufficient to suppress NLRP1-mediated caspase-1 activation. This peptide binds to NLRP1 with a high affinity (IC50 ≈ 10 nM), which is comparable to full-length loop peptide 35–83 (IC50 ≈ 12 nM), and to the full-length Bcl-2 and Bcl-XL proteins (Ki ≈ 10 nM). Thus, the loop appears to contain a peptidyl-ligand for NLRP1 that suppresses its activation. In this regard, the loop regions of Bcl-2 and Bcl-XL are unstructured in the available 3D structures of these proteins (17), but when bound to NLRP1, segments of the loop may develop secondary structure or constrained structures that adapt to the relevant interacting surface of NLRP1. Comparisons of the sequences of the 71–80 loop region of human Bcl-2 with other species reveals strong conservation in primates, with partial conservation in lower mammals, comprising the motif P-L-X-T-P-A-A-P-(X)1–3-A (Fig. S4). Conservation of this motif is also found in the loop of Bcl-XL .
Given the striking species-specific differences among NLR-family proteins within vertebrates, we speculate that the peptidyl motifs of Bcl-2-family proteins responsible for binding specific NLRs may vary to accommodate those differences. Alternatively, suppression of NLRs by Bcl-2 family proteins may be a recent evolutionary adaptation, although this explanation seems unlikely given the prior precedent set by C. elegans where Bcl-2 homolog CED-9 binds the caspase activator CED-4. Interestingly, the loops of Bcl-2 and Bcl-XL are subject to posttranslational modifications, which to date include phosphorylation, deamidation, and proteolytic cleavage (18–20). It will be important therefore in future studies to explore the functional consequences of these posttranslational modifications on the interaction of NLRP1 with Bcl-2 and Bcl-XL. Such modifications, for example, might contribute to mechanisms for releasing NLRP1 to allow subsequent caspase-1 activation. Also meriting note, somatic mutations have been reported in the loop region of Bcl-2 in lymphomas where the gene that encodes this anti-apoptotic protein is involved in chromosomal translocations with the Ig heavy-chain gene locus (21). Thus, it may also be interesting to evaluate the effects of such mutations on Bcl-2/NLRP1 interactions.
In summary, we provide insights into the molecular mechanism by which Bcl-2 and Bcl-XL suppress the NLRP1 inflammasome. Moreover, the demonstration that a peptide derived from the loop of Bcl-2 is sufficient to recapitulate the inhibitory activity of the intact protein raises the possibilities of developing therapeutic cell-permeable peptides for suppressing NLRP1 activity in inflammatory diseases and of generating high-throughput screening (HTS) assays using these peptides for identification of chemical compounds that occupy the same peptide binding site on NLRP1 and that mimic the inhibitory activity of the Bcl-2 loop peptide. Such agents may find utility for preventing NLRP1-mediated activation of pro-inflammatory cytokines and for suppressing NLRP1-induced cell death in pathological conditions.
Experimental Procedures
Protein Production and Purification.
His-6-tagged recombinant proteins, (His)6-NRLP1, (His)6-NRLP1ΔLRR, and (His)6-pro-caspase-1 were produced from Sf9 cells using recombinant baculoviruses, as previously described (7). Proteins in complex with Ni2+-Sepharose (2 mL for 2 L of culture) were washed with 300 mL of lysis buffer, 1 M NaCl, and 20 mM Imidazole. Then, proteins were eluted using an imidazole gradient (0–250 mM) and the eluted proteins were submitted to gel-filtration using Superdex 200 columns in 20 mM Hepes-KOH pH 7.5, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 150 mM NaCl, 10 mM KCl, 0.1% CHAPS, supplemented with 1 mM DTT for NLRP1 but not for pro-caspase-1. Fractions containing monomers were detected by UV absorbance at 280 nm, concentrated, and quickly frozen in dry ice/ethanol. Fraction size ≈150 kDa (His-6-NLRP1), ≈50 kDa (His-6-pro-caspase-1) were retained corresponding to monomers. Note, however, that some caspase-1 dimerization occurs in these preparations over time and results in some background protease activity (7). For expression and purification of Bcl-2 family proteins, GST-fusion proteins containing Bcl-XL, Bcl-2, and Bcl-XLΔLoop (Δ44–84) lacking their C-terminal transmembrane domains (≈last 20 aa) were expressed from pGEX4T-1 plasmid in XL-1 Blue cells (Stratagene, Inc.). GST-fusion proteins were purified by affinity chromatography using glutathione-Sepharose as described in ref. 22.
Caspase Activity Measurements.
Recombinant proteins NLRP1 and pro-caspase-1 were diluted in 20 mM Hepes pH 7.5, 150 mM NaCl, 0.1% CHAPS, 1 mM DTT, maintaining total protein constant with bovine serum albumin (BSA). Proteins were mixed in caspase-1 buffer (100 mM Hepes, pH 7.5, 10% Sucrose, 0.1% CHAPS, 1 mM DTT) in a final volume of 10 μL in 96-well plates. Various activators were added (ATP, Mg2+, MDP-LD), adjusting final volume with water. Samples were then incubated for 30 min at 37 °C and caspase-1 activity was measured using 20 μM fluorometric substrate Ac-WEHD-AMC (Calbiochem). Release of AMC product was monitored continuously by spectrofluorometry in kinetic mode over 1 h at 25–27 °C. Rates of catalysis were calculated using the initial slope and caspase activity was expressed as the change in fluorescence over time derived from the linear phase of reactions. Data were analyzed using the PRISM Statistics software, employing an unpaired t test.
Kinetic Parameters of NLRP1 Inflammasome Inhibition.
Apparent Km and Vmax were determined with or without GST-Bcl-2, GST-Bcl-XL, 71–80 or 31–50 peptides. Aliquots containing 8.5 nM NLRP1 and 8.5 nM pro-caspase-1 were incubated in caspase-1 buffer for 30 min at 37 °C in the presence of activators (MDP-LD, ATP, and Mg2+) in a total volume of 10 μL. Reaction mixtures were subsequently assayed with various concentrations of Ac-WEHD-AMC substrate in 96-well plates. Km and Vmax were determined using a nonlinear regression method to fit the Michaelis–Menten equation: V = (Vmax − V0)[S]/(Km + [S]) + V0, where V = initial catalytic rate, in nanomolar AMC per min; [S] = substrate concentration in nanomolar or micromolar; V0 = limiting value of V without [S]; Vmax = limiting value of V at saturating [S]. Competitive inhibition was determined after linearization of the Michaelis–Menten equation, using the double-reciprocal Lineweaver–Burk equation: 1/V = Km/Vmax[S] + 1/Vmax. Equilibrium constants of inhibition (Ki) were determined based on the competitive inhibition equation: KmI = Km (1 + [I]/Ki), where KmI = Km in presence of inhibitor; and [I] = concentration of inhibitor. IC50 values for loop peptides were estimated using a fitting procedure and the sigmoidal curve: V = V0 + (Vmax − V0)/(1 + 10 exp(log IC50− [I]))n, where n = Hill slope. Fitting procedures were performed using PRISM software.
Fluorescence Polarization Assays.
NLRP1 or NLRP1ΔLRR were incubated with or without ligand in ice for 10 min, and then with 10 nM fluorescein-conjugated synthetic purified ATP in 20 mM Hepes pH 7.5, 150 mM NaCl, 0.1% CHAPS, 1 mM DTT for 5 min at 4 °C. Fluorescence polarization was measured using an Analyst AD Assay Detection System (LJL Biosystem).
Statistical Analysis.
Most data were presented as the mean ± SD from at least 3 independent experiments. Statistical comparisons between different treatments were performed by unpaired t test, where P ≤ 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We thank M. Hanaii and T. Siegfried for manuscript preparation. This work was supported by National Institutes of Health Grants AI-056324, AI035789, and CA-69381, the Crohn's and Colitis Foundation of America, and the Philippe Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809414106/DCSupplemental.
References
- 1.Ting JP, et al. The NLR gene family: A standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariathasan S, Monack DM. Inflammasome adaptors and sensors: Intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS. Caspases and apoptosis. Essays Biochem. 2002;38:9–19. doi: 10.1042/bse0380009. [DOI] [PubMed] [Google Scholar]
- 4.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 5.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 6.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 7.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 8.Bruey JM, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 9.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 10.Feldmeyer L, et al. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 11.Faustin B, Reed JC. Sunburned skin activates inflammasomes. Trends Cell Biol. 2008;18:4–8. doi: 10.1016/j.tcb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: Role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 13.Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 14.Pop SM, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22(2):269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Deveraux QL, et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 17.Minn AJ, et al. Bcl-xL forms an ion channel in synthetic lipid membranes. Nature. 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 18.Cheng E, et al. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ask1/jun n-terminal protein kinase pathway normally activated at G2/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojala PM, Yamamoto K, Korsmeyer SJ, Makela TP. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates BCL-2. Nat Cell Biol. 2000;2:819–825. doi: 10.1038/35041064. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S, Louie DC, Kant JA, Reed JC. Frequent somatic mutations in translocated BcL2 genes of non-Hodgkin's lymphoma patients. Blood. 1992;79:229–237. [PubMed] [Google Scholar]
- 22.Zhai D, Jin C, Satterthwait AC, Reed JC. Comparison of chemical inhibitors of anti-apoptotic Bcl-2-family proteins. Cell Death Differ. 2006;13:1419–1421. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
- 23.Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.