Abstract
Poly(ADP-ribos)ylation is one of the longest-known but most enigmatic posttranslational modifications transducing specific signals. The enzyme responsible for the majority of poly(ADP-ribose) polymerization in cells, PARP-1, promotes DNA repair but also mediates a caspase-independent form of apoptosis in response to stressors such as irradiation. However, the biologic function of most other PARPs is not known. Macro-PARPs constitute one branch of the large family of PARP-like proteins also designated as B aggressive lymphoma proteins (BAL1, 2a/2b, 3, or PARP-9, PARP-14, and PARP-15). To elucidate biologic role(s) of a BAL-family macro-PARP, we analyzed mice deficient in PARP-14, a binding partner of the IL-4–induced transcription factor Stat6. We show here that PARP-14 plays a fundamental role mediating protection against apoptosis in IL-4–treated B cells, including that after DNA damage, and mediates IL-4 effects on the levels of gene products that regulate cell survival, proliferation, and lymphomagenesis. Collectively, the results establish that PARP-14 mediates regulation of gene expression and lymphocyte physiology by IL-4 and has a function distinct from PARP-1. Furthermore, the findings suggest mechanisms by which BAL-family proteins might influence pathologic processes involving B lymphocytes.
Introduction
The cytokine interleukin-4 (IL-4) regulates the differentiation, proliferation, and apoptosis of lymphocytes and most other hematopoietic cells.1,2 IL-4 binding to its receptor initiates Janus kinase-mediated tyrosine phosphorylation of Signal transducer and activator of transcription 6 (Stat6). Stat6 then dimerizes, translocates to the nucleus, and binds to specific DNA sequences at which it regulates gene transcription.1,2 Although these points are established, considerably less is known about transcriptional cofactors that influence Stat6 effects on target gene expression or the impact of such proteins on IL-4 regulation of B-cell physiology.
Aberrant activation of IL-4R signaling is frequently associated with the pathophysiology of leukemia and lymphoma cells in that IL-4 receptor signaling is constitutively activated in many lymphomas, and the levels of IL-4 and IL-4 target genes are high in such hematopoietic malignancies.3–8 For instance, FIG1/IL-4I1 (IL-4-induced gene 1) is activated in primary mediastinal large B-cell lymphoma,5 and target genes such as Bcl-6 (B-cell lymphoma 6) and HGAL (human germinal center-associated lymphoma) are highly expressed in germinal center B-type diffuse large B-cell lymphoma (DLBCL), the most common non-Hodgkin lymphoma.7 Much remains unknown about DLBCL pathophysiology, but DLBCLs are often associated with dysregulated apoptosis or defective DNA repair, and the IL-4–regulated transcriptional repressor Bcl-6 is strongly implicated in pathogenesis for some subsets.7,9
In addition to these processes, a protein designated as B aggressive lymphoma (BAL) was identified by expression screening for high-risk factors in DLBCL patients. BAL is expressed at significantly higher levels in more aggressive DLBCL than in low-risk tumors.10 Although it was suggested that BAL might promote malignant B-cell migration, the function and regulatory mechanisms of BAL family members are unclear. More recently, BAL protein family members have been assigned to one branch of a family of (poly-ADP ribose) polymerases (PARPs).11 BAL-family proteins encode one or several iterations of a nonhistone “macro” domain present in the histone variant macroH2A followed by C-terminal similarities to PARP catalytic domain.
Enzymatically active PARPs catalyze the transfer of ADP-ribose moieties from NAD+ to target proteins, thereby building negative charged polymers of ADP-ribose.11 This process is counterbalanced by the activity of a PAR glycohydrolase (PARG) that can remove ADP-ribose from target proteins. Use of broad-spectrum PARP inhibitors or inactivation of PARG established that poly(ADP-ribos)ylation (PARylation) plays diverse roles in molecular and cellular processes, including DNA damage detection and repair, modulation of global chromatin condensation, transcription, cell survival, and apoptosis. Protein PARylation is an immediate biochemical response to DNA damage11,12 and a probable role for recombinase-mediated DNA damage in the genesis of B-cell lymphomas has been recognized.13,14
Although originally considered as a single enzymatic function, genomic studies revealed that mammalian cells encode as many as 17 different proteins with a domain homologous to the conserved PARP catalytic region.11 Of these, only PARP-1 has been studied intensively. This enzyme constitutes approximately 70% of PARP activity in cells and accounts for much of the NAD+ depletion and activation of the caspase-independent apoptosis-inducing factor (AIF)–dependent pathways that follow after excessive DNA damage or other physiologic insults.15,16 In contrast to the large body of knowledge about PARP-1, the functions and biologic roles of other PARP families are not clear. In particular, almost nothing is known about functional roles or mechanisms of macro-PARP subfamily members. Recently, we identified a Stat6-interacting protein, Collaborator of Stat6 (CoaSt6),17 later determined to be a macro-PARP subfamily member, PARP-14, and homologous to human BAL2b, a member of the BAL family. Both mouse and human PARP-14 /CoaSt6/ BAL2b encode a triple macro domain and a C-terminal region with sequence homology to PARP; each is highly similar to the original human BAL protein. PARP-14 is expressed in lymphoid organs and lymphocyte cell lines.17 Although expressed equally in the basal state and in IL-4-treated lymphoma cells without apparent change in the fraction of PARP-14 in the nucleus, transfection assays provided evidence that PARP-14 could enhance the transcriptional activity of Stat6 in responses to IL-4 in vitro.17 However, the physiologic functions of PARP-14 and other members of the BAL (macro-PARP) family are not known. We hypothesized that, even though PARP-1 is responsible for most NAD+ conversion into PAR, PARP-14 plays precise roles in signal transduction that are distinct from those of PARP-1. We show here that absence of PARP-14 led to altered proportions of B-cell subsets in the spleen and impairment of the antigen-specific IgA response. In contrast to PARP-1, PARP-14 mediated IL-4–induced B-cell protection against apoptosis after irradiation or growth factor withdrawal. Furthermore, the induction of several B-cell survival factors (Pim-1; Mcl-1) by IL-4 depended on PARP-14.
Methods
Generation of PARP-14–deficient mice
PARP-14–deficient mice were generated from ES cells heterozygous for a disruption of the 5′ end of PARP-14 genomic sequences (Lexicon Genetics, Woodlands, TX) by standard techniques. The resultant chimeras were mated to C57BL/6 females to generate mice heterozygous for the PARP-14 mutation. Germline transmission of the mutated allele was identified and confirmed by polymerase chain reaction (PCR; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and Southern blot analyses of Eco R1-digested genomic DNA probed with a 315-bp 5′ internal fragment. Mice were maintained in microisolators in specific pathogen-free conditions in the Vanderbilt University mouse facility and used at 6 to 8 weeks of age after approved protocols. Studies involving mice were approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Measurement of antibody responses
Mice (age 6 weeks) were immunized (100 μL keyhole limpet hemocyanin [KLH], 100 μg, emulsified with 100 μL incomplete Freund adjuvant given intraperitoneally), boosted in identical manner after 14 days, followed 5 days later by collection of immune sera. Antibodies (Abs) of each isotype were quantitated by enzyme-linked immunosorbent assay (with and without antigen capture) using SBA Clonotyping System (Southern Biotechnology, Birmingham, AL).
Proliferation and S-phase assays
Triplicate samples of splenocytes (2 × 105 cells per 100 μL of media in 96-well plates) were cultured (24 hours; 37°C) in medium, anti-IgM (1 μg/mL), or anti-IgM plus IL-4 (5 ng/mL). Tritiated thymidine (1 μCi in 100 μL of media) was added to each well for the final 8 hours before counting. To measure S-phase entry rates, splenocytes cultured (16 hours) in the presence of anti-IgM or anti-IgM plus IL-4 were processed as detailed in Supplemental Materials.
Flow cytometry and apoptosis assays
Antibodies were from BD Biosciences (San Jose, CA) unless otherwise indicated. Cells were analyzed using fluorochrome-conjugated Abs, flow cytometry with a fluorescence-activated cell sorter (FACS) Calibur (BD Biosciences), and FlowJo software (TreeStar, Ashland, OR) as described.17 For apoptosis assays, cells were cultured with or without IL-4 as indicated, with or without γ irradiation (2-Gy dose from a 137Cs source). TdT-mediated dUTP nick end labeling (TUNEL) assays were performed as described previously and detailed in Document S1. To assay active caspase-3, cells were surface stained for B220, fixed with 4% paraformaldehyde, permeabilized (0.2% saponin, 1% fetal bovine serum in phosphate-buffered saline), and then stained with fluorescein isothiocyanate (FITC)–conjugated Abs specific for active caspase-3 (BD Biosciences), rinsed, and analyzed. For FACS measurement of single-cell pan-caspase activity, cells were incubated with FITC-z-VAD-FMK for 30 minutes at 37°C (BioVision, Mountain View, CA), washed, surface stained, and analyzed.
Immunoprecipitation and PARP assays
ΦNX cells were transfected with pcDNA3 or pcDNA3-FLAG-PARP-14. Lysates prepared as described previously17 were used for immunoprecipitations with monoclonal anti-FLAG (M2) (Sigma-Aldrich, St Louis, MO). Immune complexes, collected using protein G beads (Santa Cruz Biotechnology, Santa Cruz, CA), were rinsed, divided equally, and one eluted portion was analyzed by immunoblotting. The other fraction was incubated (30 minutes at room temperature) in PARP assay buffer (20 μL) containing 5 μCi (0.25 μM) 32P-labeled NAD+ (1000 Ci/mmol; GE Healthcare, Little Chalfont, United Kingdom), 12.5 μM NAD+, 50 mM Tris-Cl (pH 8.0), 4 mM MgCl2, and 0.2 mM dithiothreitol; where indicated, samples contained 1 μM PJ-34 (Calbiochem, San Diego, CA). Beads were washed with phosphate-buffered saline and eluted proteins visualized by autoradiography after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Northern blot and RT-PCR
B cells were purified (∼ 90%-95%) by depleting Thy1+ cells using biotin-anti-Thy1.2 and streptavidin paramagnetic beads (BD IMAG) according to the manufacturer's instructions, plated (5 × 106 cells/mL), and cultured at 37°C in the presence or absence of IL-4. RNA was isolated using TriZol reagent (Invitrogen, Carlsbad, CA). Northern blotting and phosphorimaging (Fuji FLA-2000) for quantitation of radioactive signals were performed as described, as were syntheses of cDNAs for reverse-transcribed PCR (RT-PCR) analyses.17 Specific cDNA templates were amplified either in semiquantitative manner, validated by probing Southern blots of resolved products with radiolabeled internal oligonucleotides (data not shown), or by real-time PCR using SYBR Green and the primer pairs detailed in Table S1.
Western blots
Proteins in whole cell extracts were separated by SDS-PAGE, transferred onto nylon membranes (Millipore, Billerica, MA), and then incubated with rabbit antibodies against PARP-14,17 phospho-Stat6 (Y641; Cell Signaling Technology, Danvers, MA), Stat6 (Santa Cruz Biotechnology), Mcl-1 (Santa Cruz Biotechnology), and Bcl-xL (Santa Cruz Biotechnology); and mouse anti-FLAG (Sigma-Aldrich) or goat anti-actin (Santa Cruz Biotechnology) Abs followed by the appropriate fluorophore-conjugated, species-specific anti-Ig antibodies (Rockland Immunochemicals, Gilbertsville, PA; and LI-COR (Lincoln, NE)). Proteins were visualized and quantitated by laser excitation and infrared imaging (Odyssey, LI-COR). To measure induction of Bcl-2 family members (Bcl-2, Bcl-xL, and Mcl-1), B cells were cultured (20 hours) in complete medium, counted, and lysates of equal numbers of viable cells were analyzed by Western blotting. For measurements of the induction of Stat6 phosphorylation by IL-4, purified B cells were cultured 2 hours in medium alone and then further cultured (0.5 and 12 hours) in the presence or absence of IL-4 (5 ng/mL). Decay experiments included samples induced with IL-4 (30 minutes), then rinsed, cultured further (6 hours), lysed, and analyzed. Confirmatory experiments measured phospho-Stat6 in freshly isolated samples and freshly isolated cells stimulated 30 minutes with IL-4 as for proliferation assays (not shown).
Results
Generation of PARP-14 KO mice
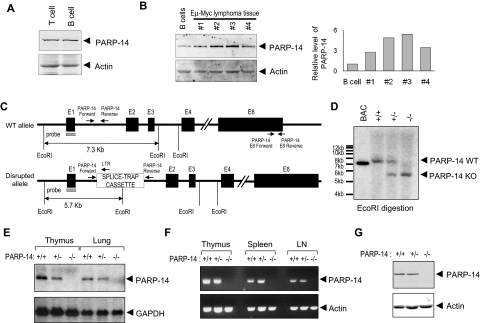
BAL2b/CoaSt6/PARP-14 expression was observed predominantly in lymphoid organs of the mouse.17 This pattern of PARP-14 expression, along with the association of another macro-PARP with human DLBCL pathophysiology, led us to evaluate PARP expression in primary lymphocytes and to compare normal B cells with primary B lymphoma samples. Both B and T cells expressed PARP-14, and levels of this macro-PARP were elevated in Myc-induced lymphoma samples compared with normal B cells (Figure 1A,B). To explore physiologic roles of BAL2b/CoaSt6/PARP-14, we analyzed PARP-14–deficient mice generated by an insertion into the germ line DNA at the 5′ end of the first exon of the PARP-14 locus (Figure 1C). Southern blotting was used to validate PCR evidence of germ line transmission (Figure 1D). PARP-14−/− mice homozygous for the disrupted allele were born in Mendelian ratios, and there were no gross defects in growth or survival. Using Northern blotting and RT-PCR, PARP-14 RNA was not detected in tissues from PARP-14 mice homozygous for the disrupted (−) allele, and PARP-14+/− mice expressed half the amount of PARP-14 RNA compared with wild-type (WT) mice (Figure 1E). To screen for alternatively spliced PARP-14 mRNA expressed after initiating transcription in the 5′-most exon, we performed RT-PCR with primer pairs specific for several regions of the PARP-14 locus. We observed no PARP-14 RNA in the lymphoid tissues of −/− mice when using primers for the 3′ region (Figure 1F) or others (not shown). In addition, the level of PARP-14 protein in heterozygotes was approximately 50% that of +/+ controls, and none was detected in −/− mice (Figure 1G).
Figure 1.
Germ line gene disruption generates a loss-of-expression PARP-14 allele in mice. (A) Western blot analysis of CoaSt6/PARP-14 levels in T and B lymphocytes. Extracts from T and B lymphocytes, purified as described in “Northern blot RT-PCR,” were subjected to immunoblot analysis using affinity-purified antipeptide antibodies against CoaSt6 (PARP-14) or actin, as indicated. (B) Equal masses of protein in extracts from normal B cells or carefully excised primary tumor masses from 4 separate mice (1-4) were analyzed by Western blotting as described in panel A. Shown to the right is a bar graph of the results from quantitating the fluorescence intensity of the Western bands and normalizing to actin (PARP-14 was reproducibly increased with or without normalization). Similar results were obtained with cultured B-lymphoma lines (Figure S1). (C) Simplified map schematizing the normal and mutated PARP-14 alleles, EcoR1 restriction sites used for Southern blot analysis, and positions of hybridization probe and PCR primers used. Black boxes represent exons 1 to 4 and 8, E1 to E4 and E8, respectively; other portions of the locus downstream from intron 4 are omitted. Gene expression was disrupted by insertion of a splice-trap element containing an EcoR1 site; PARP-14 PCR primers flanked the insertion site in intron 1 (I1), which along with a splice-trap specific primer permitted unambiguous assignment of the allele on each chromosome. E8 primers used for RT-PCR also are diagrammed (sequences in Table S1). (D) Descendants were bred from founder lines established as described in “Generation of PARP-14–deficient mice.” Shown is an autoradiograph of Southern blots of EcoR1-digested genomic DNA from mice of indicated genotype (identified by PCR using the indicated I1-specific primers, sequences in Table S1) after probing with radiolabeled PARP-14 5′ sequences (gray rectangle in panel C) to reveal approximately 7.3-kb and approximately 5.7-kb bands (WT and mutated, respectively). (E) Total RNA isolated from thymus or lung of PARP-14+/+, PARP-14+/−, and PARP-14−/− mice, analyzed by Northern blots probed with radiolabeled PARP-14 (551 bp) or GAPDH DNA fragments. (F) RT-PCR using RNA isolated from lymphoid tissues of +/+, +/−, and −/− mice. The primer pair (downstream pair in panel C) detects sequences downstream from the gene disruption. (G) Immunoblot analysis of BAL2b/CoaSt6/PARP-14 and actin in splenocytes from WT and PARP-14−/− mice. Similar data were obtained with extracts of lung, and analysis of the full gel lane disclosed no evidence of a truncated form of the polypeptide.
Role of PARP-14 in MZB/FOB differentiation and IgA response to antigen
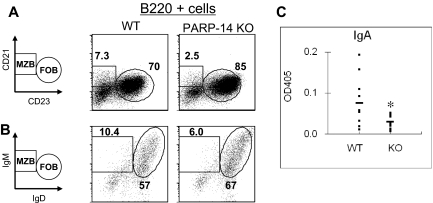
The pattern of PARP-14 expression prompted us to determine whether there would be any immune system defect in PARP-14 knockout (KO) mice. Analyses of thymic and peripheral lymphoid tissue cellularity and subset distributions showed that the overall numbers of cells in thymus, spleen, and lymph nodes of PARP-14–deficient mice were no different from controls. Among T-lineage subsets, there was an increase in the frequency of thymocytes exhibiting evidence of ongoing positive selection (TCRhi) and in CD44hi T cells in the periphery, especially for mature CD8 T cells (Table S2). Because of the association between BAL-family macro-PARP proteins and DLBCL, however, we focused on the B lymphoid lineage. Analyses of splenic B cells showed that, compared with WT (+/+) littermates, PARP-14 KO mice had less marginal zone B (MZB; WT, 9.9% ± 0.6% vs KO, 5.1% ± 0.8%, P < .01) and more follicular B (FOB) cells (WT 63% ± 2.9% vs KO 79% ± 2.5, P < .01; Figure 2A,B; Table 1). MZB cells are quantitatively greater sources of IgA than FOB.18 In exploring B-cell function by immunizing mice and measuring antibody responses, we focused particularly on antigen-specific IgA levels in WT and PARP-14 KO mice. Strikingly, the IgA anti-KLH response was substantially decreased in PARP-14–deficient mice compared with littermate controls (Figure 2C), whereas there was no significant difference in the other isotypes (data not shown). These results indicate that PARP-14 influences B-cell subset differentiation and selectively contributes to IgA generation in response to antigen.
Figure 2.
PARP-14 regulates the balance of B-cell subsets and contributes to the IgA response to antigen. (A,B) Single-cell suspensions of splenocytes were analyzed as described in “Flow cytometry and apoptosis assays.” Shown are representative FACS profiles of cells in the B220+ gate using (A) CD21 versus CD23 or (B) IgM versus IgD to define distinct marginal zone and follicular B-cell subsets (MZB and FOB, respectively). A reproducible increase in basal CD23 levels on the surface of PARP-14 FOB cells was incidentally noted. (C) Role of PARP-14 in promoting the IgA antibody response to antigen. WT and PARP-14–null mice (6 weeks; n = 12) were immunized with KLH followed by measurement of the KLH-specific IgA response using capture enzyme-linked immunosorbent assay. The results are shown as absorbance value from these assays (*P < .05).
Table 1.
Prevalence and number of B-cell subsets (MZB vs FOB)
| WT | PARP-14 KO | |
|---|---|---|
| % of MZB* | 9.9 ± 0.6 | 5.1 ± 0.8† |
| % of FOB* | 63 ± 2.9 | 79 ± 2.5† |
| No. of MZB ( × 106)* | 3.4 ± 0.62 | 2.0 ± 0.86† |
| No. of FOB ( × 106)* | 20.8 ± 2.0 | 26.4 ± 3.7‡ |
Data shown as mean (± SEM) percentage or number of cells from the 3 independent replicate experiments. MZB indicates marginal zone B cells; FOB, follicular B cells.
Total splenocytes were isolated, gated for B220+ cells, and analyzed for percentage or number of MZB (CD23hi CD21hi) or FOB (CD23lo CD21hi).
Significant difference between WT and PARP-14 KO mice (P < .01).
Significant difference between WT and PARP-14 KO mice (P < .05).
Role of PARP-14 in protection of B cells from apoptosis
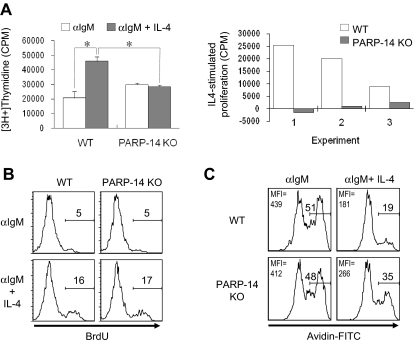
The effect of PARP-14 on B-cell subsets led us to investigate whether PARP-14–deficient mice have any defect in the proliferation or survival of B lymphocytes. Thymidine incorporation in T cells activated by αCD3 + αCD28, or PMA + IL-4, was the same for WT and PARP-14 KO samples in proliferation assays (data not shown). However, PARP-14 KO splenocytes exhibited an impaired response to IL-4 in assays of B-cell proliferation (Figure 3). Antigen receptor cross-linking induced cell proliferation, and addition of IL-4 to WT B cells enhanced this response approximately 2-fold. Although PARP-14 KO cells responded to anti-IgM with approximately 1.4-fold more proliferation than controls, they exhibited no further increase in response to IL-4, thereby exhibiting lower responses than anti-IgM + IL-4 controls (Figure 3A). The data from such assays reflect both the number of viable responding cells and their rates of S phase entry. To address whether the inability of IL-4 to enhance proliferation of anti-IgM–stimulated PARP-14 KO B cells was the result of effects on receptor expression or G1/S progression, we measured IL-4 receptor expression on and bromodeoxyuridine (BrdU) incorporation into B cells. IL-4R expression (Figure S2) and rates of G1/S progression (Figure 3B) were comparable for WT and PARP-14–null B cells activated with anti-IgM with or without IL-4.
Figure 3.
Role of PARP-14 in IL-4–stimulated proliferation. (A) Failure of anti-μ–stimulated B lacking PARP-14 to respond to IL-4 with increased proliferation. Thymidine incorporation into cells from WT and PARP-14–null mice 32 hours after stimulation with anti-IgM (1 μg/mL) or anti-IgM + IL-4. Data are shown both as mean (± SEM) cpm from an assay representative of 3 independent experiments (left panel) and as the net effect of IL-4 (IL-4–stimulated proliferation) in each of the 3 replicate experiments (right panel). Each error bar indicates SD. *P < .05, for difference between means for WT versus PARP-14–null samples in each case. (B) Normal G1/S-transition response of PARP-14 null B cells to anti-IgM plus IL-4. The same conditions for measurement of proliferation of anti-IgM + IL-4–stimulated B cells were used as in panel A, except that, instead of tritiated thymidine, cultures were pulsed with BrdU, surface stained to reveal B cells, and processed to determine the fraction of the B-cell population that incorporated BrdU. Shown are FACS profiles of anti-BrdU signal gated on B220+ cells from 1 experiment representative of 3 independent repeats; there also was no difference in the percentage of BrdU+ events in the B220(−) gate. (C) Anti-IgM–stimulated B lymphocytes depend on PARP-14 for survival signaling by IL-4. Splenocytes from WT and PARP-14 KO were cultured (20 hours) with anti-IgM (1 μg/mL) or anti-IgM + IL-4 (5 ng/mL), and analyzed by TUNEL. FACS profiles show TUNEL data (fluorescence intensity of avidin-FITC) from the B220+ gate in a representative experiment of 3 independent experiments. Inset numbers indicate both the percentage of events TUNEL-positive in the samples, and mean fluorescence intensity for the overall population.
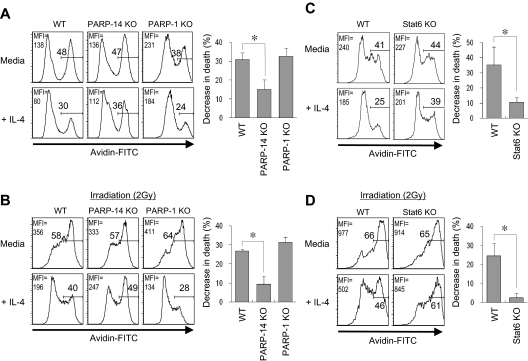
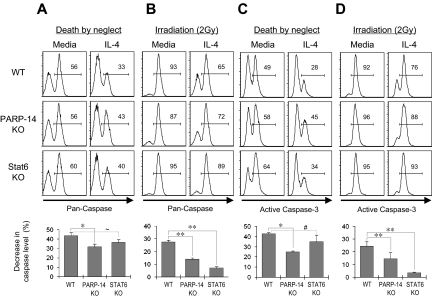
IL-4 is a survival factor for B cells, including those removed from their normal microenvironment during short-term culture ex vivo, so the proliferation results suggested that the absence of PARP-14 led to impaired IL-4–induced survival signaling. Consistent with this possibility, TUNEL assays showed that IL-4 decreased the rate of apoptosis for anti-IgM–treated WT B cells more potently than their PARP-14−/− counterparts (Figure 3C). Similarly, when B cells were cultured in the presence or absence of IL-4, stimulation decreased the amount of “death by neglect” for WT B cells, but this rescue was attenuated for PARP-14–null B cells (Figure 4A). This cytokine can protect T cells from radiation-induced death,19 but the mechanism by which IL-4 exerts this protective effect and whether it also operates in B cells are not known. We examined whether IL-4 can rescue B cells from apoptosis induced by ionizing radiation, and whether this process involves PARP-14. IL-4 induced a decrease in the apoptosis of WT B cells after γ-irradiation, and assays of PARP-14 KO B cells found this protective action of IL-4 to be impaired (Figure 4B). Intriguingly, the ability of IL-4 to protect T lymphocytes against death by neglect or irradiation was PARP-14–independent (Figure S3). PARP-1 helps signal DNA damage repair but also mediates apoptosis after irradiation and other forms of cellular stress.15,20,21 In contrast to PARP-14, PARP-1–deficient cells exhibited no defect in IL-4 rescue in either assay, ie, death by neglect or after irradiation (Figure 4A,B). Despite the dominant role of PARP-1 in global rates of protein PARylation, therefore, PARP-14 mediates IL-4–induced B-cell rescue from apoptosis, thereby playing a role in regulating lymphocyte survival distinct from that of PARP-1.
Figure 4.
PARP-14 mediates IL-4–induced B-cell rescue from apoptosis. (A,B) Role of PARP-14 in IL-4-induced B-cell protection against apoptosis. Splenocytes from WT, PARP-14 KO, and PARP-1 KO mice were cultured (20 hours) in the presence or absence of IL-4 and analyzed by TUNEL after death by neglect (A) or after irradiation (2 Gy) (B). FACS profiles show TUNEL data (avidin-FITC) from the B220+ gate in a representative experiment. Bar graphs show mean (± SEM) quantitation of the reductions in apoptosis for IL-4–treated samples of the indicated genotypes (n = 5 for WT, PARP-14–null; n = 3 for PARP-1−/−; *P < .05) compared with the matched medium-only sample [“% decrease in death” = (% TUNEL+medium-only minus% TUNEL+IL-4–treated)/% TUNEL+medium-only]. Inset numbers indicate both the percentage of events TUNEL-positive in the samples and mean fluorescence intensity for the overall population. (C,D) Stat6 mediates IL-4 protection of B cells from radiation-induced death. WT and Stat6 KO B cells were analyzed for death by neglect (C) or after irradiation (D) as in panels A and B; samples are representative of the 5 independent experiments averaged to formulate the bar graphs of mean ( ± SEM) quantitative data. Results congruent with these data were also observed by annexin V staining (not shown).
Stat6 in IL-4–mediated B-cell rescue from radiation-induced apoptosis
IL-4 receptors transduce signals through nuclear induction of Stat6.1 Stat6 binds to BAL2b/CoaSt6/PARP-14, so we investigated whether Stat6 is involved in IL-4–induced protection of B cells from apoptosis. When tested for death by neglect and γ-irradiation–induced apoptosis, Stat6-null B cells were poorly rescued by IL-4 (Figure 4C,D). Thus, Stat6 has crucial role in IL-4–induced rescue of resting B cells from apoptosis induced by irradiation, paralleling effects of PARP-14. Although the role of PARP-14 may be based in part on its interaction with the IL-4–induced transcription factor, the magnitude of the Stat6 contribution appeared larger than that of PARP-14. This difference suggests that not all of the protective molecular mechanisms of Stat6 absolutely require PARP-14.
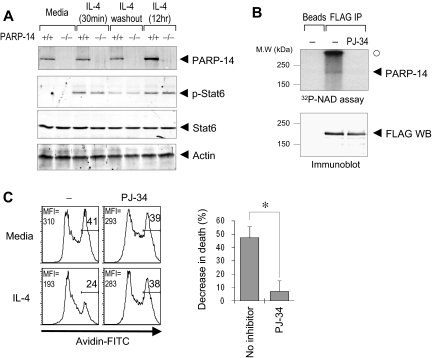
Stat6 and CoaSt6/PARP-14 physically interact, so we considered the possibility that the similarity of apoptosis assay results with knockout cells arose because PARP-14 influences Stat6 levels, induction of Stat6 phosphorylation by IL-4, or its persistence after abrogation of the IL-4 signal. Western blotting of phospho-Stat6 or Stat6 in splenocytes after IL-4 treatment, or induction followed by removal of IL-4, revealed no difference between PARP-14–null cells and WT controls (Figure 5A). To address the role of ADP-ribosylation in protection of B cells by IL-4, we next examined whether PARP activity is important for the rescue process. Experiments using a restricted segment of BAL2b, the human PARP-14 fused to GST, have provided evidence that BAL2b probably encodes a functional PARP.22 The results of in vitro ADP-ribosylation reactions, in which full-length CoaSt6/PARP-14 protein was incubated with 32P-NAD+ in the presence or absence of a cell-permeable PARP inhibitor (PJ-34), showed that the full-length PARP-14 protein has intrinsic PARP activity (Figure 5B). Furthermore, IL-4–induced protection of B cells against apoptosis was blocked by PJ-34 (Figure 5C). These results indicate that PAR polymerization mediates a survival signal in resting B lymphocytes.
Figure 5.
ADP ribosylation mediates survival signaling in B cells, and PARP-14 has intrinsic PARP activity. (A) Stat6 regulation is intact in PARP-14 KO cells. Splenocytes from WT (+/+) and PARP-14–null (−/−) mice were cultured in media, alone (−IL-4), stimulated with IL-4 (0.5 and 12 hours; +IL-4), or stimulated by IL-4 (30 minutes) followed by removal of IL-4 and further culture for 6 hours. (B) ADP-ribosyltransferase activity associated with PARP-14. Proteins from cells transfected with pcDNA3 or pcDNA3-FLAG-PARP-14 were immunoprecipitated, and bead-bound immune complexes were assayed for transfer of 32P-labeled ADP-ribose from NAD+ onto proteins in the presence or absence of the cell-permeable PARP inhibitor PJ-34 (1 μM). Shown are autoradiographs of the labeled proteins (top panel) and immunoblots to test protein expression levels (bottom panel) after resolution by SDS-PAGE. An arrowhead marks the position on the gel autoradiograph expected for full-length PARP-14 based on the Western blot; the radioactive material marked by a closed circle derived from nonspecific polymeric material generated by active PARP and trapped at the stacker/resolving gel interface. (C) Impaired survival signaling in B cells subjected to PARP inhibition. B cells were pretreated with PJ-34 (1 μM), during culture (20 hours) in the presence or absence of IL-4 and analyzed by TUNEL assays. Shown are FACS profiles in the B220+ gate (left, FACS profile) as well as a bar graph showing the mean (± SEM) percentage of cells rescued from apoptosis by IL-4 in the 3 independent replicate experiments (n = 3; *P < .05).
PARP-14 mediates IL-4–induced inhibition of caspase-3
The preceding findings suggested that PARP-14 might mediate inhibition of cell death through caspases rather than the caspase-independent pathway mobilized by PARP-1. To determine whether IL-4 decreases caspase activity in primary B cells by a PARP-14–dependent mechanism, B cells were cultured in the presence or absence of IL-4, with or without irradiation, and caspase activity was measured using a fluorophore-linked caspase substrate. In assays of death by neglect, IL-4 was less effective in suppressing total caspase activity in PARP-14–null B cells compared with WT (Figure 6A). Furthermore, IL-4 decreased levels of total caspase activity in irradiated WT B cells (P = .02), but this cytokine was less potent in decreasing caspase activation in PARP-14 KO B cells (Figure 6B). Activation of upstream caspases can be uncoupled from commitment to apoptosis by prosurvival proteins. Although also subject to inhibition by survival proteins23 and not a measurement of apoptosis or death,24 activation of “executioner caspases” such as caspase-3 is a late step in the apoptotic program and closer to death. IL-4 treatment reduced levels of activated caspase-3 in WT B lymphocytes (P < .001). However, levels of this activated form of executioner caspase were higher in IL-4–treated PARP-14–null B cells, and the caspase-inhibitory effect of IL-4 was diminished in the absence of PARP-14 (Figure 6C,D; P < .05 for both death by neglect and death after irradiation). These findings indicate that PARP-14 helps mediate IL-4 suppression of caspase-3 activation in IL-4–treated B cells. Intriguingly, although Stat6 and PARP-14 are each essential for a full protective effect of IL-4 against B-cell apoptosis (Figure 4), PARP-14 played a quantitatively more significant role in terms of IL-4 suppression of the frequency of cells positive for active caspase, especially in death by neglect assays. This disparity suggests that, as is typical for cofactors of transcription factors, PARP-14 may exert some Stat6-independent functions rather than being uniquely linked to Stat6.
Figure 6.
IL-4 inhibition of the caspase-3 pathway by a PARP-14–dependent mechanism. Splenocytes from littermate WT and PARP-14-null mice, along with age-matched Stat6-deficient counterparts, were cultured (20 hours) in medium with or without IL-4. (A) PARP-14 mediates repression of caspase activity by IL-4. Shown in top panels are FACS profiles of B220+ cells analyzed using a fluorophore-conjugated substrate of enzymatically active caspases. The bar graph in the bottom panel summarizes mean (± SEM) data on the protective effect of IL-4 in the experiments, calculated as in Figure 4. Additional statistical analyses confirmed that percentage of caspase-positive was higher in IL-4–treated PARP-14 KO samples than controls (40.3 ± 2.0 vs 31.3 ± 2.9; P < .05). (B) IL-4 acts on B cells with DNA damage to signal inhibition of caspase activity by a PARP-14–dependent mechanism. As in panel A, except that cells were γ-irradiated (2 Gy) before culture (20 hours) in the presence or absence of IL-4. (C) Suppression of caspase-3 activity in IL-4–treated B cells depends on PARP-14. As in panel A, except that the B cells were probed by intracellular staining with antibodies specific for the cleaved, activated form of caspase-3. (D) As in panel C, except that cells were irradiated (2 Gy) before culture with or without IL-4. All inset numbers represent the percentage of cells positive for the caspase signal (total activity or activated caspase-3); n = 4. *P < .05; **P < .01; ∼P = .07; #P = .14.
PARP-14 mediates IL-4 regulation of the expression of genes determining B-cell survival
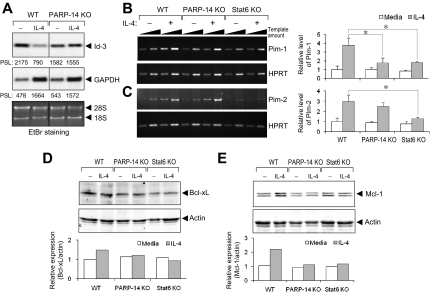
To dissect further the molecular role of PARP-14 in IL-4–induced B-cell rescue from apoptosis, we measured the expression of gene products known to determine the susceptibility of B cells to apoptosis. In addition to screening best-known survival genes, we performed microarrays on B-cell mRNAs to identify IL-4–regulated, PARP-14–dependent genes. Among differentially regulated genes of interest (Table S3), we selected Id3 (inhibitor of DNA binding-3), an inhibitor of leukemia/lymphoma-associated E2A transcription factor known to promote survival of B-cell progenitors as an additional candidate. Id3 expression was suppressed by IL-4 treatment of WT B cells but sustained in PARP-14 KO B cells under these conditions (Figure 7A), showing that IL-4 represses Id3 expression through a PARP-14–dependent pathway.
Figure 7.
PARP-14 mediates IL-4 regulation of genes that influence caspase activation, apoptosis, and B lymphomagenesis. (A) Role of PARP-14 in repression of Id-3 by IL-4. RNA from B cells of WT and PARP-14 KO mice, cultured for 20 hours with and without IL-4, was analyzed by Northern blotting for Id-3 and a GAPDH control. An autoradiograph from one such analysis is shown, representative of 3 independent experiments. Below each lane are the PhosphorImager quantitations (in arbitrary units of pixel density) of the relative radioactive signal for each band, and ethidium bromide–stained rRNA signals designated by ◀ (28S, 18S). Note that GAPDH transcription rates and mRNA levels are known to increase in response to IL-4. (B,C) PARP-14 mediates IL-4 induction of the pro-survival kinase Pim-1. B cells purified from WT, PARP-14−/−, and Stat6−/− mice were cultured in medium, alone or supplemented with IL-4. After determination of the optimal induction kinetics for each (not shown), Pim-1 (B) or Pim-2 (C) mRNA levels were assayed in RNA isolated after 4 hours (B) or 12 hours (C), using both conventional (left panel) and real-time RT-PCR (right panel). For conventional semiquantitative RT-PCR, the panels show ethidium bromide–stained results from one experiment representative of 4 independent replicates, with the linear relationship of amplifications to template amount documented by using 2 amounts of input cDNA in separate amplifications for each sample (5 × 104 and 1:3 dilution of 5 × 104 cell equivalents of recovered RNA; increasing amounts symbolized by the slope of the black triangle above each lane pair). Southern blotting probed with an internal oligonucleotide sequence was used to confirm the identity, linearity, and relative abundance of the signals (data not shown). To the right in panels B and C, bar graphs show representative data (mean ± SEM; *P < .05) from separate real-time quantitative RT-PCR performed on the independent replicate samples, with each Pim kinase cDNA signal normalized to the quantitative RT-PCR result for hypoxanthine phosphoribosyl transferase cDNA levels in each sample. (D,E) PARP-14 mediates IL-4 regulation of antiapoptotic Bcl-2-family protein. Extracts of purified B cells cultured (20 hours) in the presence or absence of IL-4 were analyzed by immunoblots using anti–Bcl-xL (D) and anti-Mcl-1 (E) Ab, along with antiactin as a loading control. Quantitation of the relative levels of Bcl-xL and Mcl-1 after normalization to the actin controls (each in arbitrary fluorescence intensity units detected by the Odyssey imager) is shown in each bar graph (one representative result from reproducible data [n = 3 independent replicate experiments]). Mcl-1 appears as a doublet because the antibody detects both long (40 kDa) and short (32 kDa) forms expressed in cells.48
Pim-1 and -2 levels in T cells are increased by IL-2, -4, and -7.25 The expression of these prosurvival kinases may be regulated by members of the Stat transcription factor family in other settings.26 Therefore, we analyzed the effect of IL-4 on expression of Pim-1 and -2 in B cells and the role of Bal2b/PARP-14 in regulating their levels. Purified B cells from WT, PARP-14 KO, and Stat6 KO were stimulated with IL-4 at times determined (not shown) to maximize induction by IL-4 in WT samples. Pim-1 was induced after IL-4 stimulation of WT but not PARP-14 or Stat6-null B cells (Figure 7B). In contrast to its regulation in T cells, Pim-2 was modestly induced at 12 hours after IL-4 treatment of control B cells, and there was no difference between WT and PARP-14 KO B cells in terms of Pim-2 induction by IL-4 (Figure 7C). Thus, PARP-14 specifically regulates Pim-1 expression, mediating IL-4 induction of this survival factor whose overexpression synergizes with oncogenic stimuli in B lymphoma pathogenesis.
In activated lymphocytes, antiapoptotic Bcl-2 family members are induced by IL-4 or other cytokines whose receptors share a common γ-chain.27,28 As for Pim-1, increased levels of these proteins cooperate with oncogenic stimuli to enhance B lymphomagenesis.29–31 We observed no role for PARP-14 in regulating levels of Bcl-2 (data not shown) and at most a modest decrease in Bcl-xL levels in IL-4–treated PARP-14–null B cells compared with WT controls (Figure 7D). In contrast, the doubling of Mcl-1 expression induced in WT B cells by IL-4 was abrogated in the absence of PARP-14 (Figure 7E); such induction was also lacking in Stat6-deficient B cells. These results establish that PARP-14 mediates IL-4 induction of Mcl-1, which is known to inhibit caspase induction and protect lymphocytes from apoptosis after cytokine withdrawal or irradiation. Collectively, our findings show that a macro-PARP coordinately yet specifically regulates expression of a set of genes that promote B-cell survival and, intriguingly, are known to cooperate with oncogenic stimuli to promote a more aggressive emergence of B lymphoma.
Discussion
We have shown that PARP-14, a member of the macro-PARP subfamily, mediates the induction and repression of key regulators of apoptosis and inhibition of an apoptotic program involving caspase-3. These contributions culminate in a requirement for PARP-14 in IL-4 induction of B lymphocyte survival after irradiation or withdrawal from an appropriate trophic environment. DNA strand breaks characterize early development in both T- and B-lymphoid lineages before differentiation into mature lymphocytes, but B cells have substantial strand breaks during class-switch recombination and somatic hypermutation, processes that can be promoted by IL-4 and are absent from T cells. Several studies reported at best a modest requirement for Stat6 in IL-4–induced protection against apoptosis of T cells.32,33 Intriguingly, we observed that the ability of IL-4 to reduce cell death was fully preserved in PARP-14–deficient T cells. Thus, the differential role of PARP-14 provides an intriguing parallel to cell type–specific differences in the role of Stat6. Finally, comparisons of these functions of PARP-14 to results with PARP-1 indicate that the macro-PARP plays at least some role in cell physiology different from the protein that biochemically is the predominant poly(ADP-ribose) polymerase.
PARP-1 is exclusively nuclear, ubiquitously expressed, and plays a pivotal role that balances the DNA damage response and cell survival. After relatively less severe damage, the functions of PARP-1 in promoting assembly and activity of DNA repair complexes appear to dominate, allowing the damaged cell to recover when its genomic stability can be maintained.34 After severe DNA damage, however, PARP-1 triggers the release of AIF from mitochondria and initiates a caspase-independent cell death pathway.15,21 AIF-dependent, caspase-independent apoptosis appears to be mediated by PARG-dependent release of PAR into the cytosol,16 suggesting that any PARP might be able to activate AIF release but that the outcome requires a high enough PARP activity. Overlapping and partially redundant roles have been identified for PARP-2, which can function in a physical complex with PARP-1.35 Although development of both B- and T-cell precursors requires double-strand breaks and unresolved dsDNA breaks are an intrinsic feature of the B lymphocyte after its generation,36,37 no significant abnormality of lymphocytes has been reported for PARP-1–deficient mice, which generated IgA+ B cells normally.38 PARP-2–deficient animals exhibited a 50% decrease in the number of double-positive (DP) thymocytes and a modest increase in their susceptibility to apoptosis.39 Our data show that PARP-14 influences the balance of B-cell subsets at steady state as well as IgA responses in vivo, and the cellular responses of B lymphocytes to IL-4 were affected differently by deficiency of PARP-14 versus PARP-1. The preferential impact on IgA responses observed in PARP-14–deficient mice is intriguing; although the mechanism is not clear, IL-4 can enhance the effect of IL-5 on B-cell production of IgA.40
Roles of ADP-ribosylation in cell death after stress or DNA damage have been identified from analyses of PARP inhibitors, leading to the emergence of potential therapeutics.41 Both ribosylation-dependent and -independent roles in these cellular processes have been worked out for PARP-1, both in gene regulation and its enhancement of responses to DNA damage and mediation of AIF activation in launching caspase-independent apoptosis. However, neither the physiologic functions of any macro-PARP nor biologic roles of most other PARP family members are known. One possibility raised by our findings is that PARP-14, and perhaps other macro-PARPs, protects B lymphocytes against apoptosis mediated by the AIF pathway. PARP-1 activity is inhibited by macro domains,42,43 and this domain in a macro-PARP appears able to serve as a PAR-binding module.43,44 DNA double-strand breaks are inevitable aspects of normal B-cell physiology, occurring both when rearranging antigen-receptor gene segments during development and later during class-switch recombination. ATM appears vital for protection of new B cells against unresolved breaks,45 and PARP-1 interacts with ATM.46 Therefore, release of ADP-ribose polymers originally generated by PARP-1 may present a risk of apoptosis from PAR-mediated AIF activation such that macro-PARPs are needed for sequestration of these ADP-ribose polymers. However, our gene induction data indicate that this speculative mechanism is not the only reason why PARP-14 is a critical regulator of caspase induction and apoptosis susceptibility in B cells.
A fundamental question about the biology of macro-PARP proteins is raised by their association with the aggressiveness of B-cell lymphomas in the DLBCL family and the elevated expression of PARP-14 in the primary tumor masses of myc-induced B lymphomas. The mechanism(s) by which macro-PARPs might influence lymphoma pathophysiology are unclear. Transfection and overexpression of PARP-9/BAL1 enhanced chemokine SDF-1α–stimulated cell migration.10 Expression of CXCR4, a receptor for this chemokine, is enhanced by IL-4,47 and increased expression of IL-4 target genes is observed in many leukemias and lymphomas. The overactivity of IL-4R signaling in lymphomas suggests that targets whose induction is regulated by PARP-14 may be involved in the pathogenesis of the disease. Although the functional impact of this regulatory network remains to be determined for lymphoid malignancies, each of 3 key targets of IL-4 and PARP-14 (ie, Id3, Pim-1, and Mcl-1) impacts cell survival and cooperates with oncogenes such as c-Myc to render B lymphomagenesis more aggressive.
The antiapoptotic Bcl-2 family member protein Mcl-1 plays a crucial role in both B- and T-cell survival.48 Transgenic mouse models with deregulated expression of Mcl-1 developed B-cell lymphoma,49 and higher expression of Mcl-1 was detected in high-grade B-cell lymphomas compared with low-grade lymphomas.50 Importantly, a 50% reduction in Mcl-1 expression resulted in reduced numbers and increased apoptotic susceptibility of peripheral B cells in Mcl-1+/− mice. Thus, the reduction in IL-4–induced Mcl-1 expression in PARP-14–null B cells may be sufficient to account for their increased susceptibility to apoptosis.
Pim kinase family members also have a critical role in cell survival and proliferation.51 Previously, it was shown that Pim-1 is competent to cooperate strongly with c-Myc in the development of murine B-cell leukemia31,52 and Pim-1 transgenic mice had a very high frequency of lymphoma induction by chemical carcinogen.53 Pim-2 also can collaborate with Myc, but increased Pim-2 is not required for Pim-1 to exert its effect.50 Accordingly, increased expression of macro-PARPs may increase the expression of Pim-1, thereby influencing lymphoma progression. The importance of Pim-1 regulation by PARP-14 is emphasized by a network of paired interactions that have been identified. Pim-1 binds to p100, cooperatively increasing activity of the proto-oncogene c-Myb,54 whereas p100 also binds to Stat655 and Stat6 interacts with PARP-14.17 Given the differences between results for Stat6- and PARP-14–null B cells in terms of several endpoints (Pim-2 induction; caspase inhibition), further studies will be required fully to elucidate in which settings Stat6 collaborates with PARP-14 to mediate specific IL-4 effects on B cells. Nonetheless, the findings presented here establish that a coordinated set of prosurvival factors and B-lymphocyte apoptosis susceptibility are regulated by the macro-PARP BAL2b/CoaSt6/PARP-14.
Acknowledgments
The authors thank B. Kee for helpful advice and generous gift of a plasmid and D. Ballard for critical reading of the manuscript.
This work was supported by the Sandler Program for Asthma Research and National Institutes of Health (grant GM071735), followed by AI068149. Microarray support came from the National Technology Agency of Finland and Sigrid Juselius Foundation. C.M.E. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.H.C. designed and performed the research, analyzed data, and wrote the paper; M.B. designed the research, analyzed the data, and wrote the paper; S.G. and C.M.E. designed and performed research; and T.H., A.R., P.G., and R.L. performed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Boothby, Department of Microbiology & Immunology, Vanderbilt University, AA-4214 MCN, Nashville, TN 37232; e-mail: mark.boothby@vanderbilt.edu.
References
- 1.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 3.Mainou-Fowler T, Proctor SJ, Taylor PR. Interleukin 4 production by peripheral blood lymphocytes in patients with classical Hodgkin lymphoma. Leuk Res. 2004;28:159–166. doi: 10.1016/s0145-2126(03)00216-9. [DOI] [PubMed] [Google Scholar]
- 4.Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood. 2003;101:433–440. doi: 10.1182/blood-2002-06-1931. [DOI] [PubMed] [Google Scholar]
- 5.Copie-Bergman C, Boulland ML, Dehoulle C, et al. Interleukin 4-induced gene 1 is activated in primary mediastinal large B-cell lymphoma. Blood. 2003;101:2756–2761. doi: 10.1182/blood-2002-07-2215. [DOI] [PubMed] [Google Scholar]
- 6.Skinnider BF, Elia AJ, Gascoyne RD, et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2002;99:618–626. doi: 10.1182/blood.v99.2.618. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Nechushtan H, Ding F, et al. Distinct IL-4-induced gene expression, proliferation, and intracellular signaling in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas. Blood. 2005;105:2924–2932. doi: 10.1182/blood-2004-10-3820. [DOI] [PubMed] [Google Scholar]
- 8.Guiter C, Dusanter-Fourt I, Copie-Bergman C, et al. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood. 2004;104:543–549. doi: 10.1182/blood-2003-10-3545. [DOI] [PubMed] [Google Scholar]
- 9.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- 11.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 12.Heale JT, Ball AR, Jr, Schmiesing JA, et al. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanasse GJ, Halbrook J, Thomas S, et al. Genetic pathway to recurrent chromosome translocations in murine lymphoma involves V(D)J recombinase. J Clin Invest. 1999;103:1669–1675. doi: 10.1172/JCI6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MT, Danska JS. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 15.Yu SW, Wang H, Poitras MF, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 16.Yu SW, Andrabi SA, Wang H, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goenka S, Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc Natl Acad Sci U S A. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski DA, Stavnezer J. Enhanced IgA class switching in marginal zone and B1 B cells relative to follicular/B2 B cells. J Immunol. 2006;177:6025–6029. doi: 10.4049/jimmunol.177.9.6025. [DOI] [PubMed] [Google Scholar]
- 19.Boise LH, Minn AJ, June CH, Lindsten T, Thompson CB. Growth factors can enhance lymphocyte survival without committing the cell to undergo cell division. Proc Natl Acad Sci U S A. 1995;92:5491–5495. doi: 10.1073/pnas.92.12.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keil C, Grobe T, Oei SL. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J Biol Chem. 2006;281:34394–34405. doi: 10.1074/jbc.M606470200. [DOI] [PubMed] [Google Scholar]
- 21.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25:259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 23.Riedl SJ, Renatus M, Schwarzenbacher R, et al. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 24.Puga I, Rao A, Macian F. Targeted cleavage of signaling proteins by caspase 3 inhibits T cell receptor signaling in anergic T cells. Immunity. 2008;29:193–204. doi: 10.1016/j.immuni.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med. 2005;201:259–266. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stout BA, Bates ME, Liu LY, Farrington NN, Bertics PJ. IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol. 2004;173:6409–6417. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- 27.Akbar AN, Borthwick NJ, Wickremasinghe RG, et al. Interleukin-2 receptor common gamma-chain signaling cytokines regulate activated T cell apoptosis in response to growth factor withdrawal: selective induction of anti-apoptotic (bcl-2, bcl-xL) but not pro-apoptotic (bax, bcl-xS) gene expression. Eur J Immunol. 1996;26:294–299. doi: 10.1002/eji.1830260204. [DOI] [PubMed] [Google Scholar]
- 28.Wurster AL, Rodgers VL, White MF, Rothstein TL, Grusby MJ. Interleukin-4-mediated protection of primary B cells from apoptosis through Stat6-dependent up-regulation of Bcl-xL. J Biol Chem. 2002;277:27169–27175. doi: 10.1074/jbc.M201207200. [DOI] [PubMed] [Google Scholar]
- 29.Moroy T, Verbeek S, Ma A, Achacoso P, Berns A, Alt F. E mu N- and E mu L-myc cooperate with E mu pim-1 to generate lymphoid tumors at high frequency in double-transgenic mice. Oncogene. 1991;6:1941–1948. [PubMed] [Google Scholar]
- 30.Stone DM, Norton LK, Magnuson NS, Davis WC. Elevated pim-1 and c-myc proto-oncogene induction in B lymphocytes from BLV-infected cows with persistent B lymphocytosis. Leukemia. 1996;10:1629–1638. [PubMed] [Google Scholar]
- 31.van Lohuizen M, Verbeek S, Krimpenfort P, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 32.Vella A, Teague TK, Ihle J, Kappler J, Marrack P. Interleukin 4 (IL-4) or IL-7 prevents the death of resting T cells: stat6 is probably not required for the effect of IL-4. J Exp Med. 1997;186:325–330. doi: 10.1084/jem.186.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aronica MA, Goenka S, Boothby M. IL-4-dependent induction of BCL-2 and BCL-x(L) in activated T lymphocytes through a Stat6- and pi 3-kinase-independent pathway. Cytokine. 2000;12:578–587. doi: 10.1006/cyto.1999.0603. [DOI] [PubMed] [Google Scholar]
- 34.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 35.Menissier de Murcia J, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 37.Puebla-Osorio N, Zhu C. DNA damage and repair during lymphoid development: antigen receptor diversity, genomic integrity and lymphomagenesis. Immunol Res. 2008;41:103–122. doi: 10.1007/s12026-008-8015-3. [DOI] [PubMed] [Google Scholar]
- 38.Wang ZQ, Stingl L, Morrison C, et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yelamos J, Monreal Y, Saenz L, et al. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coffman RL, Shrader B, Carty J, Mosmann TR, Bond MW. A mouse T cell product that preferentially enhances IgA production: I. Biologic characterization. J Immunol. 1987;139:3685–3690. [PubMed] [Google Scholar]
- 41.Curtin NJ. PARP inhibitors for cancer therapy. Expert Rev Mol Med. 2005;7:1–20. doi: 10.1017/S146239940500904X. [DOI] [PubMed] [Google Scholar]
- 42.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, et al. The histone variant mH2A1. 1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- 44.Karras GI, Kustatscher G, Buhecha HR, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callen E, Jankovic M, Difilippantonio S, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Haince JF, Kozlov S, Dawson VL, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 47.Jourdan P, Abbal C, Noraz N, et al. IL-4 induces functional cell-surface expression of CXCR4 on human T cells. J Immunol. 1998;160:4153–4157. [PubMed] [Google Scholar]
- 48.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 49.Zhou P, Levy NB, Xie H, et al. MCL1 transgenic mice exhibit a high incidence of B-cell lymphoma manifested as a spectrum of histologic subtypes. Blood. 2001;97:3902–3909. doi: 10.1182/blood.v97.12.3902. [DOI] [PubMed] [Google Scholar]
- 50.Cho-Vega JH, Rassidakis GZ, Admirand JH, et al. MCL-1 expression in B-cell non-Hodgkin's lymphomas. Hum Pathol. 2004;35:1095–1100. doi: 10.1016/j.humpath.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 51.Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood. 2005;105:4477–4483. doi: 10.1182/blood-2004-09-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen JD, Verhoeven E, Domen J, van der Valk M, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15:1133–1141. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- 53.Breuer M, Slebos R, Verbeek S, van Lohuizen M, Wientjens E, Berns A. Very high frequency of lymphoma induction by a chemical carcinogen in pim-1 transgenic mice. Nature. 1989;340:61–63. doi: 10.1038/340061a0. [DOI] [PubMed] [Google Scholar]
- 54.Leverson JD, Koskinen PJ, Orrico FC, et al. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Aittomaki S, Pesu M, et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.