Abstract
Attempts to identify coryneform isolates resembling Corynebacterium xerosis can lead clinical microbiologists to identification schemes with conflicting descriptions which result in confusing C. xerosis with Corynebacterium striatum. For the present study we purchased all available American Type Culture Collection and National Collection of Type Cultures reference cultures of C. xerosis (n = 10) and C. striatum (n = 4) and analyzed them as follows: (i) analysis of biochemical reactions in conventional tests and in the Rapid CORYNE system, (ii) whole-cell fatty acid analysis by using the gas-liquid chromatography research software of Microbial ID, Inc., and (iii) analysis of DNA homology in dot blot hybridizations. Three C. xerosis strains were indistinguishable from the C. striatum strains in whole-cell fatty acid analyses and DNA hybridizations and shared very similar biochemical reactions. The remaining seven strains of C. xerosis clustered into five groups on the basis of fatty acid patterns, DNA hybridizations, and biochemical tests. No reference strain of C. striatum fit the species description in Bergey's Manual of Systematic Bacteriology. The type strains of both C. striatum and C. xerosis fit their respective descriptions by the Centers for Disease Control and Prevention. This study suggests that the 10 commercially available reference strains of C. xerosis represent six different taxa which should be assigned to new species.
Full text
PDF
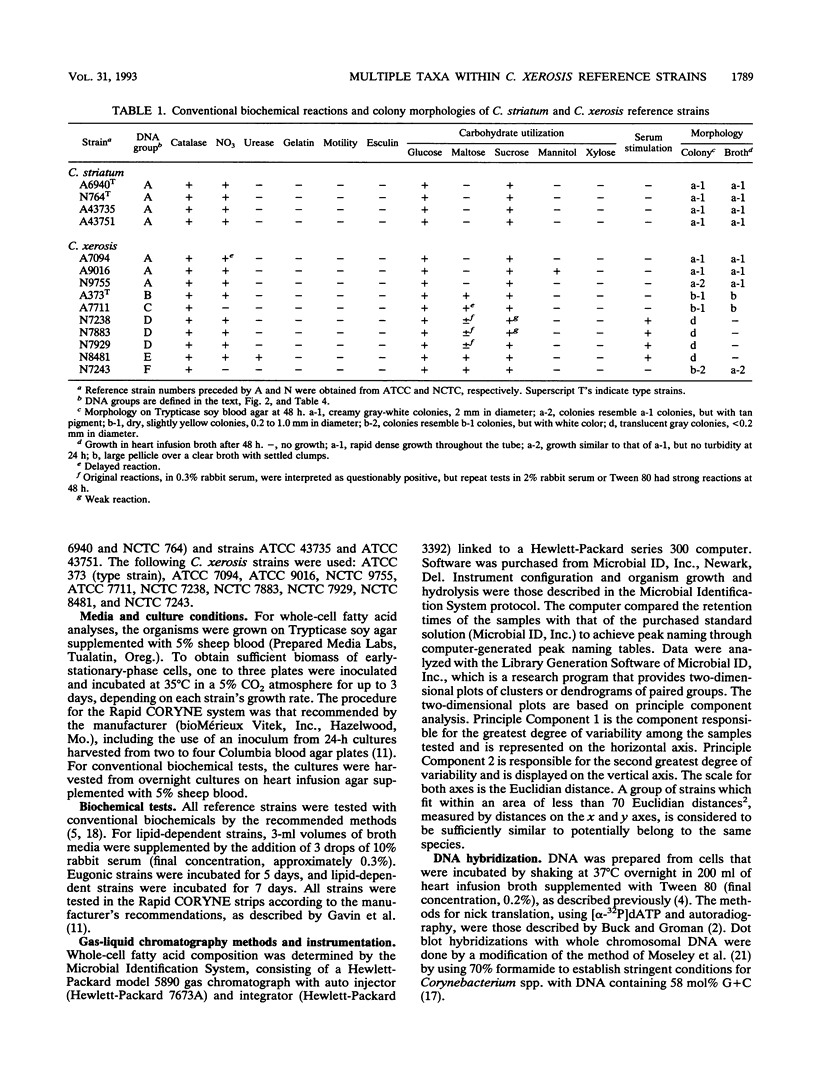

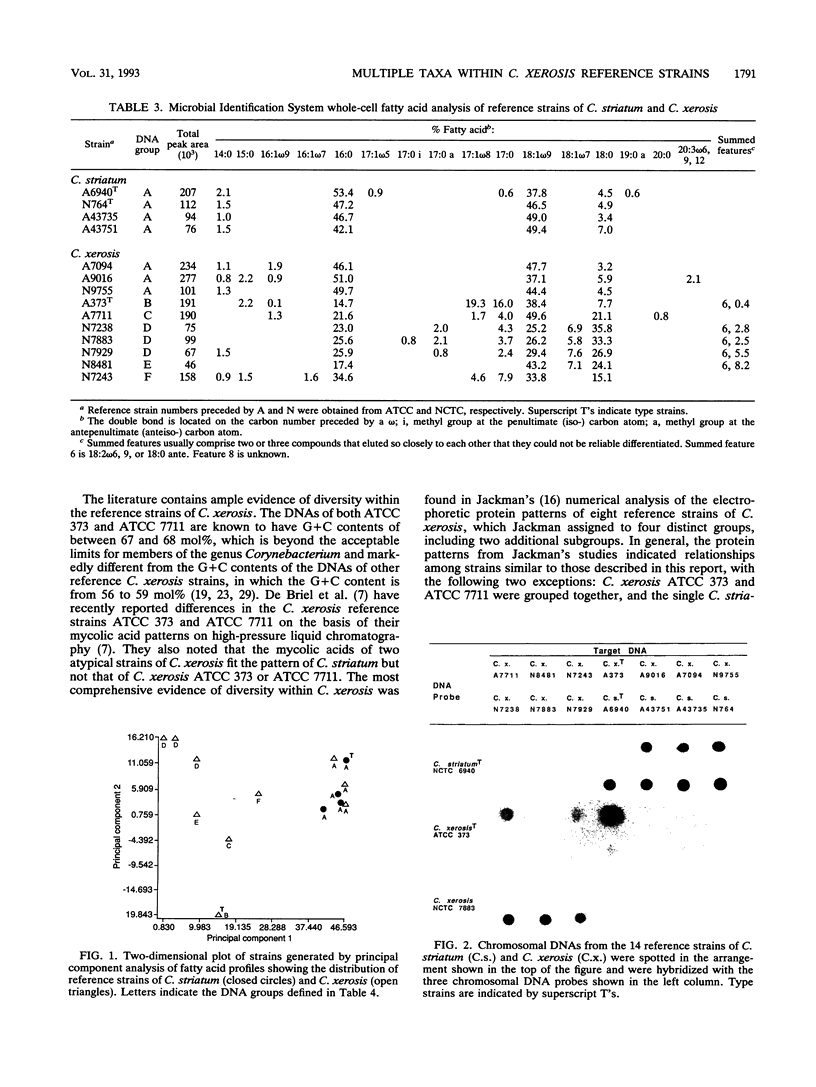

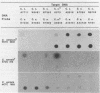
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernard K. A., Bellefeuille M., Ewan E. P. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J Clin Microbiol. 1991 Jan;29(1):83–89. doi: 10.1128/jcm.29.1.83-89.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Physical mapping of beta-converting and gamma-nonconverting corynebacteriophage genomes. J Bacteriol. 1981 Oct;148(1):131–142. doi: 10.1128/jb.148.1.131-142.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle M. B., Groman N. B., Russell J. Q., Harnisch J. P., Rabin M., Holmes K. K. The molecular epidemiology of three biotypes of Corynebacterium diphtheriae in the Seattle outbreak, 1972-1982. J Infect Dis. 1989 Apr;159(4):670–679. doi: 10.1093/infdis/159.4.670. [DOI] [PubMed] [Google Scholar]
- De Briel D., Couderc F., Riegel P., Jehl F., Minck R. High-performance liquid chromatography of corynomycolic acids as a tool in identification of Corynebacterium species and related organisms. J Clin Microbiol. 1992 Jun;30(6):1407–1417. doi: 10.1128/jcm.30.6.1407-1417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersgaard H., Justesen T. Multiresistant lipophilic corynebacteria from clinical specimens. Biochemical reactions and antimicrobial agents susceptibility. Acta Pathol Microbiol Immunol Scand B. 1984 Feb;92(1):39–43. [PubMed] [Google Scholar]
- Evans N. M. The classification of aerobic diphtheroids from human skin. Br J Dermatol. 1968 Feb;80(2):81–83. doi: 10.1111/j.1365-2133.1968.tb12264.x. [DOI] [PubMed] [Google Scholar]
- Freney J., Duperron M. T., Courtier C., Hansen W., Allard F., Boeufgras J. M., Monget D., Fleurette J. Evaluation of API Coryne in comparison with conventional methods for identifying coryneform bacteria. J Clin Microbiol. 1991 Jan;29(1):38–41. doi: 10.1128/jcm.29.1.38-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin S. E., Leonard R. B., Briselden A. M., Coyle M. B. Evaluation of the rapid CORYNE identification system for Corynebacterium species and other coryneforms. J Clin Microbiol. 1992 Jul;30(7):1692–1695. doi: 10.1128/jcm.30.7.1692-1695.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine J. E., Hill L. R., Lapage S. P. Corynebacterium spp in human disease. Lancet. 1978 Aug 12;2(8085):376–376. doi: 10.1016/s0140-6736(78)92976-8. [DOI] [PubMed] [Google Scholar]
- Jackman P. J. Classification of Corynebacterium species from axillary skin by numerical analysis of electrophoretic protein patterns. J Med Microbiol. 1982 Nov;15(4):485–492. doi: 10.1099/00222615-15-4-485. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- MUNCH-PETERSEN E. A corynebacterial agent which protects ruminant erythrocytes against staphylococcal beta toxin. Aust J Exp Biol Med Sci. 1954 Jun;32(3):361–368. doi: 10.1038/icb.1954.39. [DOI] [PubMed] [Google Scholar]
- Marples R. R. Diphtheroids of normal human skin. Br J Dermatol. 1969;81(Suppl):47+–47+. doi: 10.1111/j.1365-2133.1969.tb12833.x. [DOI] [PubMed] [Google Scholar]
- Moseley S. L., Huq I., Alim A. R., So M., Samadpour-Motalebi M., Falkow S. Detection of enterotoxigenic Escherichia coli by DNA colony hybridization. J Infect Dis. 1980 Dec;142(6):892–898. doi: 10.1093/infdis/142.6.892. [DOI] [PubMed] [Google Scholar]
- POLLOCK M. R., WAINWRIGHT S. D., MANSON E. E. D. The presence of oleic acid-requiring diphtheroids on human skin. J Pathol Bacteriol. 1949 Apr;61(2):274-6, 2 pl. doi: 10.1002/path.1700610218. [DOI] [PubMed] [Google Scholar]
- Riley P. S., Hollis D. G., Utter G. B., Weaver R. E., Baker C. N. Characterization and identification of 95 diphtheroid (group JK) cultures isolated from clinical specimens. J Clin Microbiol. 1979 Mar;9(3):418–424. doi: 10.1128/jcm.9.3.418-424.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. F. Characterization of human cutaneous lipophilic diphtheroids. J Gen Microbiol. 1969 Mar;55(3):433–443. doi: 10.1099/00221287-55-3-433. [DOI] [PubMed] [Google Scholar]
- Somerville D. A. A taxonomic scheme for aerobic diphtheroids from human skin. J Med Microbiol. 1973 May;6(2):215–224. doi: 10.1099/00222615-6-2-215. [DOI] [PubMed] [Google Scholar]