Abstract
The nematode Caenorhabditis elegans is an important model organism for the study of such diverse aspects of animal physiology and behavior as embryonic development, chemoreception, and the genetic control of lifespan. Yet, even though the entire genome sequence of this organism was deposited into public databases several years ago, little is known about xenobiotic metabolism in C. elegans. In part, the paucity of detoxification information may be due to the plush life enjoyed by nematodes raised in the laboratory. In the wild, however, these animals experience a much greater array of chemical assaults. Living in the interstitial water of the soil, populations of C. elegans exhibit a boom and bust lifestyle characterized by prodigious predation of soil microbes punctuated by periods of dispersal as a non-developing alternative larval stage. During the booming periods of population expansion, these animals almost indiscriminately consume everything in their environment including any number of compounds from other animals, microorganisms, plants, and xenobiotics. Several recent studies have identified many genes encoding sensors and enzymes these nematodes may use in their xeno-coping strategies. Here, we will discuss these recent advances, as well as the efforts by our lab and others to utilize the genomic resources of the C. elegans system to elucidate this nematode’s molecular defenses against toxins.
“Think of a place where any living organisms might survive, then go and look, and you will probably find nematodes.” (Chitwood and Chitwood, ’74). In fact, nematodes can be found in almost every environmental niche including the soil, water, and as parasites of plants and animals. Due to the large economic and health burden of parasitic nematodes, much is known about the impact of these animals on their hosts and environment. In contrast, little is known about how these animals cope with the equally diverse chemistry of their diet and habitat. During the last three decades, Caenorhabditis elegans has emerged as the most popular nematode for the study of animal behavior, physiology, and molecular biology. Among the many salient features of C. elegans as a research system is a wealth of genomic resources including a completely sequenced genome (The C. elegans Sequencing Consortium, ’98), a readily accessible bioinformatics database (Chen et al., 2005), DNA microarrays (Reinke, 2002), a gene knock-out consortium (http://celeganskoconsortium.omrf.org), and the capacity for rapid and large-scale loss-of-function experiments via RNA-mediated interference (Sugimoto, 2004). These resources, along with the recent research interest in xenobiotic metabolism and the aging process have converged to help us understand how C. elegans, and perhaps other nematodes and animals, cope with toxic compounds from their environment and internal biochemistry. These advances also point to the use of C. elegans as a biosensor for environmental quality and contamination.
C. ELEGANS NATURAL HISTORY REQUIRES EFFICIENT DETOXIFICATION
In the soil, C. elegans lives within the interstitial water and has evolved a dispersal strategy utilizing a fast generation time, large brood sizes, and rapid habitat depletion (Riddle, ’97). In a short period of time, adult hermaphrodites can quickly populate a pocket of rich microbial food resources eventually exhausting the habitat of nutrients. During these periods of rapid population expansion, the animals are not only consuming their bacterial prey, they also take in the surrounding fluid along with any dissolved or suspended substances. One line of defense against the ingestion of harmful substances is behavioral; C. elegans can sense the presence of certain chemicals and avoid them (Bargmann and Mori, ’97). C. elegans can also use chemical cues to avoid pathogenic bacteria (Pujol et al., 2001; Rodger et al., 2004) and have the capability to associate bacterial odors with pathogenicity to avoid newly encountered dangers (Zhang et al., 2005). However, nematodes are not completely selective—for instance, the first descriptions of the fate of ingested particles in C. elegans were obtained by observing ingested iron particles and mineral oil (Avery and Thomas, ’97). Further, a nematode may live in a pocket of fluid where the mobility of the animal would be confined to the geographic extent of the pocket. Studies on bacterial pathogenicity on the worm also demonstrate that C. elegans will eat deleterious bacteria (Mahajan-Miklos et al., ’99). Thus, during the “boom” periods of their “boom and bust” lifestyle, detoxification of xenobiotics is critical for efficient population expansion.
This lifestyle also presents the challenge of starvation for the progeny of founder animals as the habitat becomes depleted and crowded. To avoid starvation, C. elegans is capable of entering diapause as an alternative third larval stage called a dauer. Dauer larvae have a decreased metabolism, are resistant to desiccation, do not feed, and have behaviors that promote dispersal. Although presumably not consuming xenobiotics during the dauer stage, it is likely that many endogenous toxic compounds are still produced from the basal metabolism exhibited by dauers. In addition to a pause in post-embryonic development, dauers also temporarily suspend the aging process as passage through dauer has little effect upon the lifespan of post-dauer adults (Riddle, ’88). The link between aging, dauers, and detoxification has recently been established by Joshua McElwee and colleagues (McElwee et al., 2004; Gems and McElwee, 2005). By comparing gene expression profiles of delayed and non-aging animals, daf-2 mutants (which exhibit an increased lifespan) and dauers, they identified several classes of genes that implicate detoxification as an important mechanism in aging. In addition to known aging factors, they detected decreased expression of genes linked to nutrient uptake and increased expression of detoxifying enzymes including all the major enzyme classes of phase 1 and 2 metabolism (see below). In what they call the “green theory of aging”, they have proposed that in addition to resistance to oxidative stress, dauers and long-lived mutants limit the cellular aging process primarily through increased detoxification and increased protein repair via the small heat shock proteins (McElwee et al., 2004; Gems and McElwee, 2005). Although delayed aging does not provide an evolutionary advantage of increased fitness, the increased expression of the detoxification network in dauers would increase their longevity thus promoting dispersal and resumption of the life cycle when conditions improve. Thus, detoxification of either accumulated xenobiotics or endogenously produced toxicants is also important during the “bust” and dispersal period of C. elegans natural history.
DETOXIFICATION ENZYMES IN C. ELEGANS
The completion of the C. elegans sequencing project has allowed many researchers to characterize nematode biology at the level of an entire genome. We wished to catalog the genomic toolbox available to C. elegans for detoxification. Using iterative BLAST and text searching, we mined the C. elegans database (www.wormbase.org, release WS142, May 8, 2005) and literature for the major classes of enzymes involved in detoxification including cytochromes P450 (CYP), short-chain dehydrogenases (SDR), UDP-glucuronosyl or glycosyl transferases (UGT), and glutathione S transferases (GST). This catalog is summarized in Table 1. Additionally, C. elegans contains numerous genes encoding other potential detoxification enzymes such as sulfotranferases, methyltrans-ferases, and acetyltranferases.
TABLE 1.
C. elegans contains a large number of genes encoding the four major classes of detoxification enzymes
| Enzyme class | # genes in C. elegans |
# with documented mRNAs |
# with documented phenotypes |
|---|---|---|---|
| CYP | 86 | 84 | 9 |
| SDR | 68 | 63 | 4 |
| UGT | 72 | 71 | 2 |
| GST | 48 | 46 | 1 |
We mined the C. elegans database (www.wormbase.org, release WS142, May 8, 2005) with text and iterative BLASTP searches to find genes and predicted genes that encode likely Cytochrome P450 (CYP), short chain dehydrogenase (SDR), UDP-glucuronosyltransferase (UGT), and glutathione S transferase (GST) enzymes.
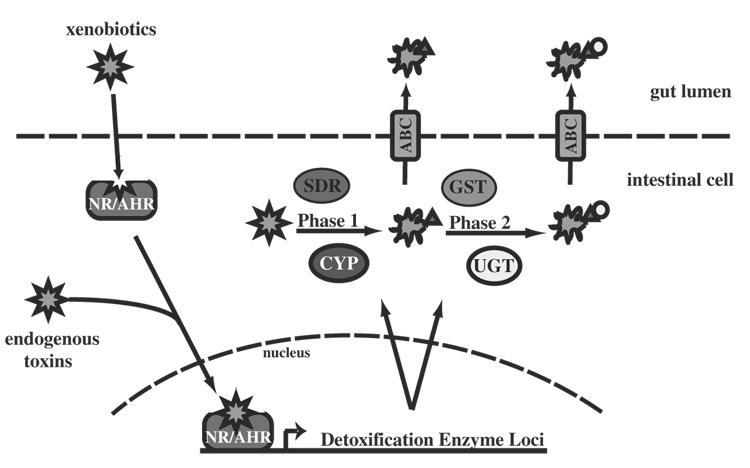
Xenobiotic metabolism is divided into two successive phases resulting in the detoxification and eventual excretion of compounds that could be detrimental to the organism (Fig. 1). Phase 1 metabolism comprises reactions that add functional groups, often hydroxyl groups, onto the offending compounds. These functional groups are normally, but not always, required for Phase 2 metabolism. The addition of the Phase 1 functional group typically results in a more polar molecule, thus making it more excretable and a more reactive substrate for phase 2 enzymes, which catalyze conjugative reactions which further increase solubility. Each phase of detoxification is catalyzed by specific enzymes, most of which reside in the smooth ER, which serves as a cellular detoxification center (Gibson and Skett, 2001).
Fig. 1.
A model for activation of the detoxification network in C. elegans. We propose that ligand activated transcription factors, such as the Nuclear Receptors (NR) and the aryl hydrocarbon receptor (AHR) activate the expression of specific detoxification enzymes for phase 1 and 2 metabolism. Phase 1 enzymes, such as the cytochromes P450 (CYP) and short chain dehydrogenases (SDR) prepare the toxic compounds for phase 2 detoxification by the UDP-glucuronosyl transferases (UGT) and glutathione S transferases (GST). Export of the modified toxins from the cell is accomplished by the ATP-binding cassette transporters (ABC).
Two classes of enzymes catalyze the reactions that make up phase 1 metabolism and allow for subsequent detoxification in phase 2: CYPs and SDRs. CYPs are the principal phase 1 enzymes and comprise a superfamily of heme-containing monoxygenases (Menzel et al., 2001). There are at least 86 genes in the C. elegans genome that encode CYP proteins; eight of these are predicted to be pseudogenes. A comprehensive list of known CYPs in C. elegans and many other species can be found on the Cytochrome P450 Homepage (drnelson.utmem.edu/CytochromeP450.html). All but two of the 86 CYP genes have documented mRNAs by their detection in SAGE (Serial Analysis of Gene Expression, Jones et al., 2001; McKay et al., 2003) and EST (Expressed Sequence Tags, Reboul et al., 2001) projects. Three large-scale RNAi projects have identified phenotypes for nine CYP genes: embryonic lethality (C01F6.3, H02I12.8, Y62E10A.15), slow and stunted growth (T10B9.1, Simmer et al., 2003), uncoordinated movement (Y80D3A.5, T10B9.1), a clear body cavity with sluggish movement and some embryonic lethality (T10B9.4, Kamath et al., 2003), and reduced fat storage (K07C6.4, K07C6.5, K09D9.2, Ashrafi et al., 2003). One CYP, DAF-9, was defined genetically and is well characterized as an important enzyme in the dauer pathway (Gill et al., 2004; Mak and Ruvkun, 2004).
The short chain dehydrogenase/reductases are found in both the smooth ER and the cytosol and catalyze the reduction of carbonyl groups in aldehydes and ketones (Gibson and Skett, 2001). At least 68 genes encode SDRs in C. elegans, 63 of which have documented mRNAs. Four of these genes have documented phenotypes, including defective growth and embryonic lethality (T02E1.5, Maeda et al., 2001) and slow growth (M03A8.1, C01G12.5, Kamath et al., 2003; Simmer et al., 2003). One gene, F01G4.2, has multiple phenotypes when disrupted including sluggish, uncoordinated movement, slow growth (Kamath et al., 2003; Simmer et al., 2003), sterile progeny, protruding vulva, and larval arrest (Simmer et al., 2003).
Phase 2 metabolism comprises the actual detoxification reactions of xenobiotic metabolism, employing the UGTs and GSTs as the main enzymatic tools for these reactions. There are at least 72 genes that code for UGT-like proteins, nine of which are likely pseudogenes (Jim Thomas, personal communication). All but one UGT have documented mRNAs while only two genes have been assigned phenotypes from RNAi experiments; T19H12.11, which causes slow growth when inactivated (Simmer et al., 2003) and C33A12.6, which when disrupted results in increased fat storage (Ashrafi et al., 2003). GSTs catalyze glutathione conjugation rendering nontoxic derivatives of poisonous compounds (Gibson and Skett, 2001). Of the 48 GST genes, 46 have documented mRNAs.
Although not detoxifying enzymes, ATP-binding cassette (ABC) transporters are also important players in xenobiotic metabolism (Staudinger et al., 2003). These transporters make up the largest family of transporters in the C. elegans genome, with 60 genes encoding likely ABC transporters (Sheps et al., 2004). Phenotypes for five of these have been elucidated, four of which show increased sensitivity to xenobiotics, bacterial toxins, or heavy metals (Broeks et al., ’95, ’96; Mahajan-Miklos et al., ’99). The fifth, ced-7, has been shown genetically to be required for proper engulfment of apoptotic cell corpses (Wu and Horvitz, ’98).
TRANSCRIPTIONAL REGULATION OF THE DETOXIFICATION NETWORK
We are primarily interested in the regulation of gene expression in response to changing environmental conditions. The transcriptional regulation of detoxification genes is an excellent example of this kind of environment–genome interaction. Rather than constitutive regulation of the suite of detoxification enzymes, which would be a monumental molecular undertaking for C. elegans given the number of loci, animals primarily regulate expression of toxin-specific enzymes via members of the nuclear receptor (NR) superfamily (Fig. 1). In mammals, two xenosensing NRs, CAR and PXR, act primarily in the liver and are responsible for detoxification of a number of known toxins that act as transactivating ligands for these receptors. Additionally, PXR and CAR, along with at least four additional NRs, are implicated in detoxification of a variety of endogenous hydrophobic compounds (Xie et al., 2004; Handschin and Meyer, 2005). Sequence comparisons of known NRs have allowed conserved NRs to be grouped based upon apparent ancestral lineage of this superfamily (Escriva et al., ’97; Laudet, ’97). Thus far, the xenosensing NRs are all members of the NR1I subfamily (Handschin and Meyer, 2005). In C. elegans, there are three members of this subfamily, NHR-8, DAF-12, and NHR-48 (Lindblom et al., 2001; Maglich et al., 2001). (In C. elegans nomenclature, “NHR” refers to Nuclear Hormone Receptor due to the similarity of nematode NRs to the vertebrate steroid hormone receptors which are founding members of the NR superfamily.)
The best studied of these nematode NRs is DAF-12 (Daf5Dauer Defective) which is a key regulator in the dauer pathway. DAF-12 integrates signals from insulin-like growth factor and TGF-β signaling systems to determine the identity of the third larval stage as developing or dauer (Antebi et al., ’98). Recently, a partially purified mix of methylated sterols called gramravali was discovered to activate DAF-12, indicating that sterols play an endocrine role in C. elegans post-embryonic development (Matyash et al., 2004). Although yet to be biochemically demonstrated, the activation of DAF-12 by such a sterol would be the first demonstrated ligand for a nematode NR. Of the C. elegans NR1I NRs, NHR-48 is the least well characterized. Although multiple transcripts for NHR-48 have been detected (Antebi et al., 2000), no phenotype has been detected or reported from RNAi experiments (Gissendanner et al., 2004) or from a deletion allele (www.wormbase.org, release WS142, May 8, 2005).
While it appears unlikely that DAF-12 is involved in detoxification and the function of NHR-48 remains unknown, in keeping with the likely ancestral function of the NR1I subfamily, NHR-8 is required for xenobiotic resistance in C. elegans. The nhr-8 locus is expressed in the intestine beginning in late embryogenesis and continuing throughout larval development. Without NHR-8, C. elegans is more susceptible to colchicine and chloroquine, two xenobiotic targets of CYP hydroxylation (Lindblom et al., 2001). This sensitivity is similar to that exhibited by animals lacking the ABC transporter, PGP-3 (Broeks et al., ’95). Thus, the pgp-3 locus may be a transcriptional target of NHR-8. However, pgp-3 mutant animals are also more sensitive to the bacterial toxin phenazine (Mahajan-Miklos et al., ’99) and nhr-8 mutants are not, indicating that the two proteins share some xenocoping responsibilities but also have distinct toxin specificities as well (Lindblom et al., 2001). This result also indicates that while NHR-8 may play some role in the transcriptional regulation of pgp-3, it cannot account for the complete production of PGP-3.
Conservation of function based upon conservation of sequence was a driving hypothesis for the characterization of NHR-8 and other C. elegans NRs (Gissendanner and Sluder, 2000; Lindblom et al., 2001). However, of the astounding number of NRs in C. elegans, 270+, only 15 show conservation with NRs from other phyla (Sluder et al., ’99). Thus, the vast majority of NRs in worms do not yield clues to their function based upon homology. These divergent or supplementary NRs can be traced back to a rapid evolutionary expansion from a single NR ancestor of HNF4. Much of the expansion of the supplementary NRs occurred on chromosome V (Robinson-Rechavi et al., in press), analogous to the expansion of chemoreceptor genes (Robertson, 2001). Several authors have indicated that the large number of NRs in C. elegans could be a mechanism for responding to changing and diverse conditions in their environment (Sluder et al., ’99; Van Gilst et al., 2002). Evidence to support this hypothesis is yet to be reported and will likely require creative high-throughput assays for examining the function of this large superfamily.
In addition to NRs, cytochrome P450 genes are also regulated by the aryl hydrocarbon receptor (AHR) in mammals in response to xenobiotics (Nebert et al., 2004). C. elegans contains one AHR gene, ahr-1, as well as a gene encoding an AHR nuclear translocator (ARNT), aha-1 (Powell-Coffman et al., ’98). The mammalian receptor is tethered in the cytoplasm by heat shock proteins and translocates to the nucleus with ARNT upon activation by ligand binding. The C. elegans receptor, AHR-1, binds to nematode heat shock proteins and is capable of binding mammalian xenobiotic response elements in cooperation with AHA-1, but does not bind to known AHR ligands (Powell-Coffman et al., ’98).
GENOMICS APPLIED TO UNDERSTANDING DETOXIFICATION IN C. ELEGANS
The advent of genome-wide resources in the C. elegans research system has prompted many large-scale analyses of genome content, gene expression, and gene function with respect to several time courses and biological functions. However, only recently have these powerful tools been applied to understanding xenocoping strategies in C. elegans. One study by Menzel et al. (Menzel et al., 2001) utilized a variety of genomic approaches to specifically probe the regulation of phase 1 enzyme expression. Menzel and his colleagues exposed C. elegans populations to 18 compounds known to induce the expression of CYP genes in other organisms. They then analyzed the resultant mRNA production with a C. elegans CYP gene filter, semi-quantitative RTPCR of selected CYP genes, whole genome DNA microarrays, and GFP reporter constructs. From this analysis, they discovered that 14 of the more than 80 CYP genes are specifically induced in response to particular CYP substrates. Additionally, they also recognized upregulated expression of UGT and GST transcription with the whole genome DNA microarray (Menzel et al., 2001). This study illustrates the responsiveness of the detoxification network but does not define the mechanism of the response. One obvious hypothesis is that these and other genes are regulated by ligand activated transcription factors such as the NRs and AHR-1. We are currently investigating this hypothesis with detoxification enzyme-specific and whole genome DNA microarrays. Alternatively, these genes could be regulated by other transcription factors such as SKN-1, which has homology to bZIP and CNC-group proteins in vertebrates (An and Blackwell, 2003), although a mechanism for ligand activation is not readily apparent.
Several other studies utilizing genomic tools have also discovered conditions that result in increased expression of detoxification enzymes. Several of these were aimed at discovering genes expressed in response to the dauer pathway. As mentioned above, by comparing data sets from DNA microarray gene expression profiles from dauers and long-lived daf-2 mutants, McElwee and colleagues revealed upregulated expression of CYP, UGT, and SDR genes in dauers and daf-2 mutants with the addition of GST genes in the long-lived mutants (McElwee et al., 2004). Similarly, Wang and Kim used DNA microarrays to analyze gene expression throughout the dauer stage while focusing on gene expression during the exit from dauer. Like the previously mentioned study, these researchers also showed that detoxification enzymes, specifically CYPs and UGTs, are expressed during dauer with decreasing expression as the larvae exit the dauer stage to continue post-embryonic development. During the early stages of dauer exit, a second distinct group of CYPs and UGTs are transiently expressed. Additionally, transient expression of drug transporters and NRs was also observed during exit from dauer (Wang and Kim, 2003). From the published data on gene expression and dauers, it is clear that the dauer stage requires the expression of a number of important components of the detoxification machinery. Presumably, this confers an increased level of toxin resistance for cellular health and longevity during diapause. However, several authors have also pointed out that these enzymes may be involved in the production of dauer inducing and/or exit hormones (Wang and Kim, 2003; Matyash et al., 2004). Indeed, one CYP, DAF-9, is thought to be a key metabolic enzyme required for the production of the dauer inducing hormone and DAF-12 ligand (Gill et al., 2004; Mak and Ruvkun, 2004).
In addition to genome-wide analysis of gene expression, the C. elegans system also offers genome-wide analysis of gene function using high-through-put RNAi. There are two typical strategies to approach the functions of large numbers of genes by RNAi—experiments to inactivate many genes with an unbiased scoring of many phenotypes and screens of large numbers of genes with a biased scoring of phenotypes to understand particular processes. The former has resulted in phenotypes for a few of the genes described in this paper and they are included above in the description of detoxification enzymes in C. elegans. The latter has also resulted in assigning function to many C. elegans genes including many detoxification genes. Several detoxification enzyme genes and NRs were shown to be involved in fat metabolism in a genome-wide RNAi analysis for fat storage and mobilization. Ashrafi et al. (2003) performed RNAi for almost every gene in C. elegans and examined the resultant RNAi animals for increased or decreased fat storage which was indicated by staining with a vital dye for fat. Among the many genes that when perturbed caused a reduction in body fat storage were two CYPs, an ABC transporter, and two NRs, daf-12 and nhr-88. Included in the genes that increase body fat content when inactivated are several NRs including nhr-8, the NR gene implicated in xenobiotic resistance (see above). Although the connection between fat storage and these genes is currently unknown, this research illustrates how the application of a whole genome approach is likely to shed light on the connections between typically isolated research objectives, including detoxification.
One of the more intriguing uses of genomics tools to understand C. elegans biology that has implications for xenobiotic detoxification is an assessment of gene clusters in the worm genome. Jim Thomas developed and applied an algorithm to the C. elegans genome to look for clusters of related genes that would indicate sequence duplication and potentially high rates of evolution. He discovered that genes encoding proteins implicated in environmental response are enriched in the list of clustered genes. These genes include members of the seven-pass transmembrane domain proteins responsible for various sensory processes, various gene families involved in innate immunity, and the four major classes of detoxification enzymes—CYP, SDR, UGT, and GST families. The nematode-specific NRs are also clustered in a similar manner. Groups of clustered genes are speculated to participate in a common biological process; therefore, duplication of a cluster would allow the daughter cluster to evolve to meet a new environmental challenge (Thomas, 2005). For C. elegans, this is particularly intriguing given the life history of the animal in the wild. Upon meeting a new xenobiotic challenge, nematodes with a duplicated “detoxification cluster” would be equipped to evolve the detoxification machinery to fit the new chemical challenge.
Given this hypothesis, one would expect broad divergence of drug metabolism capacity when comparing detoxification enzyme sequences from C. elegans and other nematodes. Brugia malayi is human parasitic nematode that causes lymphatic filariasis or elephantiasis. B. malayi passes through an intermediate insect vector for larval development and dispersal (Blaxter and Bird, ’88). Thus, the chemistry of this nematode’s habitats, while complex, remains relatively constant compared to free living species such as C. elegans and Caenorhabditis briggsae. As of May 2005, The Institute for Genomic Research has sequenced the B. malayi genome to more than eight-fold redundancy is available for BLAST searches on their website (www.tigr.org). We mined the available genome sequence of B. malayi for sequences encoding drug metabolizing enzymes and found that compared to the free living C. elegans and C. briggsae, B. malayi have remarkably few sequences encoding detoxification enzymes. Compared to C. elegans, B. malayi contains as few as 16% of the total number of sequences encoding CYP, SDR, UGT, and GST enzymes (manuscript in preparation). We speculate that the disparity between C. elegans and B. malayi is due to the differences in the natural history of the two species as free living nematodes are under selective pressure for more detoxification due to chemical flux of their habitats.
C. ELEGANS AS A BIOSENSOR
While the study of xenobiotic detoxification in C. elegans offers a glimpse into nematode and perhaps general detoxification strategies, the system has also been tapped as a metazoan biomonitoring system. Although C. elegans is a multicellular organism, it is simple enough that cellular activity can easily be monitored. Its short life cycle allows it to be utilized for rapid toxicity assessment as well as an indicator of the effects that toxicants have on subsequent generations. It is important for researchers to find test species that are sensitive enough to xenobiotics so as to provide viable data, while at the same time utilizing an appropriate endpoint that allows for these data to be readily collected. C. elegans fulfills both of these requirements.
C. elegans as a biosensor offers several endpoints that can serve as markers of cellular stress, such as lethality, gene expression, and behavioral changes. Lethality of either the exposed animals (Ura et al., 2002) or lethality in terms of reproductive disruption has been tested for a variety of xenobiotic compounds such as steroids, phenols, and a variety of xenobiotics (Tominaga et al., 2002, 2003a,b, 2004). Assessment of lethality has been used to screen a variety of antihelmintic drugs (Simpkin and Coles, ’81) and to test soil samples for availability of metal ions to resident nematodes (Boyd and Williams, 2003a).
While lethality is certainly an indicator of cellular stress, the more subtle expression of transgenes in response to stressors allows for a more sensitive detection of toxic irritation. Several studies have utilized reporter constructs to analyze gene expression in response to xenobiotics. Most of these studies have utilized the LacZ reporter gene driven by the promoter for an endogenous heat shock protein gene. In response to stressors including xenobiotics and heat shock, they demonstrate the expression of the transgene as an indicator of overall cellular stress (Jones et al., ’96; Guven et al., ’99; Easton et al., 2001). This system was used to test an English river system for contaminating heavy metals (Mutwakil et al., ’97), to detect non-thermal stress from microwaves (de Pomerai et al., 2000), and evaluate the toxicity of several kinase inhibitors (Dengg and van Meel, 2004). A variation on this theme, one that does not require histochemical staining following exposure, is the use of the heat shock promoter driving expression of green fluorescent protein (Link et al., ’99; de Pomerai et al., 2000). One study utilized a constitutive promoter to drive expression of the bioluminescent firefly luciferase and assayed a loss of luminescence in response to temperature, metal ions, and xenobiotics (Lagido et al., 2001), although it appears that the assay itself is toxic to C. elegans (Hollis et al., 2001).
A particularly sensitive approach to using C. elegans as a biosensor has been developed that uses computer-assisted videotracking of nematodes in the presence of various toxicants. This assay has been used to distinguish metals, pesticides, and solvents that are thought to be neurotoxins from those that are toxic but non-neuronally targeted (Anderson et al., 2001). One study used this system to concurrently test survival, reproduction, movement, and feeding behavior in C. elegans exposed to copper (Boyd and Williams, 2003b). As a test for neurotoxicity of 15 different xenobiotic compounds including many insecticides, this system was used to rank toxicity of the compounds for comparison with known values in mice and rats. The researchers found significant correlation between the ranked order of toxicity on C. elegans, rats, and mice indicating the suitability of this system for analysis of potential neurotoxicity in mammals (Cole et al., 2004).
In a similar way to its impact on general biological knowledge, whole genome approaches are also likely to dramatically impact the power of C. elegans as a biosensor. Custodia and colleagues demonstrated this nicely when they examined global changes in gene expression in response to vertebrate steroids. The same laboratory had previously determined that contaminating endocrine disrupters were likely responsible for the reductions in a freshwater turtle population and sought to test C. elegans as a possible biosensor for these xenobiotics. What distinguishes this study from those mentioned above is the utilization of changes in global gene expression as an endpoint. In addition to a Western analysis of changes in vitellogenin production, a known indicator of vertebrate endocrine disruption, they probed DNA microarrays with RNA purified from nematodes exposed to progesterone and estrogen. They found significant changes in gene expression for vitellogenin, metallothionein, GST, CYP, and heat shock genes. Importantly, the changes in expression, either up- or down-regulated, were specific to each hormone (Custodia et al., 2001) indicating that this test could distinguish the vertebrate steroids. Although yet to be fully realized, the use of genome-wide tools as endpoints could therefore not only be used to test the toxicity of environmental samples but may also provide a mechanism for the identification of the chemical source of the toxicity. Additionally, such a system, once fully characterized, would allow the prescreening of pharmacological compounds for potential toxicity in vertebrates.
CONCLUSION
Compared to many cellular processes, our knowledge of the molecular mechanisms of detoxification in C. elegans has lagged behind. However, as the genomics resources for this model organism have become available, the knowledge of this important aspect of nematode biology has likewise increased. Nematodes are a major component of the biosphere inhabiting almost every niche, including plant and animal hosts. As we uncover the mechanisms of detoxification, we gain insight into ways to fight parasitic nematodes, the use of free living nematodes as environmental and pharmacological sensors, aging in nematodes and perhaps animals in general, and the biochemistry of toxin metabolism.
ACKNOWLEDGMENTS
We wish to thank Ann Sluder and Ian Callard for the opportunity to write the paper. We also thank A.S. for many thoughtful discussions including the suggestion to look for sequences in the B. malayi genome. Chris Gissendanner provided much insight and a critical review of the manuscript. We are especially grateful to James Thomas for communicating unpublished data and to Brianne Orr for assisting with the database searches. We wish to acknowledge the very important database resources at Wormbase, and the Institute for Genomic Research. The International Brugia Genome Sequencing Project and is supported by an award from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Grant sponsor: National Institutes of Health grant; Grant number: #R15 ES013128-01.
LITERATURE CITED
- An JH, Blackwell TK. SKN-1 links C. elegans mesendo-dermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GL, Boyd WA, Williams PL. Assessment of sublethal endpoints for toxicity testing with the nematode Caenorhabditis elegans. Environ Toxicol Chem. 2001;20:833–838. [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Avery L, Thomas JH. Feeding and defecation. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 679–716. [PubMed] [Google Scholar]
- Bargmann CI, Mori I. Chemotaxis and thermotaxis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 717–738. [PubMed] [Google Scholar]
- Blaxter M, Bird D. Parasitic nematodes. In: Wood WB, editor. The nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. [PubMed] [Google Scholar]
- Boyd WA, Williams PL. Availability of metals to the nematode Caenorhabditis elegans: toxicity based on total concentrations in soil and extracted fractions. Environ Toxicol Chem. 2003a;22:1100–1106. [PubMed] [Google Scholar]
- Boyd WA, Williams PL. Comparison of the sensitivity of three nematode species to copper and their utility in aquatic and soil toxicity tests. Environ Toxicol Chem. 2003b;22:2768–2774. doi: 10.1897/02-573. [DOI] [PubMed] [Google Scholar]
- Broeks A, Janssen HW, Calafat J, Plasterk RH. A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J. 1995;14:1858–1866. doi: 10.1002/j.1460-2075.1995.tb07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A, Gerrard B, Allikmets R, Dean M, Plasterk RH. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 1996;15:6132–6143. [PMC free article] [PubMed] [Google Scholar]
- Chen N, Harris TW, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Canaran P, Chan J, Chen CK, Chen WJ, Cunningham F, Davis P, Kenny E, Kishore R, Lawson D, Lee R, Muller HM, Nakamura C, Pai S, Ozersky P, Petcherski A, Rogers A, Sabo A, Schwarz EM, Van Auken K, Wang Q, Durbin R, Spieth J, Sternberg PW, Stein LD. WormBase: a comprehensive data resource for Caenorhab-ditis biology and genomics. Nucleic Acids Res. 2005;33(Database issue):D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood BG, Chitwood MB. Introduction to nematology. Baltimore: University Park Press; 1974. [Google Scholar]
- Cole RD, Anderson GL, Williams PL. The nematode Caenorhabditis elegans as a model of organophosphate-induced mammalian neurotoxicity. Toxicol Appl Pharmacol. 2004;194:248–256. doi: 10.1016/j.taap.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Custodia N, Won SJ, Novillo A, Wieland M, Li C, Callard IP. Caenorhabditis elegans as an environmental monitor using DNA microarray analysis. Ann NY Acad Sci. 2001;948:32–42. doi: 10.1111/j.1749-6632.2001.tb03984.x. [DOI] [PubMed] [Google Scholar]
- de Pomerai D, Daniells C, David H, Allan J, Duce I, Mutwakil M, Thomas D, Sewell P, Tattersall J, Jones D, Candido P. Non-thermal heat-shock response to microwaves. Nature. 2000;405:417–418. doi: 10.1038/35013144. [DOI] [PubMed] [Google Scholar]
- Dengg M, Van Meel JC. Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. J Pharmacol Toxicol Methods. 2004;50:209–214. doi: 10.1016/j.vascn.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Easton A, Guven K, de Pomerai DI. Toxicity of the dithiocarbamate fungicide mancozeb to the nontarget soil nematode, Caenorhabditis elegans. J BiochemMol Toxicol. 2001;15:15–25. doi: 10.1002/1099-0461(2001)15:1<15::aid-jbt2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Escriva H, Safi R, Hanni C, Langlois MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/ IGF-1 signaling? Mech Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Gibson GG, Skett P. Introduction to drug metabolism. Cheltenham, UK: Nelson Thornes; 2001. p. 256. viii. [Google Scholar]
- Gill MS, Held JM, Fisher AL, Gibson BW, Lithgow GJ. Lipophilic regulator of a developmental switch in Caenorhabditis elegans. Aging Cell. 2004;3:413–421. doi: 10.1111/j.1474-9728.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Guven K, Power RS, Avramides S, Allender R, De Pomerai DI. The toxicity of dithiocarbamate fungicides to soil nematodes, assessed using a stress-inducible transgenic strain of Caenorhabditis elegans. J Biochem Mol Toxicol. 1999;13:324–333. doi: 10.1002/(sici)1099-0461(1999)13:6<324::aid-jbt6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Handschin C, Meyer UA. Regulatory network of lipidsensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Arch Biochem Biophys. 2005;433:387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hollis RP, Lagido C, Pettitt J, Porter AJ, Killham K, Paton GI, Glover LA. Toxicity of the bacterial luciferase substrate, n-decyl aldehyde, to Saccharomyces cerevisiae and Caenorhabditis elegans. FEBS Lett. 2001;506:140–142. doi: 10.1016/s0014-5793(01)02905-2. [DOI] [PubMed] [Google Scholar]
- Jones D, Stringham EG, Babich SL, Candido EP. Transgenic strains of the nematode C. elegans in biomonitoring and toxicology: effects of captan and related compounds on the stress response. Toxicology. 1996;109:119–127. doi: 10.1016/0300-483x(96)03316-1. [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Lagido C, Pettitt J, Porter AJ, Paton GI, Glover LA. Development and application of bioluminescent Caenorhabditis elegans as multicellular eukaryotic biosensors. FEBS Lett. 2001;493:36–39. doi: 10.1016/s0014-5793(01)02271-2. [DOI] [PubMed] [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- Lindblom TH, Pierce GJ, Sluder AE. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr Biol. 2001;11:864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, Carrick K, Kamdar K, Willson TM, Moore JT. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0029. RESEARCH0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Mak HY, Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development. 2004;131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- Matyash V, Entchev EV, Mende F, Wilsch-Brauninger M, Thiele C, Schmidt AW, Knolker HJ, Ward S, Kurzchalia TV. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004;2:e280. doi: 10.1371/journal.pbio.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJ, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Menzel R, Bogaert T, Achazi R. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch Biochem Biophys. 2001;395:158–168. doi: 10.1006/abbi.2001.2568. [DOI] [PubMed] [Google Scholar]
- Mutwakil MH, Reader JP, Holdich DM, Smithurst PR, Candido EPM, Jones D, Stringham EG, de Pomerai DI. Use of stress-inducible transgenic nematodes as biomarkers of heavy metal pollution in water samples from an English river system. Arch Environ Contam Toxicol. 1997;32:146–153. doi: 10.1007/s002449900167. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci USA. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Tzellas N, Thierry-Mieg N, Moore T, Jackson C, Shin-i T, Kohara Y, Thierry-Mieg D, Thierry-Mieg J, Lee H, Hitti J, Doucette-Stamm L, Hartley JL, Temple GF, Brasch MA, Vandenhaute J, Lamesch PE, Hill DE, Vidal M. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- Reinke V. Functional exploration of the C. elegans genome using DNA microarrays. Nat Genet. 2002;32 Suppl:541–546. doi: 10.1038/ng1039. [DOI] [PubMed] [Google Scholar]
- Riddle DL. The dauer larva. In: Wood WB, editor. The nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Riddle DL. Introduction to C. elegans. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. xvii–1222. [PubMed] [Google Scholar]
- Robertson HM. Updating the str and srj (stl) families of chemoreceptors in Caenorhabditis nematodes reveals frequent gene movement within and between chromosomes. Chem Senses. 2001;26:151–159. doi: 10.1093/chemse/26.2.151. [DOI] [PubMed] [Google Scholar]
- Rodger S, Griffiths BS, McNicol JW, Wheatley RW, Young IM. The impact of bacterial diet on the migration and navigation of Caenorhabditis elegans. Microb Ecol. 2004;48:358–365. doi: 10.1007/s00248-003-0201-1. [DOI] [PubMed] [Google Scholar]
- Sheps JA, Ralph S, Zhao Z, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, Van der Linden AM, Kuijk E, Van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkin KG, Coles GC. The use of Caenorhabditis elegans for anthelmintic screening. J Chem Tech Biotechnol. 1981;31:66–69. [Google Scholar]
- Sluder AE, Mathews SW, Hough D, Yin VP, Maina CV. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- Staudinger JL, Madan A, Carol KM, Parkinson A. Regulation of drug transporter gene expression by nuclear receptors. Drug Metab Dispos. 2003;31:523–527. doi: 10.1124/dmd.31.5.523. [DOI] [PubMed] [Google Scholar]
- Sugimoto A. High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004;72:81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Thomas JH. Analysis of homologous gene clusters in C. elegans reveals striking regional cluster domains. Genetics. 2005 doi: 10.1534/genetics.104.040030. Epub ahead of print, November 15, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga N, Tomoeda M, Kohra S, Takao U, Nagae M, Ueda K, Ishibashi Y, Kai T, Arizono K. A convenient sublethal assay of alkylphenol and organotin compounds using the nematode Caenorhabditis elegans. J Health Sci. 2002;48:555–559. [Google Scholar]
- Tominaga N, Kohra S, Iguchi T, Arizono K. A multi-generational sublethal assay of phenols using the nematode Caenorhabditis elegans. J Health Sci. 2003a;49:459–463. [Google Scholar]
- Tominaga N, Ura K, Kawakami M, Kawaguchi T, Kohra S, Mitsui Y, Iguchi T, Arizono K. Caenorhabditis elegans responses to specific steroid hormones. J Health Sci. 2003b;49:28–33. [Google Scholar]
- Tominaga N, Kohra S, Iguchi T, Arizono K. Effects fo perfluoro organic compound toxicity on nematode Caenorhabditis elegans fecundity. J Health Sci. 2004;50:545–550. [Google Scholar]
- Ura K, Kai T, Sakata S, Iguchi T, Arizono K. Aquatic acute toxicity testing using the nematode Caenorhabditis elegans. J Health Sci. 2002;48:583–586. [Google Scholar]
- Van Gilst M, Gissendanner CR, Sluder AE. Diversity and function of orphan nuclear receptors in nematodes. Crit Rev Eukaryot Gene Expr. 2002;12:65–88. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.40. [DOI] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Xie W, Uppal H, Saini SP, Mu Y, Little JM, Radominska-Pandya A, Zemaitis MA. Orphan nuclear receptor-mediated xenobiotic regulation in drug metabolism. Drug Discov Today. 2004;9:442–449. doi: 10.1016/S1359-6446(04)03061-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]