Abstract
MicroRNAs (miRNAs) are a class of non-coding RNAs involved in post-transcriptional gene silencing. A small number of striated muscle-specific miRNAs have been identified and shown to have an important role in myogenesis, embryonic muscle growth and cardiac function and hypertrophy. One of these myomiRs (myo = muscle + miR = miRNA), miR-206, is unique in that it is only expressed in skeletal muscle. The purpose of this review is to discuss what is currently known about miR-206 and its function in myogenesis as well as propose potential new roles for miR-206 in skeletal muscle biology. The review is also intended to serve as a comprehensive resource for miR-206 with the hope of encouraging further research on the role of miR-206 in skeletal muscle.
Keywords: microRNA, skeletal muscle, satellite cell, fiber-type, circadian
Introduction
The discovery that microRNA (miRNA) let-7 is expressed in a wide range of animals was the first evidence suggesting miRNAs might be a broad biological phenomenon and not just a novel regulatory mechanism restricted to the nematode Caenorhabditis elegans [1]. The finding that let-7 is phylogenetically conserved in bilateral animals built upon the pioneering work of the Ambros and Ruvkun laboratories and their identification of the first “short temporal RNA” lin-4 and its post-transcriptional regulation of the heterochronic gene lin-14 via a 3′-UTR mechanism [2–6]. For a first-hand, behind the scenes account of the journey that lead to the discovery of miRNAs, see [7,8]. A year after the let-7 discovery, three papers were published together that identified, by cloning and bioinformatics, ~30–50 new miRNAs in worm, fly and human [9–11]. These studies also hinted at the possibility that some miRNAs may be expressed in a tissue-specific manner. For example, the mature form of miRNA-1 (miR-1) was found to be expressed only in the human heart but not in the brain, kidney, liver, lung or HeLa cells [9,10]. The concept that some miRNAs might be tissue-specific was further supported by the finding that miR-1, -122a and -124a are expressed only in the muscle, liver and brain, respectively [12]. Shortly thereafter, Sempere and colleagues confirmed and expanded on these results by identifying 30 miRNAs that are enriched or specifically expressed in a particular tissue [13].
The study of Sempere et al (2004) provided the first description of the canonical myomiRs miR-1, -133a and -206 by showing that their expression is highly enriched in both human and mouse heart and skeletal muscle [13]. Subsequent microarray studies confirmed their muscle-specificity and that expression of miR-206 is primarily restricted to skeletal muscle (see Table 1) [14–17]. Recently, the designation as a tissue-specific miRNA was formally defined as a mature miRNA that is expressed at >20-fold higher level in a specific tissue as compared to the mean expression in all other tissues; tissue-enriched was defined as <20-fold higher expression in the enriched tissue compared to other tissues [18]. Expression profiling revealed that heart and skeletal muscle, along with brain and pancreas, have the most tissue-specific miRNAs [18]. In addition to the original myomiRs, potentially new myomiRs have been identified in human and zebrafish (see Table 1) [18,19].
Table 1.
MyomiRs: muscle-specific miRNAs
| Established myomiRs |
Tissue expression |
Reference |
|
|---|---|---|---|
| miR-1 | cardiac & skeletal muscle |  |
13 |
| miR-133a | cardiac & skeletal muscle | ||
| miR-206 | skeletal muscle | ||
| miR-208 | cardiac muscle | 22 | |
| Tentative myomiRs |
|||
| miR-95 | skeletal muscle |  |
18 |
| miR-128a† | skeletal muscle & brain | ||
| miR-302a† | cardiac muscle | ||
| miR-302c† | cardiac muscle | ||
| miR-302d | cardiac muscle | ||
| miR-367† | cardiac muscle | ||
| miR-499 | cardiac & skeletal muscle | 19 | |
From the time of their initial identification, there has been great interest in determining the function of myomiRs in striated muscle. The demonstration that over-expression of miR-1 in HeLa cells caused a shift toward a myogenic gene profile suggested myomiRs may have a fundamental role in promoting a muscle cell identity [20]. The importance of myomiRs in muscle development was first shown by Sokol and Ambros (2005) who reported that deletion of miR-1 in flies caused premature death from a failure of skeletal muscle to properly grow during the second instar [21]. This initial observation was more recently extended to the mouse in which myomiRs miR-1-2 and miR-208 were knocked out; miR-1-2 was found to be necessary for heart morphogenesis, proper cardiac conduction and cell cycle progression whereas miR-208 was involved in the stress-dependent regulation of β-myosin heavy chain expression [22,23].
The majority of the research on myomiRs has focused on the function of miR-1 and miR-133a in cardiac development and disease [22–30] or during skeletal muscle differentiation using an in vitro model system [31–33]. Though less studied, miR-206 is unique among the myomiR family in that it is specifically expressed in skeletal muscle, being absent or expressed at relatively low levels in other tissues. Although our current understanding of miR-206 function in skeletal muscle biology is quite limited, some intriguing possibilities can be proposed based on predicted target genes. This review will discuss what is known about the role of miR-206 in skeletal muscle development and other potential functions in myogenesis and in adult skeletal muscle.
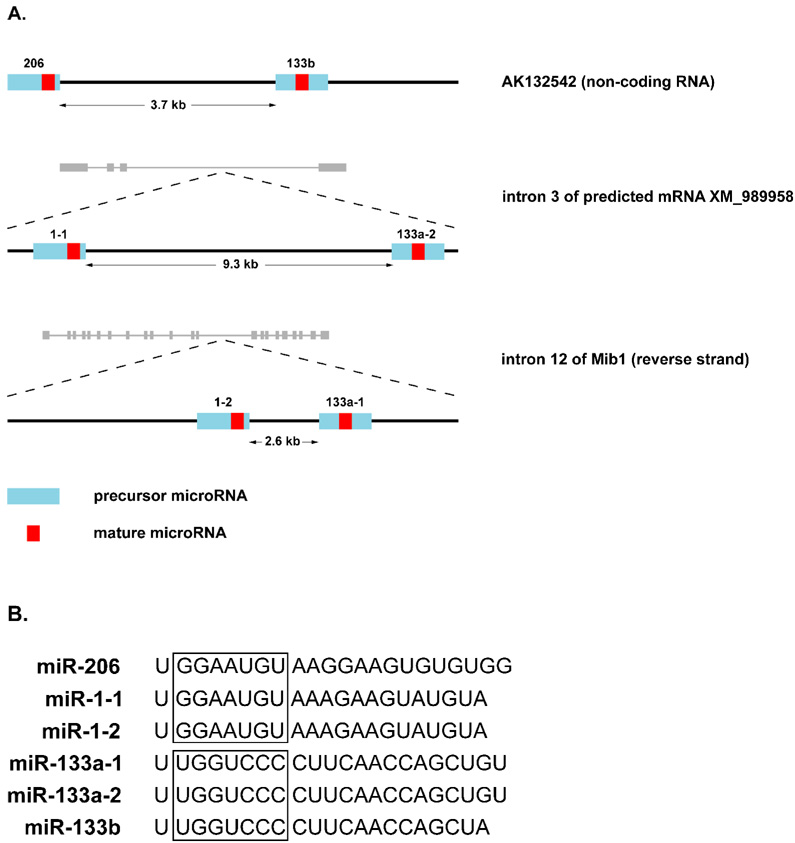
MiRNA-206 is a member of the muscle-specific miR-1 family of myomiRs. Phylogenetic analysis revealed miR-1 is an ancient miRNA, being one of only 20 miRNAs present in the last common ancestor of protostomes and deuterostomes [34,35]. The miR-1 family currently consists of six members clustered into three bicistronic pairs that most likely arose from an initial local gene duplication, giving rise to the original paralogous gene cluster (miR-1 and miR-133), followed by two “non-local” genomic duplications resulting in the new clusters being located on different chromosomes [35]. The identity of the primary miRNA transcript and location of the precursor and mature miRNAs within each transcript for each gene cluster is shown in Fig. 1A. The miR-1 family can be divided into two groups based on the sequence conservation of the seed region; either miR-1/206 or miR-133a/b (see Fig. 1B). As a result of the original local gene duplication, each bicistronic cluster consists of one member from the miR1/206 group and one from the miR-133a/b group.
Figure 1. Muscle-specific miRNAs.
A, the location of mouse precursor and mature miRNA within the primary miRNA transcript for each of the muscle-specific gene clusters: miR-206 and miR-133b are processed from the non-coding RNA AK132542A; miR-1-1 and miR-133a-2 are derived from intron three of the predicted mRNA XM_989958; miR-1-2 and miR-133a-1 are encoded in the 12th intron of the mindbomb 1 transcript on the reverse strand. B, Sequence alignment of each muscle-specific miR. The “seed” region is boxed for the miR-1/206 and the miR-133a/b groups to emphasize their respective conservation. Each gene cluster consists of a member from each of these subgroups.
Muscle-specificity
The miR-206/133b gene cluster is the most recent addition to the miR-1 family as it is only present in the vertebrate lineage whereas miR-1 and miR-133 are both conserved in the fly. Unlike other myomiR family members, miR-206 has been consistently found, by Northern blot, microarray, RNase protection assay and RT-PCR, to be highly expressed in skeletal muscle and rarely detectable in the heart (see Table S1) [13–17,36]. The skeletal muscle-specific expression of miR-206 was first clearly demonstrated by microarray analysis and later confirmed by Northern blot (see Fig. 2) [14,37]. No consistent pattern of expression for miR-206 has been shown in other non-muscle tissue (see Table S1). In contrast to the aforementioned studies, rat miR-206 was recently shown by microarray analysis to be spleen-enriched; this initial observation remains to be confirmed by RT-PCR or Northern blot [38]. MiRNA expression profiling of adult skeletal muscle from human, mouse and rat has typically found miR-206 to be one of the most abundant miRNAs expressed in skeletal muscle [13,14,16,17,36,39–41] though exceptions have been reported [15,42,43] (see Table S2). The reason for this discrepancy in the relative expression level of miR-206 is not known but could be related to the species (zebrafish) [42], the method used for profiling (cloning) [43] or possibly the muscle group as miR-206 expression has been reported to be higher in muscle composed primarily of slow-twitch/oxidative fibers i.e, soleus versus plantaris [44].
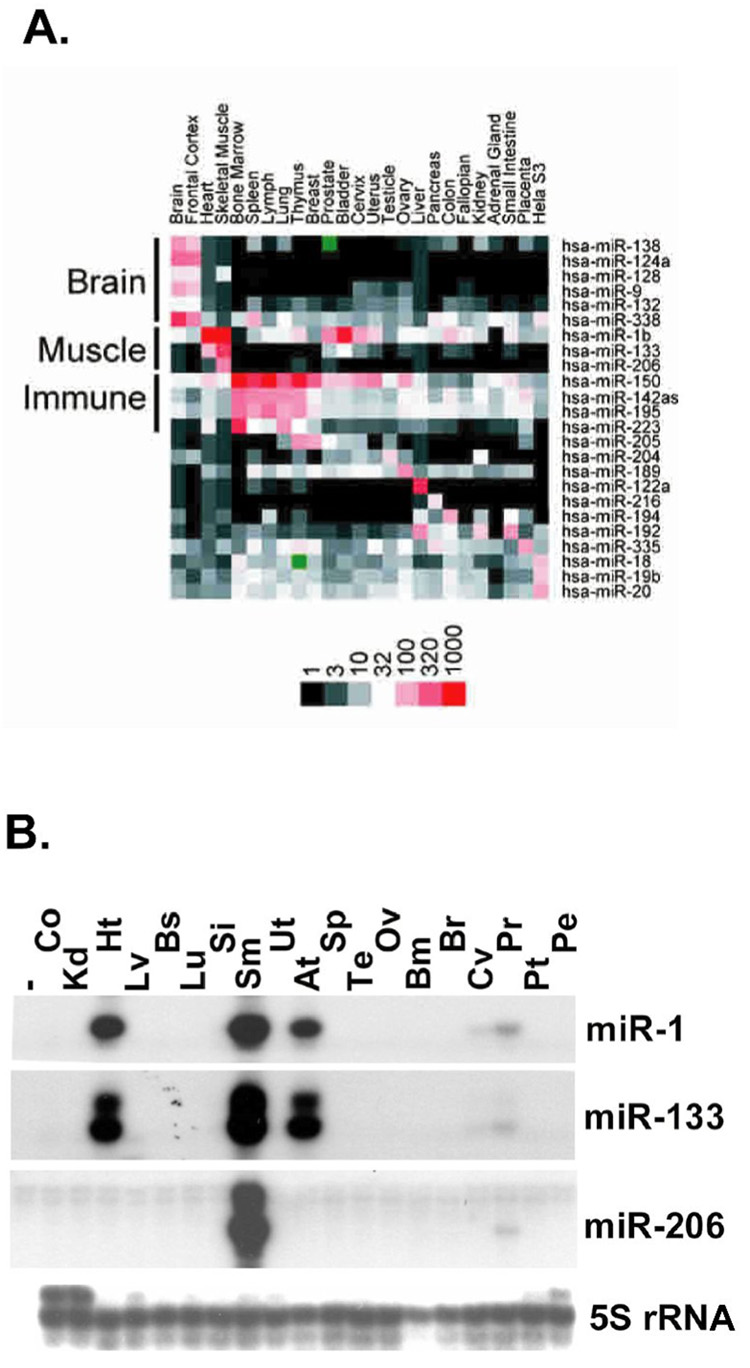
Figure 2. Skeletal muscle-specific expression of miR-206.
A, MicroRNA expression profiling of various human tissues identify the myomiRs (miR-1, -133a and -206) and show miR-206 expression is restricted to skeletal muscle. Image is reproduced from Baskerville and Bartel (2005) with permission granted from Cold Spring Harbor Laboratory Press [14].
B. RNase protection assay of different human tissues clearly shows that miR-206 is skeletal muscle specific, confirming microarray analysis presented in A; Co, colon; Kd, kidney; Ht, heart; Lv, liver; Bs, breast; Lu, lung; Si, small intestine; Sm, skeletal muscle; Ut, uterus; At, atrium; Sp, spleen; Te, testis; Ov, ovary; Bm, bone marrow; Br, brain; Cv, cervix; Pt, prostate tumor; Pe, prostate epithelium. The image is reproduced from Kim et al (2006) with permission from Rockefeller University Press [37].
Myogenesis
During embryonic development in the mouse, miR-206 is first detected at a very low level as early as 9.5 dpc and then, beginning around 11.5–12.5 dpc, miR-206 expression begins to significantly increase as determined by both Northern blot and cloning frequency [43]. Post-natal expression of miR-206 appears to peak at three days after birth and then begins to decline [45]. Similarly, in the zebrafish, miR-206 expression is low until between 12–16 hr post-fertilization at which point its expression increases over five-fold and continues to steadily increase out to 96 hr (latest time pointed examined) (see Table S3). This pattern of miR-206 expression is consistent with a global analysis of miRNA expression which found that miRNAs are generally expressed, for a given tissue, at higher levels in the embryo than in the adult [46]. Spatially, in situ hybridization studies have consistently observed miR-206 expression in the mouse, fish, frog and chick to be exclusively restricted to somites [42,47–49]. Unfortunately, to date, no studies have been published examining the expression pattern of miR-206 in adult skeletal muscle. The developmentally regulated pattern of miR-206 expression is similar to other miRNAs and is partly the basis for the hypothesis that miRNAs play an important role in cell differentiation and in the maintenance of cell identity [50].
The developmental regulation of miR-206 expression is recapitulated in vitro using the myogenic C2C12 cell line; upon initiation of differentiation there is a steady induction of miR-206 expression as well as the other myomiRs miR-1 and miR-133a [31,37,45,51]. Employing a Myod1/Myf5 null fibroblast cell line that constitutively expresses an estradiol responsive MYOD1 protein, Rosenberg et al (2006) provided multiple lines of evidence demonstrating that MYOD1 directly regulates transcription of the primary miRNA-206 transcript AK132542 and that in the absence of MYOD1, AK132542 is not expressed [52]. These same authors used chromatin immunoprecipitation (ChIP) analysis to further show MYOD1 binds to a conserved E-box located just upstream of miRNA-206, supporting earlier ChIP-chip analysis that indicated both MYOD1 and MYOGENIN bind to sequences upstream of miRNA-206 [51,52]. In the rat myogenic L6 cell line, miR-206 was found to be expressed throughout the cytoplasm and co-localized with 28S rRNA within the granular component of the nucleolus; the functional significance of the miR-206 nucleolar localization remains to be determined [53].
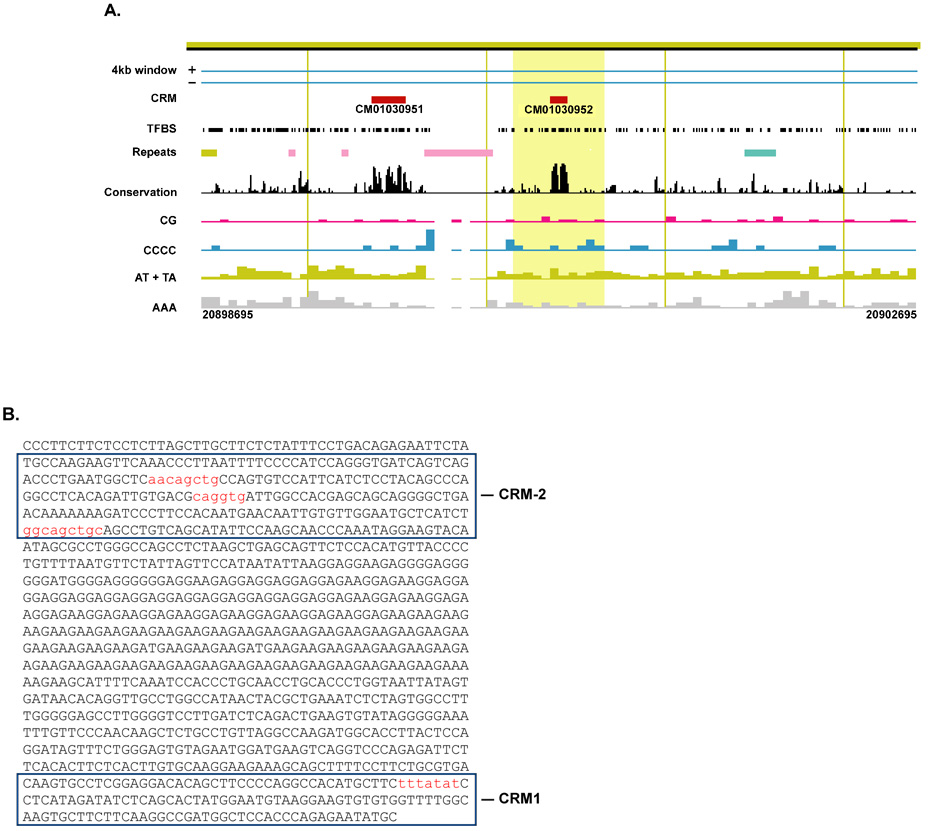
To identify cis-elements that may be involved in regulating the skeletal muscle-specific expression of miR-206, one kilobase of sequence upstream of miRNA-206 was retrieved using CisView (http://lgsun.grc.nia.nih.gov/geneindex/mm6/cisview.html). CisView analysis identified two cis-regulatory modules based on a high density of conserved cis-elements (Fig. 4). The proximal cis-regulatory module (CM01030952) spans 89 bp and is located immediately upstream of miRNA-206. Most notably this module contains highly conserved TATA-box, c-myc E-box and AML/Runx binding sites. The more distal regulatory module (CM01030951) is located approximately 800 bp upstream from miRNA-206 and is 189 bp in length. This upstream module harbors three predicted Myod1 binding sites, one of which is highly conserved. The complete set of transcription factor binding sites for both regulatory modules is provided as supplemental data (Table S6). Preliminary promoter analysis has indicated the distal upstream regulatory region is sufficient for induction during myogenic differentiation of C2C12 cells and confers Myod1 responsiveness (McCarthy, J.J. and Esser, K.A., unpublished data). These preliminary data are in good agreement with the findings of Rao and colleagues (2006) that identified a Myod1 binding site ~0.8 kb upstream of the miRNA-206 locus [51].
Figure 4. Promoter of miRNA-206 gene.
A, a screen capture from CisView schematically displays characterization of upstream sequences of miRNA-206 gene. Within this presumptive promoter sequence (~ 1kb) two highly conserved cis-regulatory modules (CRM) are identified that are separated by a stretch of low complexity repeats.
B, nucleotide sequence of miRNA-206 promoter with the two regulatory modules, CRM-1 and CRM-2, as identified by CisView boxed. Highlighted (red) is the TATA-box in CRM-1 and the three Myod1 E-boxes located in CRM-2. Using an in vitro model, preliminary data revealed CRM-2 is sufficient for miRNA-206 up-regulation during myogenic differentiation.
Established Function
Experiments designed to uncover the function of miR-206 in vivo have not been reported, however, in vitro studies using the C2C12 cell line have begun to elucidate the role of miR-206 during myogenesis. To date, four reports have used gain and loss of function experiments to rigorously test whether or not a predicted gene is an actual target of miR-206 [37,45,52,54]; the findings of each study are summarized below.
DNA polymerase alpha 1
Kim and coworkers (2006) expressed miR-206 in C2C12 myoblasts at a level comparable to that found in myotubes and then screened by microarray for genes that were predicted to be a target of miR-206 and were down-regulated [37]. The screen yielded a number of genes that fit this criteria of which the authors were able to experimentally confirm that Pola1, the largest subunit of DNA polymerase α, Rac1 interacting protein (formally known as B-ind1, butyrate induced transcript 1), Cx43 (connexin-43) and Mmd (monocyte-to-macrophage differentiation-associated protein) were direct targets of miR-206 as determined by luciferase reporter assay [37]. These authors focused on Pola1 and provided evidence showing that miR-206 inhibits DNA synthesis by repressing expression of Pola1 which, subsequently, resulted in decreased cell proliferation and the induction of cell quiescence. The major conclusion of the study was miR-206 promoted myogenesis by inducing the transition from cell proliferation to differentiation by insuring the timely down-regulation of Pola1 expression.
Connexin 43
Anderson and colleagues (2006) provided expression data and reporter gene analysis consistent with the prediction that connexin 43 (Cx43) is post-transcriptionally regulated by miR-206 during fusion of C2C12 myoblasts into myotubes [45]. Cx43 is known to be a major component of gap-junctions and has been shown to be important for muscle regeneration and in vitro differentiation [55]. It is thought that the down-regulation of Cx43 during perinatal muscle development is necessary for the proper formation of the mature neuromuscular junction (NMJ). The notion that miR-206 might have a role in NMJ formation via regulation of Cx43 is circumstantially supported by the finding that the non-coding transcript 7H4 is synaptically enriched; 7H4 was first described in 1994 and later identified as the primary miRNA for miR-206 [56,57]. Interestingly, the 7H4 transcript, in addition to be synaptically enriched, was also found to be muscle-specific and up-regulated upon denervation.
Follistatin-like 1 and Utrophin
In an effort to understand how MYOD1 activation leads to repression of follistatin-like 1 (Ftl1) expression, Rosenberg and colleagues (2006) uncovered a regulatory mechanism that involved miR-206 [52]. Employing an in vitro model, these authors showed that activation of MYOD1 increased expression of the primary miRNA-206 transcript AK132542 which in turn led to the down-regulation of Ftl1, a predicted target of miR-206. MiRNA-206 was experimentally shown to be a direct transcriptional target of MYOD1 and that Fstl1, was in turn, directly regulated via its 3′-UTR by miR-206. The function of Fstl1 is not well understood but based on its pattern of expression it is thought to have a role in early myogenesis, possibly by antagonizing myogenic regulatory factors such as Myod1 [58]. In addition to Fstl1, utrophin (Utrn) was also verified as a target gene of miR-206. Understanding the regulation of Utrn gene expression is an important area of research in the muscular dystrophy field because Utrn can rescue the dystrophic phenotype by serving as a surrogate for a defective dystrophin gene [59]. The possibility that miR-206 may be involved in repressing Utrn expression opens up a novel therapeutic strategy for modulating Utrn expression during muscular dystrophy.
Estrogen receptor alpha
A miRNA screen of estrogen receptor alpha (ERα) positive and negative breast cancer cells found that miR-206 expression was elevated in ERα negative cells [60]. A follow-up study revealed the existence of a negative feedback loop in which miR-206 and ERα repress each other’s expression [54]. Tavazoie et al., (2008), however, found a loss of miR-206 expression was associated with increased metastatic potential in human breast cancer cells and had no clear correlation to ER status [61]. Despite these contradictory findings, which may be related to the different cell types used in each study, under normal conditions ERα may function to modulate miR-206 levels in skeletal muscle. Studies in ovariectomized rats have found that changes in estrogen levels are associated with alterations in function, growth and myosin heavy chain expression in skeletal muscle [62,63]. It would be of interest to know if the responses of skeletal muscle to changes in estrogen level are in part mediated by miR-206 regulation of gene expression.
Predicted Functions of miR-206
One limitation to our understanding of miR-206 function is the small number of miR-206 target genes that have been experimentally verified. For this review, I have generated a unique list of predicted miR-206 target genes compiled from TargetScan (www.targetscan.org), PicTar (pictar.bio.nyu.edu) and Miranda (microrna.sanger.ac.uk) databases (see Table S4). The list of predicted miR-206 target genes was annotated using SymAtlas (symatlas.gnf.org/SymAtlas) and then grouped according to biological process (level 5) by DAVID (david.abcc.ncifcrf.gov/home.jsp). The results of this analysis are provided as supplemental data (Table S5) and reveal that genes associated with transcription, protein modification, transport, cytoskeleton organization and RNA metabolism are significantly over-represented. The most significantly enriched biological process was transcription, representing almost 22% of all the genes analyzed; a finding consistent with previous studies that have found miRNAs predominantly target regulators of transcription [64,65]. A closer inspection of the “regulators of transcription” list offers up some potential functions of miR-206 in skeletal muscle.
Increased skeletal muscle complexity
Among the established myomiRs, miR-206 is unique in that it is the only one specifically expressed in skeletal muscle and is the only myomiR to be exclusively expressed in vertebrates. These observations beg the question: What is different between skeletal muscle of vertebrates and invertebrates? In trying to answer this question it is important to consider the dramatic expansion in the number of vertebrate miRNAs relative to invertebrates; according to miRBase 10.0 (August 2007) there are currently 533 identified human miRNAs compared to 135 miRNAs in the worm. It has been noted that the increase in the number of miRNAs across metazoan evolution coincides with the increase in animal complexity (i.e., the number of cell types and organs) and has been proposed that miRNA regulation of gene expression may have contributed to the higher complexity of vertebrates [34,35,66]. Of the many differences that exist between vertebrate (primate, rodent and fish) and invertebrate (fly and worm) skeletal muscle, known and unknown, two major differences come to mind that can be considered as an increase in the complexity of vertebrate skeletal muscle: fly and worm skeletal muscle do not have satellite cells, the presumptive stem cell of adult skeletal muscle, nor do they possess different fiber types such as slow-twitch and fast twitch fibers as found in vertebrate skeletal muscle [67,68]. The reason for the increase in the complexity of vertebrate skeletal muscle is not definitively known but more than likely reflects the unique functional demands of the vertebrate subphylum. Though miR-206 is not proposed to be solely responsible for the emergence of satellite cells or fiber type diversity in vertebrate skeletal muscle, it is postulated that miR-206 has an important role in regulating the expression of genes involved in satellite cell specification during development and fiber type transitions in adult muscle.
Satellite cell specification
Satellite cells are the presumptive stem cell of adult skeletal muscle responsible for post-natal muscle growth and muscle regeneration from injury [69]. Satellite cells are derived from myogenic progenitor cells of the dorsal region of the somite, the dermomyotome, and require the paired box transcription factors Pax3 and Pax7 for their specification [70–74]. Furthermore, the migration of these progenitor cells to the developing limb buds requires Pax3 activation of c-Met gene expression [75–77]. Pax3 expression is subject to post-transcriptional regulation and its timely down-regulation is necessary for terminal myogenic differentiation [78]. Both Pax3 and c-Met are predicted target genes of miR-206. In particular, two highly conserved miR-206 binding sites have been identified in the 3′-UTR of Pax3 and are calculated to have the highest efficacy of any miRNA predicted to target Pax3. In addition to Pax3 and c-Met, Six4, Meox2 and Dmrt2, transcription factors also known to have a role in Pax3 regulation of early myogenesis, are predicted targets of miR-206 [79–82]. Though it is highly speculative as to whether or not miR-206 has a role in satellite cell specification, it does seem reasonable, given the number of miR-206 target genes associated with myogenesis, and the localized expression of miR-206 to the somite, that at a minimum, regulation of gene expression by miR-206 will be important for early myogenesis (see Fig. 4) [42,48]. The importance of miR-206 during myogenesis is circumstantially supported by the finding that skeletal muscle-specific Dicer-1 knockout mice have a significant reduction in muscle mass due to hypoplasia [83].
Regulation of fiber type transition
Vertebrate skeletal muscle is composed of different fiber types which possess unique contractile and metabolic properties. The fiber type composition of a given muscle is primarily determined during peri- and post-natal development but can be altered in adulthood in response to changes in contractile activity [84,85]. The plasticity of adult skeletal muscle fiber type is known to be regulated by calcium-dependent signaling pathways [86]. MiRNA-206 is predicted to regulate the expression of downstream effectors (Mef2a and Hdac4) of one branch of the calcium-dependent signaling pathway as well as a modulator the calcineurin/NFAT branch (Dscr1l1/Rcan2). Additionally, miR-206 is predicted to regulate the expression of Sox6, Purβ and Sp3 – all known transcriptional repressors of the slow myosin heavy chain gene [87,88]. In two different models in which slow myosin expression is upregulated, expression of miRNA-206 was increased; a pattern consistent with a model in which miR-206 down-regulates the expression of the aforementioned repressors of slow myosin transcription [44,89]. The idea that miRNAs may have a role in regulating skeletal muscle fiber type is supported by the finding that loss miR-214 expression leads to a reduction in slow fiber type [90]. Furthermore, Nishi and coworkers (2007) reported that over-expression of miR-206 in neonatal rat cardiomyocytes decreased the α/β myosin heavy chain ratio through regulation of retinoid X receptor alpha (RXRα) [91].
Polymorphisms
There is a growing recognition that single nucleotide polymorphisms (SNPs) which alter miRNA regulation of gene expression may be a source of phenotypic variation [92]. One of the most dramatic demonstrations of how a SNP involving a miRNA can result in an alteration in phenotype came from a study trying to identify the genetic basis for the increased muscle of mass of the Texel sheep [93]. Clop and colleagues (2006) identified a SNP within the myostatin 3′-UTR that created a functional miR-206 binding site which resulted in decreased myostatin protein expression [93]. As with the myostatin knockout mouse, the loss of myostatin expression in the sheep caused a significant increase in muscle mass [93,94]. In addition to providing a clear example of how potent miR-206 regulation can be in affecting skeletal muscle phenotype, these findings provided the first example of the degree to which a SNP involving miRNA can generate phenotypic variation [93]. Unexpectedly, a SNP (rs17578796) located within 100 bp of miR-206 was one of two miRNA SNPs identified as having a significant allelic association with schizophrenia [95]. It is difficult to reconcile these findings with the muscle-specific expression of miR-206 but, as the authors suggested, the association may be caused by a linkage disequilibrium between the miRNA SNP and some other unknown at-risk alleles [95]. A search of the human genome for polymorphisms within miRNAs themselves as well as in miRNA binding sites identified SNPs associated with miR-206 binding sites of Slc16a9, Sdc4, Polr2k and Klhdc5 genes [96]. A query of the PolymiRTS database (http://compbio.utmem.edu/miRSNP/) identified a SNP (SNPid: rs5031032) that destroys a predicted miR-206 binding site within the 3′-UTR of human insulin-like growth factor 1 (IGF-1) gene [97]. Given IGF-1’s demonstrated ability to induce muscle hypertrophy, it would be of great interest to know if the rs5031032 polymorphism is associated with increased muscle mass in humans [98]. Unfortunately, as intriguing as these SNPs may be, there currently is insufficient population allele frequency data to assess their functional significance. In addition to PolymiRTS, a second, searchable miRNA SNP database called Patrocles is available (http://www.patrocles.org/Patrocles_targets.htm) [99]. A list of human and mouse genes with SNPs that alter miR-206 binding sites, as identified by PolymiRTS and Patrocles databases, is provided as supplemental data (Table S7 and Table S8).
Splicing
The muscle-specific miR-133a has been shown to regulate alternative splicing during myogenic differentiation by repressing expression of the neural homolog of polypyrimidine tract-binding protein (nPTB) [33]. Though nPTB is a predicted target of miR-206, current evidence suggests miR-206 plays a minor role in comparison to miR-133a in regulating the expression of nPTB during differentiation [33]. Although miR-206 does not appear to regulate splicing per se, alternative splicing may impact whether or not a predicted target gene is subject to miR-206 regulation. Similar to SNPs, alternative splicing can eliminate a potential miRNA binding site located within a spliced 3′-UTR sequence. Computational analysis identified predicted miRNA binding sites which could be affected by alternative splicing events that change the 3′-UTR sequence [100]. A list of miR-206 target genes that could potentially be affected by splicing is provided as supplemental data (Table S9). Of particular interest was identification of IGF-1 as a gene in which alternative splicing could eliminate a miR-206 binding site (Table S9). It would be of interest to know if such a splice variant of IGF-1 is present in skeletal muscle and, if so, does its expression change with contractile activity and age.
Aging
The original miRNA lin-4, and its target gene lin-14, has been shown to influence life span in C. elegans through a DAF-16 dependent mechanism [101]. A decrease in lin-14 expression by lin-4 over-expression caused an increase in life span whereas inhibition of lin-4, and thus an increase in lin-14 expression, reduced life span by accelerating the rate of aging [101]. In a separate study on aging in C. elegans, the muscle-specific miR-1, along with lin-4 and let-7, showed a steady decline in expression as the worm aged [102]. The authors noted this progressive loss of miR-1 expression with age was qualitatively similar to the loss in body-wall muscle integrity observed with age in C. elegans and suggested miR-1 may have a role in this process [103]. Given that miR-206 is the vertebrate-specific homolog of miR-1, it is reasonable to speculate that its expression may also decline with age in vertebrate skeletal muscle and be involved in sarcopenia, the age-associated loss of muscle mass. Conversely, findings from a recent study found an increase in the expression of the myomiRs with muscle hypertrophy [44].
Muscle hypertrophy
MiRNA-206 is one of the most abundant miRNAs in adult skeletal muscle (Table S2). The function of miR-206 in adult skeletal muscle remains to be determined but given the importance of the muscle-specific miRNAs in muscle development, it is reasonable to propose an important role for myomiRs in skeletal muscle plasticity in the adult animal. We determined the expression of precursor and mature forms of the established myomiRs in the soleus and plantaris muscles and in the plantaris during the initial stage of muscle hypertrophy [44]. In control muscle, there was a direct correspondence in precursor and mature miRNA abundance for each of the myomiRs in both the soleus and plantaris muscles [44]. Interestingly, expression of miR-206 was 7-fold higher in the soleus muscle in comparison to the plantaris muscle suggesting miR-206 may have some role in setting fiber type [44]. During skeletal muscle hypertrophy induced by synergist ablation, transcript level of precursor miRNA-206 was elevated 18.3-fold whereas expression of miR-206 did not significantly change. The reason for this post-transcriptional regulation of miR-206 during muscle hypertrophy is not known but the authors suggested it might be the result of competitive inhibition of Drosha by ribosomal RNA (rRNA) [44]. Whatever the reason for the discordant expression of pri-miRNA-206 and miR-206 during hypertrophy, it would be of interest to determine if at a later time point, when the fast-to-slow fiber type transition is known to occur, if there is an increase miR-206 expression comparable to pri-miRNA-206 levels. If this scenario was found to be the true, it would provide further evidence to support the idea that miR-206 is involved in regulating fiber type. Determining the role of miR-206 in skeletal muscle hypertrophy and the associated phenotypic transition represent a fruitful area of research for future investigation.
Muscular dystrophy
The muscular dystrophies are a broad class of inherited myopathies distinguished by debilitating muscle wasting that often results in death as a consequence of respiratory complications [104]. Expression profiling of the established myomirs revealed a 4.5-fold increase in the expression of miR-206 in the diaphragm of mdx mouse and not hindlimb muscle; the mdx mouse is an animal model of Duchenne muscular dystrophy (DMD) [89]. This finding is significant because the diaphragm of the mdx mouse, unlike muscles of the hindlimb, exhibits the pathological characteristics of DMD thus suggesting miR-206 may have a role in the dystrophic process [105]. McCarthy and coworkers (2007) proposed that increased miR-206 expression may contribute to the chronic pathology observed in the mdx diaphragm by repressing expression of genes that otherwise would serve a compensatory function, limiting the severity of the disease as observed in the hindlimb musculature [89].
Utrophin (Utrn) is a verified target gene of miR-206 that can compensate for dystrophin and is known to be subject to a 3′-UTR-dependent post-transcriptional mechanism perfectly amenable to miRNA regulation [52,59,106,107]. In agreement with previous reports, McCarthy et al (2007) found Utrn was post-transcriptionally regulated; Utrn transcript level increased 69% whereas utrophin protein abundance increased 3.7-fold relative to control [89]. The relationship between Utrn transcript and protein levels, however, is opposite of what would be expected if Utrn expression was regulated by miR-206. What could account for the discrepancy in the findings of Rosenberg et al (2007) and McCarthy et al (2007) with respect to miRNA regulation of Utrn expression [52,89]? One possible explanation is the effect of the different experimental conditions under which Utrn expression was assessed. The dystrophic cellular environment is characterized by a metabolic crisis that leads to a heightened susceptibility to oxidative stress [108,109]. The cellular stress associated with muscular dystrophy may inhibit miRNA repression of translation as has been shown in human hepatocarcinoma cells subjected to starvation (amino acid depletion) or oxidative stress (exposure to 0.1mM sodium arsenite) [110]. It may be that under dystrophic conditions, miR-206 is not able to effectively repress translation of Utrn mRNA.
The importance of miR-206 regulation of gene expression in muscular dystrophy needs to be carefully considered in the light of a recent study by Eisenberg and colleagues [41]. Expression profiling of 10 different human dystrophies, including DMD, did not find miR-206 differentially expressed in dystrophic muscle in comparison to control muscle [41]. One possible explanation for this discrepancy is that different muscles were used in surveying miRNA expression; Eisenberg et al (2007) collected muscle samples from limb muscle (bicep and quadricep) whereas McCarthy et al (2007) found a difference in miR-206 only in the diaphragm and not the limb muscles. Alternatively, the muscle samples from the two studies were more than likely collected at different time points in the progression of the disease which is an important factor given that DMD involves cycles of degeneration-regeneration. Though no myomiRs were found to be differentially expressed in any of the dystrophies surveyed, the author did identify a set of miRNAs (miR-146b, -221, -155, -214 and -222) that were consistently dysregulated in all but one of the dystrophies, suggesting miRNAs may indeed have a role in pathology associated with the disease [41].
Circadian rhythms
The first identified miRNA, lin-4, was shown to negatively regulate expression of the heterochronic gene lin-14 [4]. Heterochronic genes act as timers during C. elegans development to insure the proper temporal specification of cell fate in somatic tissue. A RNAi screen of circadian gene homologs in C. elegans revealed that three of these homologs functioned as heterochronic genes, suggesting the intriguing possibility that circadian rhythms may be subject to regulation by miRNAs [111]. The novel idea that miRNAs may control circadian rhythms was confirmed by the demonstration that miR-132 and miR-219-1 regulate the circadian clock of the suprachiasmatic nucleus (SCN) [112]. MiRNA-219-1 was shown to be a direct transcriptional target of the CLOCK:BMAL1 heterodimer, i.e. a clock-controlled gene and is involved in setting the period length of the circadian cycle. On the other hand, the circadian expression of miR-132 was regulated by the light-inducible transcription factor CREB and shown to be involved in photic entrainment of the molecular clock. The additional identification of 12 circadianly expressed miRNAs in the retina suggests there may be a significant number of miRNAs whose expression oscillates over the course of 24 hours [113]; whether such proposed circadian miRNAs modulate the timing of the circadian clock, as do miR-219-1 and miR-132, remains to determined.
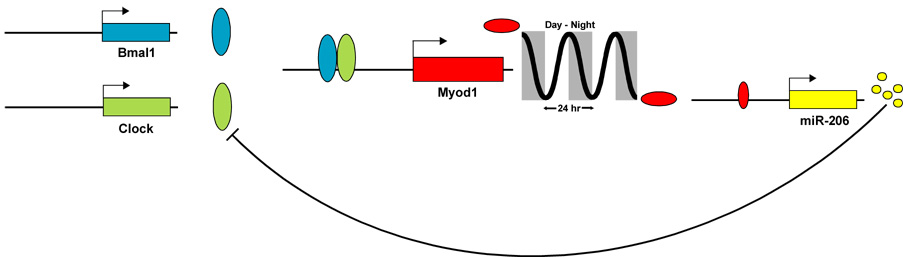
A predicted target of miR-206 is the Clock gene, a critical component of the core circadian clock. The Myod1 gene, which is known to regulate miR-206 expression, was recently identified as a constituent of the skeletal muscle circadian transcriptome in addition to being a clock-controlled gene [52,114]. Based on these observations, a model is proposed involving a Clock-Myod1-miR-206 feedback loop controlling the timing of the circadian cycle in skeletal muscle (see Fig. 5). The model proposes the CLOCK:BMAL1 heterodimer drives circadian expression of Myod1 and, in turn, Myod1 activates miR-206 expression. MiRNA-206 subsequently represses expression of Clock. Inhibition of Clock protein expression by morpholino oligonucleotide showed Clock expression is necessary to maintain normal circadian rhythmicity through regulation of Period 2 levels [115]. Preliminary results indicate that over-expression of miR-206 in C2C12 cells can reduce Clock protein abundance, without altering Clock mRNA level, as well as decrease expression of a reporter gene containing the 3′-UTR of Clock (McCarthy, J.J. and Esser, K.A., unpublished data). This model is appealing because it provides a plausible mechanism through which tissue-specific factors such as Myod1 and miR-206 can convey unique tissue requirements to the circadian clock. The exciting possibility that miR-206 regulates the circadian clock in skeletal muscle awaits experimental validation.
Figure 5. MiRNA regulation of circadian rhythms in skeletal muscle.
A diagram of a proposed model in which miR-206 is involved in a negative feedback loop that regulates Clock expression. Evidence to support the model are as follows; Expression profiling identified Myod1 as a clock-controlled gene with a circadian pattern of expression [114]. In turn, Myod1 has been shown to drive expression of primary miRNA-206 transcript AK132542 leading to increased expression of miR-206 [52]. MiRNA-206 is predicted to target Clock gene by three different databases (TargetScan, Miranda and PicTar). The model provides a novel mechanism by which skeletal muscle-specific factors (Myod1 and miR-206) can regulate circadian rhythms in a tissue-specific manner.
Concluding remarks
MiRNA-206 is unique among the muscle-specific miRNAs in that it is exclusively and highly expressed in skeletal muscle. Regulation of gene expression by miR-206 has a profound effect on skeletal muscle phenotype as shown by the work of Clop et al.,[93]. The characterization of miR-206 expression has provided a clear picture of the timing and localization of miR-206 during embryogenesis. Functional studies have validated target genes and demonstrated a role for miR-206 in myogenesis. In adult skeletal muscle, the function of miR-206 is unknown but circumstantial evidence strongly suggests a potential role in hypertrophy, fiber type switching and the pathophysiology of muscular dystrophy. Based on predicted target genes, additional roles for miR-206 in satellite cell specification, sarcopenia and circadian rhythms have been proposed. The key question that remains to be addressed is why is miR-206 expressed only in vertebrate skeletal muscle? Using this basic question to guide future research is anticipated to uncover a fundamental role for miR-206 in skeletal muscle biology.
Supplementary Material
Table S1. Expression level of miR-206 in different tissues.
Table S2. Expression level of miRs in skeletal muscle.
Table S3. Developmental pattern of expression of miR-206.
Table S4. Annotation of predicted target genes of miRNA-206.
Table S5. Gene ontology of predicted target genes of miRNA-206.
Table S6. Transcription factor binding sites within CRM1 and CRM2.
Table S7. SNP associated with microRNA-206 according to Patrocles database.
Table S8. SNP associated with microRNA-206 according to PolymiRTS database.
Table S9. MicroRNA-206 predicted target genes that are alternatively spliced.
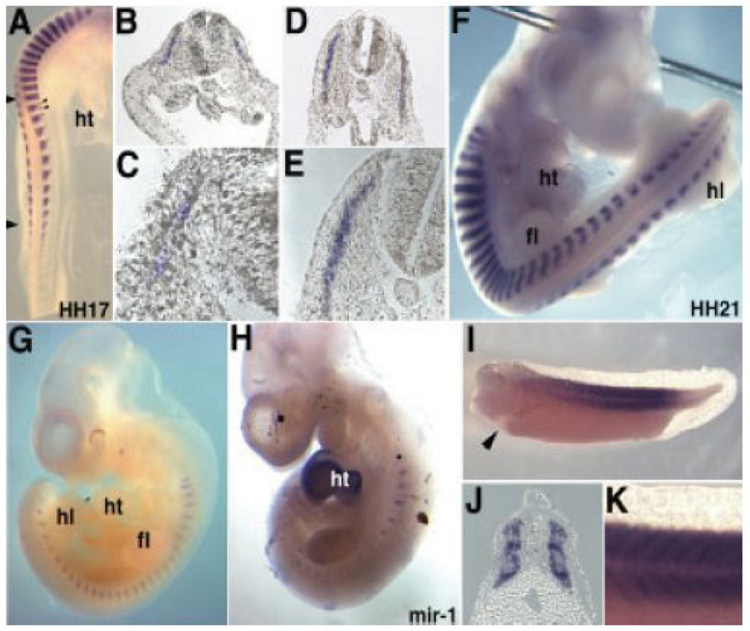
Figure 3. Expression of miR-206 is restricted to somites in mouse, chick and frog embryos.
A, miR-206 expression in somite of chick embryo (stage HH17) with small arrows at axial level indicating expression in lateral myotome; B–E, different magnifications of transverse sections through trunk of chick embryo (stage HH17) reveal miR-206 expression in myotome; F, mir-206 expression in chick embryo at stage HH21; G, somite-specific expression of miR-206 mouse embryo at E10; H, in contrast to miR-206, miR-1 is expressed in the heart and somites in the mouse embryo at E10; I-K, expression of miR-206 in Xenopus laevis tadpole (stage 35) is restricted to somites with arrow in (K) indicating lack of expression in heart, (J) transverse section through trunk show somite staining and (K) is higher magnification of (I); fl; forelimb, ht, heart; hl, hind limb. Image is reproduced from Sweetman et al (2006) with permission from John Wiley & Sons, Inc. [48].
ACKNOWLEDGEMENTS
I would like to thank Drs. Esser, Andrade and Shroeder for their careful reading of the manuscript. A special thanks to Dr. Sempere for providing raw data and insightful comments on the manuscript. This publication was made possible by grants from NIAMS/NIH to JJM (AR053641) and KAE (AR45617).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 2.Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3'-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Ruvkun G, Wightman B, Ha I. The 20 years it took to recognize the importance of tiny RNAs. Cell. 2004;116:S93–S96. doi: 10.1016/s0092-8674(04)00034-0. 92 p following S96. [DOI] [PubMed] [Google Scholar]
- 8.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–S92. doi: 10.1016/s0092-8674(04)00035-2. 81 p following S96. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 13.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, Dengler U, Hall J, Filipowicz W, Weiler J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 2007;35:e52. doi: 10.1093/nar/gkl1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E. An optimized isolation and labeling platform for accurate microRNA expression profiling. Rna. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EJ, Baek M, Nuovo GJ, Chen C, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines and tumors. RNA. 2008;14:1–8. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloosterman WP, Steiner FA, Berezikov E, de Bruijn E, van de Belt J, Verheul M, Cuppen E, Plasterk RH. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 21.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 25.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 27.Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Zhang Y, Yang B, Wang Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 29.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 31.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima N, Takahashi T, Kitamura R, Isodono K, Asada S, Ueyama T, Matsubara H, Oh H. MicroRNA-1 facilitates skeletal myogenic differentiation without affecting osteoblastic and adipogenic differentiation. Biochem Biophys Res Commun. 2006;350:1006–1012. doi: 10.1016/j.bbrc.2006.09.153. [DOI] [PubMed] [Google Scholar]
- 33.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C.C. Sempere LF, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 35.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. Rna. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Weng T, Gou D, Chen Z, Chintagari NR, Liu L. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics. 2007;8:29. doi: 10.1186/1471-2164-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of Micro Ribonucleic Acid 29, Highly Up-Regulated in Diabetic Rats, Leads to Insulin Resistance in 3T3-L1 Adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 43.Takada S, Berezikov E, Yamashita Y, Lagos-Quintana M, Kloosterman WP, Enomoto M, Hatanaka H, Fujiwara S, Watanabe H, Soda M, Choi YL, Plasterk RH, Cuppen E, Mano H. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 45.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler G, Ntounia-Fousara S, Granda B, Rathjen T, Dalmay T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006;580:2195–2200. doi: 10.1016/j.febslet.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 48.Sweetman D, Rathjen T, Jefferson M, Wheeler G, Smith TG, Wheeler GN, Munsterberg A, Dalmay T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev Dyn. 2006;235:2185–2191. doi: 10.1002/dvdy.20881. [DOI] [PubMed] [Google Scholar]
- 49.Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 50.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 51.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci U S A. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 55.Araya R, Eckardt D, Maxeiner S, Kruger O, Theis M, Willecke K, Saez JC. Expression of connexins during differentiation and regeneration of skeletal muscle: functional relevance of connexin43. J Cell Sci. 2005;118:27–37. doi: 10.1242/jcs.01553. [DOI] [PubMed] [Google Scholar]
- 56.Velleca MA, Wallace MC, Merlie JP. A novel synapse-associated noncoding RNA. Mol Cell Biol. 1994;14:7095–7104. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amthor H, Connolly D, Patel K, Brand-Saberi B, Wilkinson DG, Cooke J, Christ B. The expression and regulation of follistatin and a follistatin-like gene during avian somite compartmentalization and myogenesis. Dev Biol. 1996;178:343–362. doi: 10.1006/dbio.1996.0223. [DOI] [PubMed] [Google Scholar]
- 59.Miura P, Jasmin BJ. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we? Trends Mol Med. 2006;12:122–129. doi: 10.1016/j.molmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 61.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piccone CM, Brazeau GA, McCormick KM. Effect of oestrogen on myofibre size and myosin expression in growing rats. Exp Physiol. 2005;90:87–93. doi: 10.1113/expphysiol.2004.028373. [DOI] [PubMed] [Google Scholar]
- 63.McCormick KM, Burns KL, Piccone CM, Gosselin LE, Brazeau GA. Effects of ovariectomy and estrogen on skeletal muscle function in growing rats. J Muscle Res Cell Motil. 2004;25:21–27. doi: 10.1023/b:jure.0000021398.78327.39. [DOI] [PubMed] [Google Scholar]
- 64.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and Local Architecture of the Mammalian microRNA-Transcription Factor Regulatory Network. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R.D. Prochnik SE, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- 67.Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- 68.Lecroisey C, Segalat L, Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J Muscle Res Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- 69.Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- 70.Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Somitic origin of limb muscle satellite and side population cells. Proc Natl Acad Sci U S A. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 72.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Relaix F, Rocancourt D, Mansouri A, Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 75.Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126:1621–1629. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- 76.Yang XM, Vogan K, Gros P, Park M. Expression of the met receptor tyrosine kinase in muscle progenitor cells in somites and limbs is absent in Splotch mice. Development. 1996;122:2163–2171. doi: 10.1242/dev.122.7.2163. [DOI] [PubMed] [Google Scholar]
- 77.Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 78.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 79.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mankoo BS, Collins NS, Ashby P, Grigorieva E, Pevny LH, Candia A, Wright CV, Rigby PW, Pachnis V. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature. 1999;400:69–73. doi: 10.1038/21892. [DOI] [PubMed] [Google Scholar]
- 81.Seo KW, Wang Y, Kokubo H, Kettlewell JR, Zarkower DA, Johnson RL. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev Biol. 2006;290:200–210. doi: 10.1016/j.ydbio.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 82.Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 83.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biressi S, Molinaro M, Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol. 2007;308:281–293. doi: 10.1016/j.ydbio.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- 86.Bassel-Duby R, Olson EN. Signaling Pathways in Skeletal Muscle Remodeling. Annu Rev Biochem. 2006 doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 87.Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 88.Ji J, Tsika GL, Rindt H, Schreiber KL, McCarthy JJ, Kelm RJ, Jr., Tsika R. Puralpha and Purbeta collaborate with Sp3 to negatively regulate beta-myosin heavy chain gene expression during skeletal muscle inactivity. Mol Cell Biol. 2007;27:1531–1543. doi: 10.1128/MCB.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293:C451–C457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 90.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton Zebrafish JG. miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O.K. Nishi H, Iwanaga Y, Horie T, Kinoshita M, Nagao K, Hasegawa K, Kita T. MicroRNAs Controls Expression of α- and β-myosin Heavy Chain Genes in Cardiac Myocytes. Journal of Cardiac Failure. 2007:S39. [Google Scholar]
- 92.Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–176. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 93.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 94.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 95.Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I, Hall H, Timm S, Wang AG, Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, Cui Y. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 99.Georges M, Clop A, Marcq F, Takeda H, Pirottin D, Hiard S, Tordoir X, Caiment F, Meish F, Bibe B, Bouix J, Elsen JM, Eychenne F, Laville E, Larzul C, Milenkovic D, Tobin J, Charlier AC. Polymorphic microRNA-target interactions: a novel source of phenotypic variation. Cold Spring Harb Symp Quant Biol. 2006;71:343–350. doi: 10.1101/sqb.2006.71.056. [DOI] [PubMed] [Google Scholar]
- 100.Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3' untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 102.Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 103.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 104.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 105.Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, Petrof B, Narusawa M, Leferovich JM, Sladky JT, Kelly AM. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 106.Weir AP, Burton EA, Harrod G, Davies KE. A- and B-utrophin have different expression patterns and are differentially up-regulated in mdx muscle. J Biol Chem. 2002;277:45285–45290. doi: 10.1074/jbc.M205177200. [DOI] [PubMed] [Google Scholar]
- 107.Gramolini AO, Belanger G, Jasmin BJ. Distinct regions in the 3' untranslated region are responsible for targeting and stabilizing utrophin transcripts in skeletal muscle cells. J Cell Biol. 2001;154:1173–1183. doi: 10.1083/jcb.200101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Porter JD, Merriam AP, Leahy P, Gong B, Feuerman J, Cheng G, Khanna S. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Genet. 2004;13:257–269. doi: 10.1093/hmg/ddh033. [DOI] [PubMed] [Google Scholar]
- 109.Rando TA, Disatnik MH, Yu Y, Franco A. Muscle cells from mdx mice have an increased susceptibility to oxidative stress. Neuromuscul Disord. 1998;8:14–21. doi: 10.1016/s0960-8966(97)00124-7. [DOI] [PubMed] [Google Scholar]
- 110.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 111.Banerjee D, Kwok A, Lin SY, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 2005;8:287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 112.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 114.McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Allen GC, Farnell Y, Bell-Pedersen D, Cassone VM, Earnest DJ. Effects of altered Clock gene expression on the pacemaker properties of SCN2.2 cells and oscillatory properties of NIH/3T3 cells. Neuroscience. 2004;127:989–999. doi: 10.1016/j.neuroscience.2004.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Expression level of miR-206 in different tissues.
Table S2. Expression level of miRs in skeletal muscle.
Table S3. Developmental pattern of expression of miR-206.
Table S4. Annotation of predicted target genes of miRNA-206.
Table S5. Gene ontology of predicted target genes of miRNA-206.
Table S6. Transcription factor binding sites within CRM1 and CRM2.
Table S7. SNP associated with microRNA-206 according to Patrocles database.
Table S8. SNP associated with microRNA-206 according to PolymiRTS database.
Table S9. MicroRNA-206 predicted target genes that are alternatively spliced.