Abstract
Damage to RNA from ultraviolet light, oxidation, chlorination, nitration, and akylation can include chemical modifications to nucleobases as well as RNA-RNA and RNA-protein crosslinking. In vitro studies have described a range of possible damage products, some of which are supported as physiologically relevant by in vivo observations in normal growth, stress conditions, or disease states. Damage to both messenger RNA and noncoding RNA may have functional consequences, and work has begun to elucidate the role of RNA turnover pathways and specific damage recognition pathways in clearing cells of these damaged RNAs.
Keywords: oxidative stress, UV, alkylation, RNA photoproducts, RNA crosslinking, RNA decay
Introduction
Both exogenous and endogenous agents can damage cellular DNA, RNA, proteins and lipids. Cellular components come into contact with ultraviolet light, reactive oxygen species and other oxidants, hypohalous acids, nitric oxide, and alkylating agents, all of which are known from in vitro experiments to cause chemical modification or crosslinking of nucleic acids. For RNA, modification can in theory impact the function of both messenger and noncoding RNAs in numerous ways, such as interfering with base-pairing interactions in the transcription and translation of mRNAs or by altering the chemical properties of ribosomal RNA nucleotides necessary for RNA-driven catalysis in the ribosome. As studies have begun to demonstrate that many types of RNA damage that were first described in vitro also occur in vivo under physiologic conditions, so too have observations begun to indicate that the damage interferes with RNA function. It is also becoming clear that the cell may respond to RNA damage with a number of different defense mechanisms, including repair mechanisms specific for certain forms of damage as well as normal turnover pathways. This review focuses on the emerging understanding of the scope of RNA damage within the cell and its consequences for cellular function. In addition, this review considers both described and candidate response mechanisms for RNA damage.
Ultraviolet damage
Ultraviolet (UV) irradiation can cause several types of damage to RNA: photochemical modification, crosslinking, and oxidative damage. Much of the work describing UV damage to RNA has been carried out in vitro, with a few studies suggesting damage may also occur in vivo under physiologic conditions. Additionally, the extensive study of UV damage to DNA is useful in suggesting the effects that UV damage could have on RNA, as the photochemistry of nucleic acids occurs primarily at the nucleobase (DNA photodamage is reviewed in Cadet et al., 2005; Ravanat et al., 2001).
In vitro evidence for UV-induced chemical modification of RNA
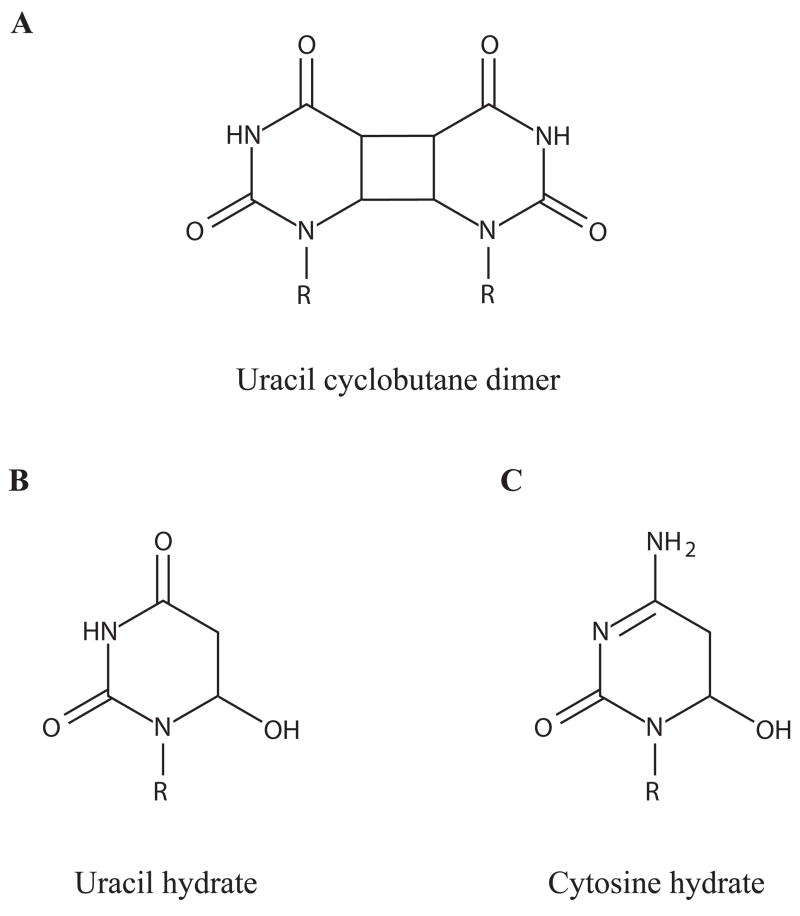
In early experiments to detect RNA photoproducts, isolated RNA was irradiated, hydrolyzed enzymatically, and then separated by chromatography to resolve photoproducts such as modified nucleotides and dimers (Small et al.,1968). With this technique, in vitro irradiation of synthetic poly(U) and poly(C) or isolated tobacco mosaic virus (TMV) RNA was observed to induce cyclobutane pyrimidine dimers, uridine hydrate, and cytidine hydrate (Singer, 1971; Miller and Cerutti, 1968). These studies identified commonly formed photoproducts (Figure 1) as well as general principles about the photoreactivity of RNA (Gordon et al., 1976). In particular, single-stranded RNA is more likely to form photoproducts than double-stranded RNA, as seen in the comparision of poly(U) to a complex of poly(U) and poly(A) (Pearson and Johns, 1966). The presence of Mg2+, which can promote the folding of RNAs, also leads to decreased susceptibility of RNA to photodamage (Singer, 1971). Additionally, studies on purified RNAs and viral particles have shown that protein binding can alter photoreactivity (Remsen et al., 1970; Gordon et al., 1976).
Fig. 1.
UV photoproducts. UV modification of bases can occur to free nucleobases (R = H) or in ribonucleosides, ribonucleotides, or in the context of an oligoribonucleotide (R = ribose). Figures produced with MarvinSketch (ChemAxon).
Evidence for UV-induced chemical modification of RNA under physiologic conditions
It is not clear whether these RNA photoproducts observed in vitro are formed in vivo under physiologic conditions. One important consideration is the wavelength of UV. The early in vitro studies of RNA photodamage primarily used UVC (200–290 nm), which is the highest energy form of UV irradiation but is not environmentally relevant as it is absorbed by the ozone layer in Earth’s atmosphere. However, the lower energy UVB (290–320 nm) wavelengths do reach the Earth’s surface. Additionally, UVA (320–400 nm) wavelengths are of even lower energy, but reach the Earth’s surface at levels ~10–100 times that of UVB and can travel farther into tissue (Kaminer, 1995). Direct absorption of UVB by nucleic acids only very inefficiently creates photoproducts, and UVA absorption by nucleic acids is even weaker. Thus, the formation of photoproducts at these wavelengths, particularly for UVA, may also involve the action of photosensitizer molecules within the cell. Although this is an area of ongoing investigation, it is speculated that molecules such as NADPH, flavins, or porphyrins could absorb UV and then in turn react with nucleic acids (Cadet et al., 1992; Cadet et al., 2005; Mitchell, 2006; Mouret et al., 2006).
Further, the doses of UV used in experiments must be evaluated. A useful reference point for UV doses is the minimal erythema dose (MED), or the dose where sunburn is first apparent. The UVB MED has a mean of 704 ± 188 J/m2, and the MED for UVA has a median of 207,000 J/m2 as measured on Caucasian subjects (Gambichler et al., 2006); these doses can be reached within an hour during the summer in Europe (Ambach and Blumthaler, 1993).
Considering these criteria, some observations have been made that are consistent with RNA photoproduct formation as a result of physiologically relevant irradiation. Early studies in insect eggs used chromatography to detect pyrimidine dimers in RNA following irradiation at a UVB wavelength of 295 nm (Jäckle and Kalthoff, 1978). Conversion of ~0.15% of pyrimidines to dimers occurred at a dose of 110 J/m2, which although strong enough to cause inactivation of the eggs, is within a relevant exposure range for humans.
Additionally, a report on rRNA damage caused by UV in cultured mammalian cells is consistent with formation of RNA photoproducts (Iordanov et al., 1998). At either a UVB dose of 600 J/m2 or high doses of UVC, lesions in the 28S rRNA were detected by a primer extention assay. The nature of the lesions was not determined, but the lesions occur primarily at adjacent pyrimidine nucleotides, which is consistent with cyclobutane pyrimidine dimer formation. Interestingly, the UV-induced lesions occurred in a site-specific, biased pattern within the 28S rRNA, particularly affecting the active site of the ribosome, and a decrease in translation activity was observed (Iordanov et al., 1998). Further, this damage to actively translating ribosomes is associated with the activation of a kinase-mediated stress response.
Although the relative susceptibility of RNA and DNA to UV-induced lesion formation in vivo is unknown, there is evidence that environmentally relevant doses of UVB and UVA lead to photoproduct formation in DNA at levels that could cause cellular consequences. High performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) quantification of pyrimidine dimer formation in human skin after low doses of UVB demonstrated a dimer formation rate of ~0.0520 lesions per 106 normal bases per J/m2. Additionally, exposure to low doses of UVA results in a rate of pyrimidine dimer formation of ~0.000008 lesions per 106 normal bases per J/m2 (Mouret et al., 2006). Using MED values to evaluate these rates of lesion formation in DNA, it appears that relevant levels of UVA and UVB exposure could cause damage on the order of 105 lesions per cell. It is thus possible that significant RNA damage also occurs, as average prokaryotic and eukaryotic cells contain ~4–6 times more RNA than DNA, most of which is noncoding RNA with a half-life on the order of days.
In vitro evidence for UV-induced RNA crosslinking
In addition to modification of nucleobases, UV can induce long-range covalent crosslinks involving RNA. This was first suggested from the experimental use of UV irradiation to probe ribosome structure, where both RNA-RNA and RNA-protein crosslinks were observed. RNA-RNA crosslinks can be induced both by UVC and UVB irradiation (Zwieb et al., 1978; Wilms et al., 1997). The frequency of crosslinking between nucleotides is correlated with the distance and angle between nucleotides as well as the ability of the nucleotides to undergo transient conformational change to a conformation that would allow crosslinking within the timescale of photoexcitation (reviewed in Huggins et al., 2005).
For RNA-protein crosslinks, the likelihood of crosslinking is also determined by the geometry and photoreactivity of the nucleotide and amino acid (Smith, 1976). As with RNA-RNA crosslinks, there is a long history of using RNA-protein crosslinking experimentally to study ribonucleoprotein (RNP) complexes including the ribosome (Möller et al., 1978). These studies provide insight into the types of crosslinks that are possible. For example, it has been noted that crosslinking does not normally occur in double-stranded regions (Noah et al., 2000). Thus, certain cellular RNPs may be more or less pre-disposed by RNA structure to UV crosslinking damage. Above all, the experimental use of UV to generate RNA-protein crosslinks reflects the inherent photoreactivity of RNPs.
Evidence for UV-induced RNA crosslinking under physiologic conditions
RNA-RNA crosslinking occurs within tRNAs in Escherichia coli treated with relevant doses of broad spectrum UVA light (320–405 nm). The observed crosslinks occur between cytidine and the naturally modified pseudouridine nucleotide (Ramabhadran et al., 1976). In vitro experiments showed that tRNAs containing this crosslink had lower rates of aminoacylation and caused inefficient amino acid incorporation in translation assays (Ramabhadran et al., 1976; Chaffin et al., 1971).
RNA-protein crosslinks have been described in UVB-irradiated insect eggs, although the RNAs involved were not identified (Jäckle and Kalthoff, 1979), and in maize (Casati and Walbot, 2004). In the maize study, environmentally relevant doses of UVB caused the formation of covalent crosslinks between protein and RNA within the ribosome. The observed crosslinks occurred at locations where there is close proximity between RNA and protein in the 3D structure of the ribosome. Further, this crosslinking damage may have functional consequences for the cell. As the duration of UVB exposure increased, the accumulation of crosslinked products increased, and the total cellular level of protein synthesis decreased. This correlation suggests that the protein-RNA crosslinks may have contributed to the loss of ribosome function, although other UV-induced mechanisms for inhibition of translation are also possible.
Oxidative damage
Oxidative damage to RNA has been reported in animal models of aging, human neurodegeneration and UV-irradiated cells. There is also growing evidence that oxidative RNA damage can lead to defects in protein synthesis, including decreased rates of protein synthesis and the production of aggregated and truncated peptides.
Under most conditions, the bulk of cellular oxidative damage is a result of reactive oxygen species (ROS) formation due to metabolic reactions. Multiple steps in the electron transport chain involve a free radical semiquinone anion (•Q−) that can react with oxygen to form the superoxide radical (O2−). The cell eliminates this superoxide radical through the action of superoxide dismutase to form hydrogen peroxide (H2O2), followed by enzymatically catalyzed reduction of H2O2 to H2O and O2 (reviewed in Finkel and Holbrook, 2000). However, the reaction of H2O2 with intracellular iron in the form of Fe2+ and Fe3+ can lead to a cascade of radical chemistry through the Fenton and Haber-Weiss reactions and the production of ROS, including the highly reactive hydroxyl radical (•OH) (reviewed in Halliwell and Gutteridge, 1984).
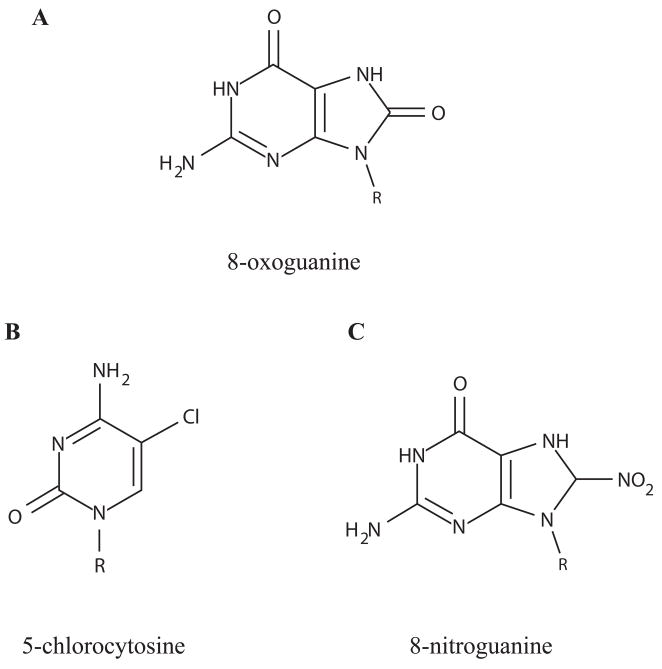
The reaction of ROS with free nucleobases, nucleosides, nucleotides or oligonucleotides can create numerous distinct modifications in RNA (Figure 2; Barciszewski et al., 1999). In studies of DNA, guanine bases have been shown to be particularly reactive, and a major product observed in vitro and in vivo is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) (Cadet et al., 2007; Ravanat et al., 2001). In vivo studies of RNA oxidation (discussed below) have also detected 8-oxo-7,8-dihydroguanosine (8-oxoG) in RNA through the use of HPLC-MS/MS and immunocytochemistry, indicating that this common oxidative modification to DNA occurs in RNA as well. The formation of 8-oxoG occurs by reaction of guanine with the •OH radical followed by oxidation, or by reaction of guanine with singlet oxygen 1O2 followed by reduction (Cadet et al., 2007). Oxidation of other nucleobases is also possible (Cadet et al., 2007), and at least two other modifications have been identified in yeast RNA (Yanagawa et al., 1992).
Fig. 2.
Chemically modified bases. R = H for free nucleobases or R = ribose for ribonucleosides.
In vivo measurement of oxidative damage to RNA
Oxidized RNA has been measured in vivo in a growing number of contexts, both as a result of experimental treatment of cells with oxidative agents and in diseased tissue. Treatment of E. coli cultures with H2O2 has been shown to cause 16S rRNA damage that can be detected by a reverse transcription polymerase chain reaction (RT-PCR) assay and is correlated with an increase in the 8-oxoG content in the cell (Gong et al., 2006). Experimental treatment has also been shown to result in RNA damage in animals. Exposure of rats to ethanol is associated with increased 8-oxoG RNA immunostaining in the pituitary gland (Ren et al., 2005), while ammonia exposure triggers RNA oxidation in the rat brain (Görg et al., 2008). Additionally, in a study of the carcinogen 2-nitropropane (2-NP), intraperitoneal injection of rats with 2-NP led to a significant increase in the levels of 8-oxoG in liver RNA (Fiala et al., 1989). In addition to increased 8-oxoG levels, HPLC-electrochemical detection (HPLC-EC) also detected other forms of unidentified modified ribonucleotides, indicating that oxidative damage to RNA in vivo may lead to the formation of numerous chemical modifications.
Measurement of oxidative damage to RNA under physiologic conditions
Oxidative damage to RNA has also been measured from natural biological processes. Oxidative RNA damage to cells under normal physiologic conditions has been suggested by detection of the oxidized ribonucleoside 8-oxoG in the urine of rats and humans as well as in human blood plasma (Park et al., 1992; Weimann et al., 2002). Further, because metabolic reactions are major sources of ROS, metabolic rate as well as mitochondrial dysfunction can be significant factors in oxidative damage. One model system is aging rats, which display mitochondrial dysfunction. In the brains of such rats, levels of 8-oxoG in RNA are significantly higher than levels in young rats (Liu et al., 2002). Indeed, treatment of old rats with metabolites that boost mitochondrial function resulted in oxidized RNA levels comparable to those of younger rats, demonstrating a link between mitochondrial function and intracellular RNA damage. Additionally, studies of aging muscle displaying deregulation of iron homeostasis are consistent with the role of intracellular metals in generating ROS through Fenton chemistry, and in turn oxidative damage (Hofer et al., 2008). Muscle from 32-month old rats was shown to have higher levels of 8-oxoG levels in total cellular RNA as well as higher free, non-heme iron levels compared to 6-month old rats. Additionally, imposing disuse atrophy on the older rats further elevated both the levels of free, non-heme iron and 8-oxoG. Thus, both the misregulation of metabolism and the loss of metal homeostasis that occur with age are correlated with increased oxidative damage to RNA.
Other biological processes may also generate reactive oxygen species that can contribute to RNA damage. For example, as a part of the immune response to pathogens, neutrophils and eosinophils use NADH oxidase to produce O2− and heme proteins to generate hypohalous acids, both of which can then lead to •OH formation. In a study designed to approximate conditions generated at sites of inflammation by the activation of neutrophils and eosinophils, fibroblasts were treated with the hypohalous acid HOCl in combination with hyperoxia (Shen et al., 2000). Such treatment resulted in increased 8-oxoG to levels of 7–8 per 105 G in poly(A) RNA and in the free nucleotide pool, as compared to 1 per 105 G in control cells. Yet, most treated cells remained viable, indicating that cells are able to survive this level of damage. Notably, other tissues also express NADH oxidases. The catalytic subunit of the phagocytic NADH oxidase has been shown to be expressed in a variety of tissues, including colon, reproductive tissues, and muscle (Suh et al., 1999), while the kidney expresses an NADH oxidase called Renox that is proposed to be an oxygen-sensing protein (Geiszt et al., 2000). Thus, many tissues may have a heightened requirement for cellular systems that respond to oxidative RNA damage.
UV-induced oxidation of RNA
UV is also a source of oxidative damage to RNA. Numerous pathways of excitation and oxidation are possible for nucleobases, as has been studied extensively in research on DNA damage (reviewed in Ravanat et al., 2001). Radical anion species can be formed from nucleobases due to the direct absorption of UVB light by a nucleobase; the radical anion species can then undergo further reaction to generate oxidized nucleotides. Oxidative damage can also occur from UVA, mediated either by photosensitizer molecules or by (ROS) formation (Cadet et al., 2007). For DNA, both UVB- and UVA-induced oxidation most commonly result in the formation of (8-oxodG), although photooxidation of adenine, cytosine, and thymine bases is also dectected in DNA from irradiated cells (Cadet et al., 2005). Importantly, much of the chemistry described for photooxidation of DNA is also applicable for RNA, as is borne out by measurement of UV-induced oxidative damage to RNA in cells.
UV-induced oxidative damage to RNA has been demonstrated in experiments with human skin fibroblasts. Exposure of fibroblasts to UVA at sublethal doses led to significantly increased levels of (8-oxoG) in RNA as measured by (HPLC-EC) analysis of total RNA (Wamer and Wei, 1997). Increasing intensity of UVA irradiation caused a dose-dependent increase in 8-oxoG. Interestingly, as seen in other examples of oxidative stress (discussed below), the RNA damage was greater than the DNA damage observed. Importantly, this study used UVA at doses comparable to the MED, supporting the physiologic relevance of UVA-induced oxidative damage to RNA.
Oxidative damage to RNA observed in disease
There is a growing body of evidence correlating oxidative damage to disease states, especially for neurodegenerative disorders (Moreira et al., 2008; Nunomura et al., 2006). In Alzheimer’s disease (AD), high levels of 8-oxoG RNA levels in the cytoplasm and nucleoli of neurons have been seen by immunostaining on DNase-treated postmortem brain sections from AD patients, as compared to the levels seen for control patients (Nunomura et al., 1999). Indeed, quantification of oxidized poly(A) RNA from frontal cortices by immunoprecipitation of 8-oxoG-containing RNAs indicated that 52.3 ± 6.15% of mRNAs contained 8-oxoG in AD patients compared to 1.78 ± 0.56% in age-matched controls (Shan and Lin, 2006). Elevated levels of oxidatively damaged RNA also have been observed in neurons from the degenerating substantia nigra of patients with Parkinson’s disease, as compared to unaffected regions of the same patient brains (Zhang et al., 1999). Similar findings are reported for patients with dementia with Lewy bodies, where high levels of 8-oxoG RNA were seen by immunostaining in neuronal populations susceptible to degeneration (Nunomura et al., 2002). Oxidation of poly(A) RNA has also been observed in neurons from affected regions of the brain and spinal cord of patients with amyotrophic lateral sclerosis (ALS) (Chang et al., 2008).
It has even been suggested that RNA oxidation may be an early event in the progression of neurodegenerative diseases, as in one study of familial AD, elevated 8-oxoG RNA staining was seen in cells that had not developed α-synuclein aggregation, a hallmark of degeneration (Nunomura et al., 2004). Further, in a mouse model of familial ALS, mRNA oxidation was observed to have preceded neurodegeneration and then to have subsided by the onset of neurodegenerative symptoms (Chang et al., 2008). The role of mRNA oxidation in disease progression appears complex, as vitamin E treatment blocked mRNA oxidation and delayed disease onset but did not alter mean life-span.
In addition to neurodegenerative diseases, RNA damage has also been noted in atherosclerosis. In atherosclerotic artery samples from carotid endarterectomies, the 8-oxoG RNA immunostaining levels were found to be elevated compared to non-diseased mammary arteries (Martinet et al., 2004).
Underlying causes of the susceptibility of RNA to oxidative damage
In many studies of oxidative damage, a common finding is that damage levels are higher in total RNA than in total DNA. This observation has been made by comparing the level of 8-oxoG immunostaining after RNase or DNase treatment, such as in carcinogen treatment of rats (Fiala et al., 1989) and studies of neurodegeneration (Nunomura et al., 1999). Several experiments have also sought to quantify nucleic acid damage. In one study on livers of rats treated with doxorubicin to induce oxidative stress, RNA and DNA was isolated simultaneously and quantified by HPLC-EC (Hofer et al., 2006). This approach found that in untreated animals, the ratio of 8-oxoG:guanosine in RNA was 1.4-fold higher than the ratio of 8-oxodG:2′-deoxyguanosine in DNA. Further, RNA damage increased significantly upon doxorubicin treatment while DNA damage did not. Quantification of 8-oxoG content in RNA versus 8-oxodG content in DNA has also been performed by in vitro 18O-labeled H2O2 treatment of human lung epithelial cells followed by HPLC-MS/MS (Hofer et al., 2005). In this context, the level of RNA damage per nucleoside was measured to be 14–25 times greater than that in DNA.
One explanation for the higher levels of oxidized nucleosides in RNA compared to DNA under normal cellular conditions is that there is a higher incidence of damage to molecules in closer proximity to mitochondria. This is supported by observations that levels of oxidized nucleosides are higher in mitochondrial DNA than in nuclear DNA (Shen et al., 2000). Indeed, in experimental treatment of cells with oxidizing reagents, this effect may be reproduced because exogenous oxidizing agents reach the cytosol first. Differential susceptibility of RNA and DNA to oxidation could also result from differences in protein association and in the degree of single-strandedness. The higher levels of oxidization observed for RNA could also be a consequence of different rates of removal of RNA and DNA damage.
RNA susceptibility to oxidative damage may also vary between noncoding and coding RNAs, among different classes of noncoding RNAs, and among different mRNA species. One suggested factor is the degree of protein association, which may serve to protect RNAs from damage. Indeed, different RNAs have varying degrees and temporal patterns of association with proteins, from the stable coating of rRNAs with ribosomal proteins to the progressive association of mRNAs with transcriptional machinery, splicing and export factors, and eventually polyribosomes. While the basis behind the differences in damage is not yet fully understood, it has been reported that different mRNA transcripts show differing levels of oxidative damage. For example, in ammonia-treated rat astrocytes, rRNAs and some mRNAs are oxidized while oxidation of other mRNAs is not detected (Görg et al., 2008). This issue has also been addressed by a study of oxidized RNAs in the brains of Alzheimer’s patients (Shan et al., 2003). Starting from purifed poly(A) RNA, oxidized mRNAs were isolated by immunoprecipitation with an antibody against 8-oxoG. RT-PCR for mRNAs of interest found that not all mRNAs were present in the pool of oxidized RNA, independent of transcript abundance. Further, even for mRNAs identified as oxidized, the prevalence of damage differed for particular mRNAs, in the range of ~54–75% damage-containing transcripts (Shan and Lin, 2006). Therefore, different cellular RNAs may be unequally susceptible to oxidative damage or certain damaged transcripts may be preferentially degraded or repaired. The consequences of such patterns of damage on disease states remain to be further elucidated.
Another factor that may render some RNAs particularly susceptible to oxidative damage is association with iron, as iron catalyzes Haber-Weiss and Fenton reactions that produce ROS (Halliwell and Gutteridge, 1984). This hypothesis has been raised particularly in regard to ribosomal RNAs. In studies of neurons from Alzheimer’s patients, rRNA was found to be oxidatively damaged at levels higher than for control patients (Ding et al., 2005; Honda et al., 2005). Intriguingly, purified ribosomes from Alzheimer’s patients showed elevated levels of associated redox-active iron (Honda et al., 2005). In vitro experiments correlated binding of redox-active iron with oxidative damage to rRNAs: rRNA was found to have higher in vitro iron-binding than tRNA or mRNA, and this iron-rich rRNA had a 13-fold greater formation of 8-oxoG in Fenton reaction oxidation experiments as compared to the iron-poor tRNA. Thus, iron-binding properties of certain classes of RNAs may correlate with vulnerability to oxidative damage under basal conditions and especially in disease states involving perturbation of intracellular metal homeostasis.
Consequences of oxidative RNA damage
The consequences of oxidative RNA damage include impairment of translation due to damage to mRNAs and due to decreased ribosome function. For example, oxidation of some mRNAs in affected regions of AD patient brains is correlated with low protein expression for those genes (Shan et al., 2007). The effect of oxidatively damaged mRNA on translation has been tested by introducing in vitro oxidized luciferase mRNA into cultured human HEK293 cells (Shan et al., 2003). The transfected oxidized mRNA was not degraded over the time course of the experiment, but compared to undamaged controls, luciferase activity and protein level from the damaged mRNA were strongly reduced. Additionally, proteins translated from the oxidized mRNA were shown to form aggregates suggestive of misfolding. In a similar study, transfected oxidized luciferase mRNA was shown to have normal polysome association, yet the expression of full-length protein decreased with a dose-dependent relationship to level of mRNA oxidation (Tanaka et al., 2007). In that study, use of protease inhibitors revealed that short polypeptides result from the translation of the oxidized mRNAs, perhaps due to both premature termination and protease degradation of aberrant full-length protein. Similar effects were seen when HEK293 cells were treated with paraquat to induce oxidative damage, demonstrating that following cellular oxidative stress, mRNAs are translated yet often generate truncated proteins that must be degraded by the proteasome (Tanaka et al., 2007). Another consequence of mRNA oxidation may be ribosome stalling, as the size of polyribosome complexes formed on in vitro oxidized mRNAs increases in rabbit reticulocyte translation systems, consistent with a slowed rate of elongation (Shan et al., 2007).
Considerable evidence exists that translation defects after oxidative stress may also result from damage to the ribosome. In vitro, oxidative damage to purified ribosomes has been shown to result in decreased translation of an undamaged poly(U) mRNA in a translation assay (Honda et al., 2005). Similarly, in vivo defects in ribosome function are suggested by the finding that for AD patients, polyribosomes isolated from regions of the brain known to incur oxidative damage show a reduced rate of protein synthesis in an in vitro assay compared to polyribosomes from nondiseased regions within AD patient brains or from control patients (Ding et al., 2005). At least two types of damage have been observed in ribosomes under oxidizing conditions. First, studies of 8-oxoG levels in rRNAs from affected regions of AD brains have revealed that rRNA is oxidized under conditions of oxidative stress (Ding et al., 2006). A second form of oxidative damage to ribosomes is the crosslinking of rRNA to ribosomal proteins. This has been observed in H2O2-treated yeast cells, where 85% of ribosomal proteins were seen to be oxidized and HPLC-MS/MS identified numerous sites of ribosomal protein-nucleotide crosslinking (Mirzaei and Regnier, 2006).
Chlorinating and nitrating damage
Radical chlorine and nitrogen species arise from multiple intracellular sources and can also cause damage to RNA. Phagocytic cells produce a range of radical species including hypochlorous acid (HOCl), nitric oxide (NO•), and peroxynitrate (ONOO−) to fight infections, and many cell types use nitric oxide as a signaling molecule. However, these radical species can lead to the chlorination, nitration, and oxidation of biological molecules including RNA (Hawkins et al., 2007).
In vitro evidence for chlorinating and nitrating damage to RNA
Reaction of HOCl with DNA nucleotides in stop-flow experiments shows that guanine and thymine are especially reactive, most likely due to chlorination at the heterocyclic nitrogen (Prütz, 1996). In cellular RNA from a human monocytic cell line treated with HOCl, HPLC-MS/MS has been used to detect 5-chlorocytidine, 8-choloroguanosine and to a lesser extent, 8-chloroadenosine and to show that chlorination of RNA occurs at higher levels than for DNA (Figure 2; Badouard et al., 2005). Interestingly, denaturation of double-stranded DNA is also seen in vitro upon HOCl treatment, suggesting that RNA base pairing and folding could also be disrupted by chlorination (Prütz, 1996).
The types of damage described for DNA from nitration include base deamination leading to depurination and strand breakage, DNA-DNA and DNA-protein crosslinking, and numerous base modifications including 8-oxodG and 8-nitro-2′-deoxyguanosine; these lesions and modifications are chemically possible for RNA as well (Reiter, 2006; Ohshima et al., 2006). Indeed, in vitro deamination of purified adenine, guanine, yeast tRNA, and bovine liver tRNA by nitric oxide has been measured (Nguyen et al., 1992). Additionally, in vitro exposure of purified total RNA to nitrogen radical species including ONOO− leads to stable 8-nitroguanosine and 8-oxoG formation (Masuda et al., 2002). Treatment of cultured human lung cancer cells with ONOO− also is associated with the formation of 8-nitroguanosine (Masuda et al., 2002).
Measurement of nitrating damage to RNA under physiologic conditions
In vivo, nitric oxide is generated as a part of the inflammatory response, and this has been observed to cause a concomitant increase in 8-nitroguanosine levels in the RNA from lungs of mice infected with virus (Akaike et al., 2003). Cytoplasmic immunostaining for 8-nitroguanosine is seen both in the infection of hamsters with parasites (Pinlaor et al., 2003) and in human patients with Helicobacter pylori infection (Ma et al., 2004), although controls were not performed in these experiments to conclusively identify RNA rather than DNA as the source of the cytoplasmic staining. These results suggest a pattern of damage to RNA from nitration in cases of infection, and further studies may extend the current understanding of the scope of chlorination and nitration under physiologic conditions.
Alkylation damage
RNA, like DNA, can undergo alkylation by SN1 or SN2 nucleophilic reactions with methylating agents (reviewed in Sedgwick, 2004). Endogenous sources of methylation are still under investigation but include methyl halides, compounds arising from the nitrosation of amines, and S-adenosylmethionine. Methylation can occur at nucleophilic N and O atoms on all of the ribonucleobases as well as at the O atoms of the phosphate backbone. As single-stranded nucleic acids are more vulnerable to methylation, many RNAs may be more susceptible than DNA to this damage.
While methylation is a naturally occurring rRNA modification, only specific, largely conserved positions are methylated in a process requiring a large class of small nucleolar ribonucleoproteins (snoRNPs) to guide methylation. Indeed, experimental re-engineering of snoRNPs to direct methylation of normally unmethylated sites in yeast rRNA can cause growth defects and inhibition of protein synthesis (Liu and Fournier, 2004). When alkylation occurs at random, one source of deleterious effects is the inhibition of base-pairing interactions. For example, methylation can interfere with tRNA-rRNA interactions in translation (Yoshizawa et al., 1999) and mRNA-tRNA interactions in translation (Ougland et al., 2004) and can theoretically interfere with any other RNA function that relies on base pairing, such as siRNA and miRNA function. Additionally, loss of base pairing interactions can cause structured RNAs to misfold. Protein recognition of RNA bases, such as in the aminoacylation of tRNAs, can also be inhibited by methylation (Ougland et al., 2004).
Cellular handling of specific forms of RNA damage
The current understanding of turnover or repair mechanisms for damaged RNA varies for different forms of damage. For some, such as alkylation, a clear mechanism has been identified. For most other types of damage, the roles of normal RNA turnover mechanisms and alternate pathways are still under investigation.
Mechanisms for handling of RNA with alkylative damage
Alkylated nucleic acids can be repaired by enzyme-catalyzed oxidative demethylation. The E. coli AlkB enzyme and one of the human oxidative demethylases, hABH3, demethylate 1-methyladenine and 3-methylcytosine in RNA in addition to having activity on DNA (Aas et al., 2003). In vivo, the action of AlkB and hABH3 can lead to the restoration of RNA function, as exemplified by reactivation of alkylated RNA bacteriophages (Aas et al., 2003), and tRNA (Ougland et al., 2004).
Interestingly, because these oxidative demethylases only function to remove methyl groups from 1-methyladenine and 3-methylcytosine, certain classes of RNAs may be more likely to accumulate alkylation damage that cannot be repaired. Alternatively, demethylases with other specificities may remain to be identified. The efficacy of AlkB in restoring tRNA function as measured by in vitro assays is less than that for mRNA, consistent with the more double-stranded nature of tRNA, which would lead to alkylation damage predominately at positions not repaired by AlkB (Ougland et al., 2004). This suggests that in vivo certain classes of RNAs may show higher capacity for repair, while others may be targeted by other mechanisms for decay.
Finally, it has been noted that AlkB has only weak affinity for DNA and may interact with other DNA-binding proteins in order to increase its affinity in vivo (Yang et al., 2008). This may be true for RNA as well, and it will be of interest to identify proteins that function with these demethylases.
Mechanisms for handling UV-induced RNA photoproducts and crosslinks
Cyclobutane pyrimidine dimers are actively repaired in some organisms by photolyase enyzmes that catalyze light-dependent dimer splitting. While many prokaryotes and eukaryotes have DNA photolyases, the physiological relevance of DNA photolyases in RNA repair is most likely minimal, as exemplified by the poor RNA binding of the E. coli photolyase (Kim and Sancar, 1991). To date, RNA photolyase activity has only been observed in plants (Gordon et al., 1976) and insects (Jäckle and Kalthoff, 1980). Therefore, it is unclear to what extent most organisms actively repair cyclobutane pyrimidine dimers in RNA.
For the rRNA-protein crosslinking observed in maize subjected to UVB irradiation (Casati and Walbot, 2004), a cellular handling mechanism is implied by the disappearance of the crosslinked products in a recovery period following UVB exposure. However, the mechanism that recognizes and degrades the crosslinked RNAs and proteins is not yet known.
Mechanisms for handling oxidized nucleotides
A mechanism has been described for the turnover of oxidized free nucleotides. Clearing oxidatively damaged free nucleotides can prevent incorporation of these nucleotides into newly synthesized RNAs. Besides triggering problems caused by the presence of oxidized nucleotides in a particular RNA, 8-oxoG can be incorporated opposite adenosine by RNA polymerase and thus cause mutations at the level of transcription (Taddei et al., 1997). Such mutations can cause errors in protein synthesis if they occur in mRNAs or affect the folding or function of noncoding RNAs. In E. coli, the MutT protein hydrolyzes 8-oxoguanosine triphospate (8-oxoGTP) to 8-oxoguanosine monophosphate (8-oxoGMP) in addition to catalyzing the same hydrolysis reaction on 8-oxo-2′-deoxyguanosine triphosphate (Taddei et al., 1997). In human cells, the MutT-related protein MTH1 has hydrolytic activity on 8-oxoGTP, and the resulting 8-oxoGMP is prevented from re-entering the nucleoside triphosphate pool because guanylate kinase lacks activity on 8-oxoGMP (Hayakawa et al., 1999). A second human MutT-related protein, NUDT5, hydrolyzes 8-oxoGTP to 8-oxoGDP, which can be further hydrolyzed by MTH1 (Ishibashi et al., 2005). Such enzymatic activity is important in ensuring transcriptional fidelity despite oxidative damage to nucleotides both by ongoing metabolic generation of ROS and by oxidative stress conditions. In kainate-induced oxidative stress within the rat hippocampus, MTH1 expression is upregulated (Kajitani et al., 2006). Further, MTH1-null mutant rats show significantly higher levels of 8-oxoG immunostaining in RNA and DNA in kainate-sensitive regions of the hippocampus and return to basal 8-oxoG levels with a slower time course after induced oxidative stress. Thus, hydrolysis of 8-oxoG by MutT-related proteins functions to protect the nucleotide pool both under normal growth and in stress conditions.
Evidence that oxidatively damaged RNAs are degraded
There is also evidence for the turnover of damaged mRNAs and noncoding RNAs. The quantification of 8-oxoG in RNA after oxidative treatment of cultured human lung epithelial cells shows that approximately a third of the 8-oxoG in RNA is cleared by 3 hours after removal of oxidative stress (Hofer et al., 2005). As 95% of cellular RNA is noncoding RNA with a normal half-life much greater than 3 hours, mechanisms must exist to rapidly degrade some damaged RNAs. Interestingly, after the 3 hour time point, the turnover of 8-oxoG slows, leading to an overall half-life of ~12.5h for 8-oxoG in RNA. Similar observations have been made for Hela cells treated with sub-lethal doses of H2O2 (Wu and Li, 2008). In that study, 8-oxoG levels in RNA rapidly peaked following a pulse of oxidative stress, were reduced by half within 30 minutes of recovery, and then slowly reached basal levels by 24h. Additionally, turnover or degradation has been reported in total RNA isolated from rat astrocytes subjected to ammonia stress (Görg et al., 2008). However, elevated levels of oxidized RNA measured in studies of aging and neurodegeneration (Liu et al., 2002; Nunomura et al., 1999) imply that at least some fraction of damaged RNA accumulates in those contexts.
For rRNA in particular, there are reports of degradation following oxidative damage. In human atherosclerotic plaques, reduction of 18S and 28S rRNA levels coincides with high 8-oxoG RNA levels (Martinet et al., 2004). Similarly, isolation of total RNA and ribosomes from affected regions of AD patient brains has shown decreased levels of all four individual rRNAs (5S, 5.8S, 18S, and 28S) and in turn decreased levels of the 40S and 60S ribosomal subunits and mature 80S ribosomes (Ding et al., 2005; Ding et al., 2006). Degradation of the 25S and 5.8S rRNAs is also seen in yeast under oxidative stress (Mroczek and Kufel, 2008; Thompson et al., 2008). However, for these observations, it is unclear to what extent the rRNA degradation was an effect of ongoing apoptosis, rather than a specific mechanism to degrade damaged RNAs.
Finally, oxidative stress has also been noted to induce the accumulation of cleaved tRNA halves in yeast, Arabidopsis thaliana, and human cells (Thompson et al., 2008). As mature tRNA levels are not reduced, it is unclear whether these cleavages affect cell function.
Cytoplasmic domains implicated in the handling of UV- and oxidatively damaged RNAs
Conditions that cause RNA damage can also lead to translational arrest and induction of two sites of mRNA accumulation within the cytoplasm, stress granules and processing bodies (P-bodies) (reviewed in Anderson and Kedersha, 2008). The role of these foci in the turnover of damaged RNA is just beginning to be understood. The first step in the formation of stress granules occurs through pathways that converge on eukaryotic initiation factor 2α (eIF2α) to stall translation. For example, arsenite-induced oxidative stress leads to heme-regulated initiation factor 2α kinase (HRI) inactivation of eIF2α, the inhibition of protein synthesis, and the formation of stress granules (McEwen et al., 2005). UV irradiation also leads to stress granule formation, possibly because UV irradiation activates protein kinase R, which is a regulator of eIF2α (Kedersha et al., 1999; Anderson and Kedersha, 2008). Ribosomes blocked at initiation accumulate on mRNA as 48S structures, which then begin to nucleate stress granules through the binding of a variety of aggregate-forming RNA-binding proteins (Kedersha et al., 1999). Translational arrest of mRNAs that had been undergoing translation during normal growth conditions may allow cells to degrade damaged RNAs and alter the profile of mRNA translation in response to stress.
mRNAs are thought to exit stress granules by reinitiation of translation or transfer to P-bodies (Kedersha et al., 2005). P-bodies are sites of mRNA decapping and decay and are found in cells under normal growth conditions but are further induced by stress (reviewed in Eulalio et al., 2007). Decay of a deadenylated mRNA via the 5′-3′ pathway is initiated by removal of the 5′ cap structure by the decapping complex Dcp1-Dcp2. Numerous activators of decapping also act at this step, including the Sm-like (Lsm) 1–7 complex, Pat1, and the helicase Dhh1. After decapping, the mRNA is degraded by the 5′-3′ exoribonuclease Xrn1. The proteins involved in the 5′-3′ decay pathway are all localized to P-bodies (Sheth and Parker, 2003). Indeed, the decay of mRNA has been demonstrated to occur within these foci, as yeast strains that lack Xrn1 have enlarged P-bodies that contain accumulating mRNA. Interestingly, at least one type of mRNA quality control, nonsense-mediated decay (NMD), is thought to use P-bodies as a site of degradation of defective mRNAs. Several factors essential for NMD are localized to P-bodies (Fukuhara et al., 2005; Unterholzner and Izaurralde, 2004) and mRNAs containing premature stop codons are recruited to P-bodies by the NMD protein Upf1 (Sheth and Parker, 2006). The finding that NMD may occur in P-bodies raises the possibility that P-bodies may be sites of turnover for other types of aberrant mRNAs.
The function of P-bodies in the handling of damaged RNA is supported by the increased sensitivity of cells lacking certain P-body proteins to conditions that cause RNA damage. Notably, a mutant xrn1 strain displays UV sensitivity (Tishkoff et al., 1991). This is consistent with the UV-sensitive phenotypes for strains lacking Pat1 and strains lacking Lsm1 (Birrell et al., 2001; Wang et al., 1999). Interestingly, in addition to the UVC sensitivity of the lsm1Δ deletion strain, both the lsm1Δ and lsm6Δ deletion strains are sensitive to the alkylating agent methyl methanesulfonate (MMS) (Chang et al., 2002). These phenotypes suggest that P-bodies may be important for the decay of several different types of damaged mRNAs and that failure to degrade damaged RNAs can impact cell growth.
Moreover, in addition to these stress sensitive phenotypes of P-body proteins, the role of P-bodies in conditions that cause RNA damage is indicated by findings that while P-bodies exist under normal cellular conditions, they enlarge or change subcellular localization in response to certain stress conditions. Yeast cells exposed to UV light show intensified localization of the RNA helicase Dhh1 and the decapping protein Dcp2 to P-bodies during recovery following treatment (Teixeira et al., 2005). Following arsenite-induced oxidative stress of mammalian cells, stress granules form and P-bodies localize adjacent to stress granules, possibly to expedite the degradation of damaged mRNAs accumulating in stress granules (Yu et al., 2005; Kedersha et al., 2005).
Additional, distinct cytoplasmic bodies named UV-induced mRNA granules (UVG) have also been proposed (Gaillard and Aguilera, 2008). Following UV treatment of yeast cells, overexpressed reporter mRNAs were observed to be stabilized and to accumulate in UVGs, which do not colocalize with P-body or stress granule components. It is possible that such localization serves to sequester mRNAs from translation, but it is not yet known how sorting to UVGs occurs or whether these RNAs contain UV damage.
Additional proteins implicated in the handling of oxidatively damaged RNAs
Another candidate for involvement in turnover of oxidatively damaged RNA that is also localized to P-bodies and stress granules (Yang and Bloch, 2007) is the Y-box-binding protein (YB-1). A member of the cold-shock domain superfamily, YB-1 participates in a wide range of processes, including transcription regulation, translation regulation, and DNA repair (reviewed in Kohno et al., 2003). In vitro experiments suggest that YB-1 may have specific binding for 8-oxoG-containing oligonucleotides (Hayakawa et al., 2002). A role for YB-1 in removal of oxidatively damaged RNAs is also consistent with the observation that expression of human YB-1 in E. coli increases resistance to paraquat-induced oxidative stress (Hayakawa et al., 2002) while YB-1 knockdown induces UV sensitivity in human epidermoid cells (Ohga et al., 1996). Further, YB1−/− mouse embryonic fibroblasts have increased sensitivity to oxidative stress and senesce prematurely in hyperoxic conditions (Lu et al., 2005). Interpretation of these phenotypes is complicated by the numerous roles of YB-1, as stress resistance could also be a result of YB-1 regulation of transcription. One additional indication of a role for YB-1 in handling damaged nucleic acids is that YB-1 has the capacity to stimulate the base excision repair activity of the NEIL2 protein, which removes oxidized bases from DNA (Das et al., 2007). While oxidatively damaged RNAs are not thought to be repaired, this finding suggests a model wherein YB-1 binding to 8-oxoG-containing RNAs may assist turnover of these RNAs by other proteins.
Much research has focused on the human polynucleotide phosphorylase protein (hPNPase), a homolog of the well-characterized bacterial 3′-5′ exoribonuclease. In bacterial cells, PNPase participates in mRNA turnover as well as the degradation of some noncoding RNAs, including aberrant precursor tRNAs and rRNAs (Deutscher, 2006). hPNPase emerged as a candidate for turnover of oxidatively damaged RNAs when in vitro binding experiments suggested hPNPase has some specificity for oxidizatively damaged RNA (Hayakawa et al., 2001). Additionally, overexpression of hPNPase lowers levels of 8-oxoG RNA while siRNA knockdown of hPNPase raises 8-oxoG levels (Wu and Li, 2008). However, a possible role for hPNPase in degrading oxidized RNAs must be reconciled with the localization of hPNPase to the mitochondrial intermembrane space, which is not known to contain RNA (Chen et al., 2006). While hPNPase mobilization into the cytosol is seen in conditions that induce apoptosis, it is not clear whether hPNPase is also released into the cytosol under other stress conditions.
Proteins and pathways that may have a general function in handling damaged RNAs
Exoribonucleases
In eukaryotes, an important candidate for degradation of damaged RNA is the exosome complex of 3′-5′ exonucleases. As this complex acts in the nuclear degradation of aberrant precursor mRNAs, tRNAs, and rRNAs as well as in the cytoplasmic degradation of mRNAs that contain premature stop codons (reviewed in Houseley et al., 2006), the exosome may also be recruited to damaged RNAs. Interestingly, yeast strains mutant for exosomal proteins show increased sensitivity to 5-fluorouracil (5FU) treatment (Fang et al., 2004), which may be consistent with a role for the exosome in turning over RNAs containing that unnatural base.
In bacteria, several exoribonucleases have been implicated in the decay of damaged RNAs. In Deinococcus radiodurans, PNPase is important for viability following UV irradiation and for growth on H2O2-containing media, indicating a potential role for that nuclease in degradation of damaged RNAs (Chen et al., 2007). Another processive 3′-5′ exoribonuclease, RNase R, which is remarkable for its ability to processively degrade through regions of significant secondary structure, shows increased activity in E. coli in a number of stresses such as growth in minimal media, starvation, and cold shock (Chen and Deutscher, 2005). Additional evidence that PNPase and RNase R function in the turnover of aberrant RNAs has been provided by experiments in E. coli. At 42°C, the PNPtsR− strain accumulates fragments of the 16S and 23S rRNAs and is deficient in 70S ribosome assembly (Cheng and Deutscher, 2003). This suggests that these two nucleases function to degrade defective rRNAs that cannot be incorporated into ribosomes. Further, PNPase has been shown to degrade mutant precursor tRNAs (Li et al., 2002). These findings suggest that these nucleases may also act to clear the cell of defective noncoding RNAs after RNA damage.
While different nucleases are known for having varying tolerance for RNA secondary structure (Deutscher, 2006), it is unclear whether different nucleases may show particular tolerance or sensitivity toward unusual structures formed by RNA damage such as RNA-RNA or RNA-protein crosslinks or modified nucleotides. This point has not been addressed directly, but interestingly, the RNase R homolog from Mycoplasma genitalium stalls at sites of 2′-O-methylation, while the E. coli RNase R does not show this sensitivity (Lalonde et al., 2007). Thus, the activity of different nucleases in degrading through crosslinks or other nucleotide modifications resulting from RNA damage may determine what nucleases are most active in the turnover of damaged RNAs.
Finally, it is notable that both prokaryotes and eukaryotes have mechanisms for large-scale turnover of rRNA. In E. coli, starvation induces degradation of most of the rRNA in the cell, presumably in order to scavenge nucleotides, although the mechanism is not well understood (Jacobson and Gillespie, 1968). In S. cerevisiae, starvation causes ribosomes to be targeted to the lysosome for degradation in a process termed ‘ribophagy’ (Kraft et al., 2008). While the extensive rRNA degradation seen during starvation has not been reported for conditions known to damage RNA, it is possible that cells use the same underlying mechanisms to target damaged RNAs for degradation.
Poly(A) polymerases and helicases
The activities of some prokaryotic nucleases and the eukaryotic exosome are enhanced by poly(A) polymerases and helicases. In prokaryotes, the degradation of mRNAs and noncoding RNAs is stimulated by the addition of poly(A) tails by poly(A) polymerase (Deutscher, 2006). In eukaryotes, polyadenylation of RNAs by the TRAMP complex promotes degradation by the nuclear exosome (LaCava et al., 2005; Vanácová et al., 2005). The TRAMP complex consists of the poly(A) polymerase Trf4 as well as an RNA helicase Mtr4 and one of two closely related RNA-binding proteins, Air1 and Air2. Polyadenylation may play a role in targeting certain RNAs for degradation, as the TRAMP proteins have been reported to preferentially polyadenylate incorrectly folded tRNAs in vitro (Vanácová et al., 2005). It is speculated that this specificity arises from greater accessibility of the 3′ end (Reinisch and Wolin, 2007), but it is unclear how this preference may extend to many types of damaged RNAs. Interestingly, polyadenylated rRNA precursors accumulate in yeast treated with 5FU, especially in strains mutant for the exosomal subunit Rrp6 (Fang et al., 2004). As 5FU-containing RNAs can form stable RNA-protein adducts with the modifying enzyme pseudouridylase, the accumulating defective rRNA precursors are hypothesized to require TRAMP/exosome-mediated degradation (Hoskins and Butler, 2008).
Helicases are thought to aid degradation by facilitating translocation of the nucleases over the RNA, removal of RNA secondary structure, and removal of proteins from the RNA. The helicase activity of the TRAMP complex is required for the degradation of substrates including aberrant tRNAs (Wang et al., 2008), while the cytoplasmic exosome interfaces with the Ski complex, of which Ski2 has helicase activity (Houseley et al., 2006). In bacteria, PNPase can function with at least two different helicases, one of which may be a cold adaptation (Prud’homme-Généreux et al., 2004), suggesting that different accessory proteins may function to adapt nuclease activity to different environmental stresses.
mRNA quality control pathways
Cells have multiple mRNA quality control pathways that eliminate faulty transcripts, including nonsense-mediated decay of mRNAs containing premature stop codons, non-stop decay of mRNAs lacking stop codons, and no-go decay of mRNAs stalled in translation (Doma and Parker, 2007; Isken and Maquat, 2007). These mechanisms may also participate in the handling of some forms of RNA damage. For example, the production of truncated protein products by the translation of oxidized mRNAs (Tanaka et al., 2007) could potentially trigger NMD, and likewise, stalling of elongation in the translation of oxidatively damaged mRNAs (Shan et al., 2007) could lead to mRNA degradation by no-go decay.
There is evidence that certain types of stress may activate NMD activity. One regulator of NMD activity is hSMG-1 kinase, which phosphorylates hUpf1 to promote NMD activity. Interestingly, activity of the hSMG-1 kinase increases with UV or ionizing radiation (IR), and higher NMD activity was observed after IR (Brumbaugh et al., 2004). However, it has not yet been demonstrated that the increased NMD activity is required for the degradation of damaged RNA, rather than another function.
Other RNA binding proteins
The Ro autoantigen, a ring-shaped RNA binding protein, also has roles both in RNA quality control and cell stress response. In vertebrate cells, the Ro protein associates with both misfolded pre-5S rRNAs (O’Brien and Wolin, 1994) and aberrant U2 snRNAs (Chen et al., 2003), and is thought to act in quality control for these RNAs, most likely by a scavenger mechanism of binding to RNAs that fail to properly associate with processing factors or incorporate into mature RNPs (Fuchs et al., 2006). Further, in the bacterial species D. radiodurans, the Ro homolog Rsr immunoprecipitates with PNPase and acts with the exoribonucleases RNase PH and RNase II in 23S rRNA maturation (Chen et al., 2007). This suggests that Ro proteins could act with nucleases in other processes as well, such as the turnover of damaged RNAs. Consistent with this hypothesis, Ro contributes to survival after UV in D. radiodurans (Chen et al., 2000) and in mammalian cells, where an accumulation of Ro is seen in the nucleus during recovery from UVC irradiation (Chen et al., 2003). Ro also exhibits genetic interactions with PNPase in both UV and oxidative stress in D. radiodurans (Chen et al., 2007). It will be of interest to determine what RNAs are bound by Ro during recovery from UV and other stresses, how Ro binding may lead to RNA degradation, and how Ro activity and subcellular localization are regulated in stress conditions.
The host factor I (Hfq) protein is another bacterial ring-shaped RNA binding protein and, like components of the Lsm1–7 ring, is a member of the Sm-like family of proteins. Hfq has a variety of functions, including modulation of several steps in RNA decay. Hfq protects some RNAs from RNase E cleavage, which often functions as an initiating event in RNA degradation (Moll et al., 2003). Hfq also promotes polyadenylation by poly(A) polymerase I (PAPI), which in turn increases mRNA turnover by making mRNAs better substrates for PNPase-mediated degradation (Mohanty, Maples & Kushner 2004). Such activities could be critical to promoting or regulating the turnover of damaged RNAs. Indeed, hfq insertion mutants show sensitivity to UV (Tsui et al., 1994) and H2O2 (Muffler et al., 1997), among other growth phenotypes. However, H2O2 sensitivity was correlated with another Hfq function, the translation of the RNA polymerase σs subunit responsible for transcription of stress-response genes (Muffler et al., 1997), and could also result from the action of Hfq in the post-transcriptional regulation of some mRNAs (Zhang et al., 2002). Therefore, it is not currently possible to conclude whether Hfq modulation of RNA decay is important for survival following stress.
Additionally, multiple ribosomal proteins have secondary activities that suggest roles in response to RNA damage. In Arabidopsis, one part of the UV-induced stress response is the rapid degradation of bulk mRNA, but not noncoding RNA (Revenkova et al., 1999). This extreme turnover of mRNA may allow more responsive upregulation of stress response genes as well as function to degrade damaged mRNAs. Disruption of the promoter of one isoform of ribosomal protein S27 inhibits this mRNA degradation and causes UVC sensitivity. Additionally, in Drosophila melanogaster, the small ribosomal subunit protein S3 and the ribosomal stalk protein P0 have been reported to have activities that are a part of DNA base excision repair (Yacoub et al., 1996; Yacoub et al., 1996). S3 was shown to have endonucleolytic activity on 8-oxodG-containing DNA, and both S3 and P0 were reported to have cleavage activity on abasic sites. The activity of P0 on single-stranded as well as double-stranded DNA raises the possibility that these proteins might act on RNA lesions as well.
Conclusions and Perspectives
A growing collection of findings suggests that RNA damage occurs in cells during normal growth and also during stress. In some cases, the functional consequences of RNA damage have been observed, including decreased protein synthesis as a result of ribosomal crosslinking and/or mRNA oxidation. As RNA damage is increased in models of aging and is in some instances correlated with disease, such as neurodegeneration and atherosclerosis, it will be important to elucidate in more detail how RNA damage inhibits cell function. In particular, a better understanding is needed of the in vivo susceptibility of particular RNAs to damage and how the varying levels of damage seen for different mRNAs and noncoding RNAs impact cellular function and disease progression. In addition, as most of the experimental work on the functional consequences of RNA damage has focused on mRNAs and rRNAs, future efforts will likely focus on the consequences of damage to more recently described classes of small noncoding RNAs including miRNAs, siRNAs, and piRNAs.
As knowledge of the turnover of damaged RNA has expanded, it has become clear that RNA turnover has critical significance to cellular stress responses. For example, mutant phenotypes for proteins that act in RNA turnover, such as Xrn1, Lsm1, Lsm6, and Pat1, are seen in stress conditions that cause RNA damage. Such phenotypes indicate that failure to clear damaged RNAs may negatively affect cell viability. These phenotypes, combined with the identification of other mechanisms to handle specific types of damage, such as MutT hydrolysis of oxidized free RNA nucleoside triphosphates and AlkB-catalyzed repair of alkyation, demonstrate the existence of cellular machinery for clearing RNA damage. Nonetheless, important questions remain regarding the extent to which turnover of damaged RNAs is a result of mechanisms that specifically recognize certain damage products or whether damaged RNAs are handled by general turnover pathways. Additionally, for the accumulation of damaged RNAs seen in aging and disease, it is unclear whether RNA degradation mechanisms become impaired or simply overwhelmed. Future studies will likely identify roles for known and novel factors in the handling of damaged RNAs and address the function of RNA turnover mechanisms in disease states.
Acknowledgments
We thank A. Alexandrov, D. Brash, and N. Kolev for comments on the manuscript. Work in our laboratory on RNA damage is supported by NIH grant GM073863 and a grant from the Ellison Medical Foundation.
Contributor Information
Elisabeth J. Wurtmann, Department of Cell Biology, Yale University, New Haven, CT
Sandra L. Wolin, Department of Cell Biology and Department of Molecular Biochemistry and Biophysics, Yale University, New Haven, CT
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Akaike T, Okamoto S, Sawa T, Yoshitake J, Tamura F, Ichimori K, Miyazaki K, Sasamoto K, Maeda H. 8-nitroguanosine formation in viral pneumonia and its implication for pathogenesis. Proc Natl Acad Sci USA. 2003;100:685–90. doi: 10.1073/pnas.0235623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambach W, Blumthaler M. Biological effectiveness of solar UV radiation in humans. Experientia. 1993;49:747–53. doi: 10.1007/BF01923543. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Badouard C, Masuda M, Nishino H, Cadet J, Favier A, Ravanat JL. Detection of chlorinated DNA and RNA nucleosides by HPLC coupled to tandem mass spectrometry as potential biomarkers of inflammation. Journal of Chromatogr B. 2005;827:26–31. doi: 10.1016/j.jchromb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Barciszewski J, Barciszewska MZ, Siboska G, Rattan SIS, Clark BFC. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol Biol Rep. 1999;26:231–238. doi: 10.1023/a:1007058602594. [DOI] [PubMed] [Google Scholar]
- Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc Natl Acad Sci USA. 2001;98:12608–13. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Cadet J, Anselmino C, Douki T, Voituriez L. New trends in photobiology: photochemistry of nucleic acids in cells. Journal Photoch Photobio B. 1992;15:277–298. doi: 10.1016/1011-1344(92)85135-h. [DOI] [PubMed] [Google Scholar]
- Cadet J, Douki T, Badouard C, Favier A, Ravanat J-L. Oxidatively generated damage to cellular DNA: mechanistic aspects. In: Evans MF, Cooke MS, editors. Oxidative Damage to Nucleic Acids. Landes Bioscience; Austin, T. X: 2007. pp. 1–13. [Google Scholar]
- Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res-Fund Mol M. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Casati P, Walbot V. Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol. 2004;136:3319–32. doi: 10.1104/pp.104.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin LJ, Omilianowski DR, Bock RM. Cross-linked transfer RNA functions in all steps of the translational process. Science. 1971;172:854–5. doi: 10.1126/science.172.3985.854. [DOI] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA. 2002;99:16934–9. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CG. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J Biol Chem. 2005;280:34393–6. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, et al. Mammalian polynucleotide phosphorylase in intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Gene Dev. 2000;14:777–82. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol. 2003;13:2206–11. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Gene Dev. 2007;21:1328–39. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci USA. 2003;100:6388–93. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, Hazra TK. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007;282:28474–84. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Markesbery W, Cecarini V, Keller J. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–8. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fang F, Hoskins J, Butler JS. 5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol Cell Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala ES, Conaway CC, Mathis JE. Oxidative DNA and RNA damage in the livers of Sprague-Dawley rats treated with the hepatocarcinogen 2-nitropropane. Cancer Res. 1989;49:5518–22. [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nature Struct Mol Biol. 2006;13:1002–9. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gaillard H, Aguilera A. A novel class of mRNA containing cytoplasmic granules are produced in response to UV-irradiation. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-02-0193. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambichler T, Moussa G, Tomi NS, Paech V, Altmeyer P, Kreuter A. Reference limits for erythema-effective UV doses. Photoch Photobiol. 2006;82:1097–102. doi: 10.1562/2006-02-06-RA-796. [DOI] [PubMed] [Google Scholar]
- Geiszt M, Kopp JB, Varnai P, Leto TL. Identification of Renox, an NAD(P)H oxidase in kidney. Proc Natl Acad Sci USA. 2000;97:8010–8014. doi: 10.1073/pnas.130135897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tao R, Li Z. Quantification of RNA damage by reverse transcription polymerase chain reactions. Anal Biochem. 2006;357:58–67. doi: 10.1016/j.ab.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Gordon MP, Huang C, Hurter J. Photochemistry and photobiology of ribonucleic acids, ribonucleoproteins, and RNA viruses. In: Wang SY, editor. Photochemistry and Photobiology of Nucleic Acids Volume II: Biology. Academic Press; New York: 1976. pp. 265–308. [Google Scholar]
- Görg B, et al. Ammonia induces RNA oxidation in cultured astrocytes and brain in vivo. Hepatology. 2008;48:567–79. doi: 10.1002/hep.22345. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem Journal. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CL, Pattison DI, Whiteman M, Davies MJ. Chlorination and nitration of DNA and nucleic acid components. In: Evans MF, Cooke MS, editors. Oxidative Damage to Nucleic Acids. Landes Bioscience; Austin, T. X: 2007. pp. 14–39. [Google Scholar]
- Hayakawa H, Hofer A, Thelander L, Kitajima S, Cai Y, Oshiro S, Yakushiji H, Nakabeppu Y, Kuwano M, Sekiguchi M. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry. 1999;38(12):3610–4. doi: 10.1021/bi982361l. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Kuwano M, Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40:9977–82. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Kuwano M, Sekiguchi M. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002;41:12739–44. doi: 10.1021/bi0201872. [DOI] [PubMed] [Google Scholar]
- Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–7. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- Hofer T, Marzetti E, Xu J, Seo AY, Gulec S, Knutson MD, Leeuwenburgh C, Dupont-Versteegden EE. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp Gerontol. 2008;43:563–70. doi: 10.1016/j.exger.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer T, Seo AY, Prudencio M, Leeuwenburgh C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol Chem. 2006;387:103–11. doi: 10.1515/BC.2006.014. [DOI] [PubMed] [Google Scholar]
- Honda K, Smith MA, Zhu X, Baus D, Merrick WC, Tartakoff AM, Hattier T, Harris PL, Siedlak SL, Fujioka H, et al. Ribosomal RNA in Alzheimer disease Is oxidized by bound redox-active iron. J Biol Chem. 2005;280:20978–20986. doi: 10.1074/jbc.M500526200. [DOI] [PubMed] [Google Scholar]
- Hoskins J, Butler JS. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics. 2008;179:323–30. doi: 10.1534/genetics.107.082727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Huggins W, Ghosh SK, Nanda K, Wollenzien P. Internucleotide movements during formation of 16S rRNA-rRNA photocrosslinks and their connection to the 30S subunit conformational dynamics. J Mol Biol. 2005;354:358–374. doi: 10.1016/j.jmb.2005.09.060. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Magun BE. Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J Biol Chem. 1998;273:15794–803. doi: 10.1074/jbc.273.25.15794. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Hayakawa H, Ito R, Miyazawa M, Yamagata Y, Sekiguchi M. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 2005;33:3779–84. doi: 10.1093/nar/gki682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–56. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Gillespie D. Metabolic events occurring during recovery from prolonged glucose starvation in Escherichia coli. J Bacteriol. 1968;95:1030–9. doi: 10.1128/jb.95.3.1030-1039.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäckle H, Kalthoff K. Photoreactivation of RNA in UV-irradiated insect eggs (Smittia sp., Chironomidae, Diptera) I. Photosensitized production and light-dependent disappearance of pyrimidine dimers. Photochem Photobiol. 1978;27:309–15. doi: 10.1111/j.1751-1097.1978.tb07605.x. [DOI] [PubMed] [Google Scholar]
- Jäckle H, Kalthoff K. Photosensitized formation of RNA-protein crosslinks in an insect egg (Smittia spec., Chironomidae, Diptera) Photochem Photobiol. 1979;29:1039–40. doi: 10.1111/j.1751-1097.1979.tb07811.x. [DOI] [PubMed] [Google Scholar]
- Jäckle H, Kalthoff K. Photoreversible UV-inactivation of messenger RNA in an insect embryo (Smittia Spec., Chironomidae, Diptera) Photochem Photobiol. 1980;32:749–761. doi: 10.1111/j.1751-1097.1980.tb04052.x. [DOI] [PubMed] [Google Scholar]
- Kajitani K, Yamaguchi H, Dan Y, Furuichi M, Kang D, Nakabeppu Y. MTH1, an oxidized purine nucleoside triphosphatase, suppresses the accumulation of oxidative damage of nucleic acids in the hippocampal microglia during kainate-induced excitotoxicity. J Neurosci. 2006;26:1688–1698. doi: 10.1523/JNEUROSCI.4948-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminer MS. Photodamage: magnitude of the problem. In: Gilchrest BA, editor. Photodamage. Blackwell Science; Cambridge, M. A: 1995. pp. 1–11. [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Sancar A. Effect of base, pentose, and phosphodiester backbone structures on binding and repair of pyrimidine dimers by Escherichia coli DNA photolyase. Biochem. 1991;30:8623–30. doi: 10.1021/bi00099a019. [DOI] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 2003;25:691–8. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lalonde MS, Zuo Y, Zhang J, Gong X, Wu S, Malhotra A, Li Z. Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA. 2007;13:1957–1968. doi: 10.1261/rna.706207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–8. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fournier MJ. Interference probing of rRNA with snoRNPs: A novel approach for functional mapping of RNA in vivo. RNA. 2004;10:1130–1141. doi: 10.1261/rna.7190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM, Cotman CW, Ames BN. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc Natl Acad Sci USA. 2002;99:2356–61. doi: 10.1073/pnas.261709299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–37. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Adachi Y, Hiraku Y, Horiki N, Horiike S, Imoto I, Pinlaor S, Murata M, Semba R, Kawanishi S. Accumulation of 8-nitroguanine in human gastric epithelium induced by Helicobacter pylori infection. Biochem Bioph Res Co. 2004;319:506–510. doi: 10.1016/j.bbrc.2004.04.193. [DOI] [PubMed] [Google Scholar]
- Martinet W, de Meyer GRY, Herman AG, Kockx MM. Reactive oxygen species induce RNA damage in human atherosclerosis. Eur J Clin Invest. 2004;34:323–7. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- Masuda M, Nishino H, Ohshima H. Formation of 8-nitroguanosine in cellular RNA as a biomarker of exposure to reactive nitrogen species. Chem-Biol Interact. 2002;139:187–97. doi: 10.1016/s0009-2797(01)00299-x. [DOI] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- Miller N, Cerutti P. Structure of the photohydration products of cytidine and uridine. Proc Natl Acad Sci USA. 1968;59:34–38. doi: 10.1073/pnas.59.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Regnier F. Protein-RNA cross-linking in the ribosomes of yeast under oxidative stress. J Proteome Res. 2006;5:3249–59. doi: 10.1021/pr060337l. [DOI] [PubMed] [Google Scholar]
- Mitchell D. Revisiting the photochemistry of solar UVA in human skin. Proc Natl Acad Sci USA. 2006;103:13567–68. doi: 10.1073/pnas.0605833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Nucleic acid oxidation in Alzheimer disease. Free Radical Bio Med. 2008;44:1493–505. doi: 10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci USA. 2006;103:13765–70. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller K, Zwieb C, Brimacombe R. Identification of the oligonucleotide and oligopeptide involved in an RNA-protein crosslink induced by ultraviolet irradiation of Escherichia coli 30 S ribosomal subunits. J Mol Biol. 1978;126:489–506. doi: 10.1016/0022-2836(78)90055-4. [DOI] [PubMed] [Google Scholar]
- Mroczek S, Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 2008;36:2874–88. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell. 2008;31:278–86. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Brunson D, Crespi CL, Penman BW, Wishnok JS, Tannenbaum SR. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc Natl Acad Sci USA. 1992;89:3030–4. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah JW, Shapkina T, Wollenzien P. UV-induced crosslinks in the 16S rRNAs of Escherichia coli, Bacillus subtilis and Thermus aquaticus and their implications for ribosome structure and photochemistry. Nucleic Acids Res. 2000;28:3785–3792. doi: 10.1093/nar/28.19.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A, Chiba S, Kosaka K, Takeda A, Castellani RJ, Smith MA, Perry G. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies. Neuroreport. 2002;13:2035–9. doi: 10.1097/00001756-200211150-00009. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Chiba S, Lippa CF, Cras P, Kalaria RN, Takeda A, Honda K, Smith MA, Perry G. Neuronal RNA oxidation is a prominent feature of familial Alzheimer’s disease. Neurobiol Dis. 2004;17:108–113. doi: 10.1016/j.nbd.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Honda K, Takeda A, Hirai K, Zhu X, Smith MA, Perry G. Oxidative Damage to RNA in neurodegenerative diseases. J Biomed Biotechnol. 2006;2006:82323. doi: 10.1155/JBB/2006/82323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer’s disease. J Neurosci. 1999;19:1959–64. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Wolin SL. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Devel. 1994;8:2891–903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, Kuwano M, Kohno K. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- Ohshima H, Sawa T, Akaike T. 8-nitroguanine, a product of nitrative DNA damage caused by reactive nitrogen species: formation, occurrence, and implications in inflammation and carcinogenesis. Antioxid Redox Sign. 2006;8:1033–45. doi: 10.1089/ars.2006.8.1033. [DOI] [PubMed] [Google Scholar]
- Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes Pl. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–16. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Park E, Shigenaga MK, Degan P, Korn TS, Kitzler JW, Wehr CM, Kolachana P, Ames BN. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivative from biological fluids with a monoclonal antibody column. Proc Natl Acad Sci. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M, Johns HE. Suppression of hydrate and dimer formation in ultraviolet-irradiated poly (A plus U) relative to poly U. J Mol Biol. 1966;20:215–29. doi: 10.1016/0022-2836(66)90061-1. [DOI] [PubMed] [Google Scholar]
- Pinlaor S, Yongvanit P, Hiraku Y, Ma N, Semba R, Oikawa S, Murata M, Sripa B, Sithithaworn P, Kawanishi S. 8-nitroguanine formation in the liver of hamsters infected with Opisthorchis viverrini. Biochem Bioph Res Co. 2003;309:567–71. doi: 10.1016/j.bbrc.2003.08.039. [DOI] [PubMed] [Google Scholar]
- Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–21. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- Prütz WA. Hypochlorous acid interactions with thiols, nucleotides, DNA, and other biological substrates. Arch Biochem Biophys. 1996;332:110–20. doi: 10.1006/abbi.1996.0322. [DOI] [PubMed] [Google Scholar]
- Ramabhadran TV, Fossum T, Jagger J. In vivo induction of 4-thiouridine-cytidine adducts in tRNA of E. coli B/r by near-ultraviolet radiation. Photochem Photobiol. 1976;23:315–21. doi: 10.1111/j.1751-1097.1976.tb07254.x. [DOI] [PubMed] [Google Scholar]
- Ravanat J, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobio B. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Reinisch KM, Wolin SL. Emerging themes in non-coding RNA quality control. Curr Opin Struc Biol. 2007;17:209–214. doi: 10.1016/j.sbi.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Reiter TA. NO* chemistry: a diversity of targets in the cell. Redox Rep. 2006;11:194–206. doi: 10.1179/135100006X116718. [DOI] [PubMed] [Google Scholar]
- Remsen JF, Miller N, Cerutti PA. Photohydration of uridine in the RNA of coliphage R17. II. The relationship between ultraviolet inactivation and uridine photohydration. Proc Natl Acad Sci USA. 1970;65:460–6. doi: 10.1073/pnas.65.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Banan A, Keshavarzian A, Zhu Q, LaPaglia N, McNulty J, Emanuele NV, Emanuele MA. Exposure to ethanol induces oxidative damage in the pituitary gland. Alcohol. 2005;35:91–101. doi: 10.1016/j.alcohol.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 1999;18:490–9. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- Shan X, Tashiro H, Lin C. The identification and characterization of oxidized RNAs in Alzheimer’s disease. J Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chang Y, Lin CG. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- Shan X, Lin CG. Quantification of oxidized RNAs in Alzheimer’s disease. Neurobiol Aging. 2006;27:657–62. doi: 10.1016/j.neurobiolaging.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Shen Z, Wu W, Hazen S. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry. 2000;39:5474–5482. doi: 10.1021/bi992809y. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. Chemical modification of viral ribonucleic acid: IX. The effect of ultraviolet irradiation on TMV-RNA and other polynucleotides. Virology. 1971;45:101–107. doi: 10.1016/0042-6822(71)90117-6. [DOI] [PubMed] [Google Scholar]
- Small GD, Tao M, Gordon MP. Pyrimidine hydrates and dimers in ultraviolet-irradiated tobacco mosaic virus ribonucleic acid. J Mol Biol. 1968;38:75–87. doi: 10.1016/0022-2836(68)90129-0. [DOI] [PubMed] [Google Scholar]
- Smith KC. The radiation-induced addition of proteins and other molecules to nucleic acids. In: Wang SY, editor. Photochemistry and photobiology of nucleic acids. Academic Press; New York: 1976. pp. 187–218. [Google Scholar]
- Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, Lambeth JD. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- Taddei F, Hayakawa H, Bouton M, Cirinesi A, Matic I, Sekiguchi M, Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–30. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci USA. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Johnson AW, Kolodner RD. Molecular and genetic analysis of the gene encoding the Saccharomyces cerevisiae strand exchange protein Sep1. Mol Cell Biol. 1991;11:2593–608. doi: 10.1128/mcb.11.5.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:1–9. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith Gr, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamer WG, Wei RR. In vitro photooxidation of nucleic acids by ultraviolet A radiation. Photochem Photobiol. 1997;65:560–3. doi: 10.1111/j.1751-1097.1997.tb08605.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–16. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watt PM, Borts RH, Louis EJ, Hickson ID. The topoisomerase II-associated protein, Pat1p, is required for maintenance of rDNA locus stability in Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:831–40. doi: 10.1007/s004380050027. [DOI] [PubMed] [Google Scholar]
- Weimann A, Belling D, Poulsen HE. Quantification of 8-oxo-guanine and guanine as the nucleobase, nucleoside and deoxynucleoside forms in human urine by high-performance liquid chromatography-electrospray tandem mass spectrometry. Nucleic Acids Res. 2002;30:e7. doi: 10.1093/nar/30.2.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilms C, Noah JW, Zhong D, Wollenzien P. Exact determination of UV-induced crosslinks in 16S ribosomal RNA in 30S ribosomal subunits. RNA. 1997;3:602–612. [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Bioph Res Co. 2008;372:288–92. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub A, Augeri L, Kelley MR, Doetsch PW, Deutsch WA. A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 1996;15:2306–12. [PMC free article] [PubMed] [Google Scholar]
- Yacoub A, Kelley M, Deutsch W. Drosophila ribosomal protein PO contains apurinic/apyrimidinic endonuclease activity. Nucleic Acids Res. 1996;24:4298–4303. doi: 10.1093/nar/24.21.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa H, Ogawa Y, Ueno M. Redox ribonucleosides. Isolation and characterization of 5- hydroxyuridine, 8-hydroxyguanosine, and 8-hydroxyadenosine from Torula yeast RNA. J Biol Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]
- Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13:704–12. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–5. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–9. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Ross A, Rinke J, Meinke M, Brimacombe R. Evidence for RNA-RNA cross-link formation in Escherichia coli ribosomes. Nucleic Acids Res. 1978;5:2705–20. doi: 10.1093/nar/5.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]