Abstract
Class-A scavenger receptors (SR-A) and TLR mediate early immune responses against pathogenic bacteria. SR-A and TLR molecules are expressed on phagocytes and interact with common ligands from Gram-negative and Gram-positive bacteria; however, the contribution of TLR activity to SR-A-mediated phagocytosis has not been assessed directly. Herein, we provide genetic and functional evidence that ligand- and TLR-specific stimuli synergize with SR-A to mediate bacterial phagocytosis. Although complete loss of SR-A (SR-A−/−) is known to impair bacterial clearance, here we identify the first deficiency attributable to SR-A heterozygosity: SR-A+/−TLR4+/− cells and mice are impaired significantly in the clearance of Gram-negative Escherichia coli. This phenotype is specific to the TLR signaling event, as SR-A+/−TLR4+/− cells are not deficient for the clearance of Gram-positive Staphylococcus aureus bacteria, which contain cell-surface TLR2 ligands but lack TLR4 ligands. We demonstrate that this is a global, phagocytic mechanism, regulated independently by multiple TLRs, as analogous to the SR-A+/−TLR4+/− deficit, SR-A+/−TLR2+/− cells are impaired for S. aureus uptake. In support of this, we show that SR-A+/−MyD88+/− cells recapitulate the phagocytosis defect observed in SR-A+/−TLR4+/− cells. These data identify for the first time that TLR-driven innate immune responses, via a MyD88 signaling mechanism, regulate SR-A-dependent phagocytosis of bacteria. These findings provide novel insights into how innate immune cells control SR-A-mediated trafficking and are the first demonstration that subtle changes in the expression of SR-A and TLRs can substantially affect host bacterial clearance.
Keywords: innate immunity, dendritic cells, E. coli, trafficking
INTRODUCTION
Phagocytic cells mediate the early immunological responses to bacterial infections. The innate immune response by these phagocytes, which include macrophages and dendritic cells (DC), is orchestrated by a plethora of pattern-recognition receptors (PRR) that recognize specific molecular signatures that are shared amongst extracellular microbes [1, 2]. During bacterial infection, microbes engage a variety of PRR on the surface of macrophages and DC which initiates the process of phagocytosis, whereby phagocytic cells engulf pathogenic microbes and degrade them [3,4,5]. Amongst these PRR is the scavenger receptor family [6, 7], which binds and traffics a variety of endogenous and microbial ligands, and the TLR family, which stimulates phagocyte activation, maturation, and the release of proinflammatory cytokines [8, 9]. TLR and Class-A scavenger receptor (SR-A) are members of these separate PRR families. In the context of bacterial recognition, SR-A binds bacterial cell-wall components [10,11,12], including LPS on Gram-negative bacteria and lipoteichoic acid (LTA) on Gram-positive bacteria, and is thought to be predominantly a trafficking receptor. In contrast, LPS interaction with the TLR4 complex and LTA interaction with TLR2 lead to NF-κB activation through MyD88 signaling [13,14,15]. However, the functional relationship between SR-A and TLR during the host response to bacterial infection is poorly understood.
Loss of SR-A in mice causes two major impairments in host defense against pathogenic infection. First, SR-A-deficient (SR-A−/−) mice are considerably more susceptible to Gram-negative and Gram-positive bacterial infections than wild-type (WT) controls as a result of the impaired ability of these mice to clear bacterial burdens early during infections [16, 17]. Second, in Gram-negative bacterial infections, SR-A−/− mice exhibit an increased susceptibility to endotoxic shock [11, 18, 19]. This is thought to be caused by decreased clearance of LPS in the absence of SR-A, thus leaving more LPS to activate TLR4 signaling and induce hyperinflammation. Since SR-A and TLR4 are expressed on the same cells and bind the same microbial molecules [20], and murine knockouts for either gene are compromised in their ability to respond to Gram-negative bacterial challenge [16, 17, 21, 22], we hypothesized that these molecules may functionally synergize to mediate bacterial clearance.
In these studies, we investigate the relationship between SR-A and TLR4 in the process of phagocytosis of bacteria by murine phagocytes. In particular, we assess how subtle changes in these two cell-surface receptors impacted the process of bacterial recognition and clearance by innate immune cells. Here, we provide genetic evidence for TLR4 and SR-A synergy in bacterial phagocytosis, showing that phagocytes heterozygous for TLR4 and SR-A are deficient in vitro and in vivo in the phagocytosis of the Gram-negative bacteria Escherichia coli, and cells singly heterozygous for either gene are fully competent in this regard. We show that this phenotype is specific to Gram-negative bacteria, but cells heterozygous for TLR2 and SR-A are deficient in the phagocytosis of Gram-positive bacteria. Furthermore, we demonstrate that MyD88-dependent signaling processes affect the rate of SR-A-driven phagocytosis as opposed to cell-surface binding of bacteria. These studies provide new insight into how multiple PRR control pathogen-specific phagocytosis by DC. This is the first report of a defective phenotype and function associated with SR-A heterozygosity and the first evidence that TLRs modulate SR-A-mediated phagocytosis.
MATERIALS AND METHODS
Mice
C57BL/10J and TLR-deficient (TLR2−/−) mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). C57BL/10ScNCr mice, which have a naturally occurring deletion of the TLR4 gene (TLR4−/−), and C57BL/6 mice were obtained from the National Cancer Institute (Frederick, MD, USA). SR-A−/− mice (C57BL/6 background) were a generous gift of Drs. Tatsuhiko Kodama (Tokyo University, Japan) and Mason W. Freeman (Massachusetts General Hospital, Boston, MA, USA) [23, 24]. MyD88−/− mice were generated by Adachi et al. [25]. Mice heterozygous for a single gene (TLR4+/−, SR-A+/−, or MyD88+/−) were bred using TLR4, SR-A, or MyD88 null animals mated to C57BL/6 WT mice. Double heterozygote mice—SR-A+/−TLR4+/−, SR-A+/−TLR2+/−, and SR-A+/−MyD88+/− genotypes—were bred by mating SR-A−/− mice to TLR4−/−, TLR2−/−, and MyD88−/− mice, respectively. Double knockout mice (SR-A−/−TLR4−/−) were bred by mating double heterozygote mice (SR-A+/−TLR4+/−). Litters from these mice were screened using gel electrophoresis analysis of PCR products from primer sets described previously for SR-A [26]. TLR4 genotypes were determined using forward primer 5′ ACATTCCTGTAAGTTACCTGCATATTT 3′ [27] and reverse primer 5′ CAATGGTCACATCACATAGTCC 3′. Mice that screened as double knockout mice by PCR were subsequently confirmed to be SR-A−/−TLR4−/− by FACS analysis of TLR4 expression and Western analysis of SR-A expression. SR-A heterozygous TLR4 knockout mice (SR-A+/−TLR4−/−) were generated by breeding double knockout mice (SR-A−/−TLR4−/−) with SR-A knockout mice (SR-A−/−). For FACS analysis of SR-A expression levels, SR-A+/− and SR-A+/−TLR4+/− mice were bred by mating C57BL/6 SR-A−/− mice or SR-A−/−TLR4−/−, respectively, to WT Balb/C mice to allow for staining with the 2F8 mAb, which does not recognize SR-A in C57BL/6 mice as a result of a genetic polymorphism [28].
Bone marrow-derived DC (BMDC) culture
The DC culture protocol is described previously [29] and is a modification of Inaba et al. [30]. In summary, BMDC were resuspended at 106 cells/ml in DC culture media (RPMI-1640 medium, 10% heat-inactivated FBS, 100 units/ml penicillin/streptomycin, 50 mM β-ME, 5% cell culture supernatant from x63 cells secreting GM-CSF [31]) and plated in six-well tissue-culture plates. On Days 2 and 4, the cells were washed and re-fed, and nonadherent cells were removed. On Day 6, wells were washed vigorously with culture medium to collect semiadherent cells, which were then phenotypically confirmed to be immature DC.
Antibodies
Anti-SR-A antibody used for Western blot confirmation of genotypes (data not shown) was from R&D Systems (Minneapolis, MN, USA); 2F8 anti-SR-A antibody for FACS analysis was purchased from Serotec (Raleigh, NC, USA); anti-CD11c and anti-F4/80 were from eBioscience (San Diego, CA, USA); and anti-TLR4 was purchased from Abcam (Cambridge, MA, USA).
Endocytic uptake/bacterial-binding assays
Acetylated low-density lipoprotein (AcLDL) uptake and trafficking assays were performed as described previously [32, 33]. Alexa488-AcLDL was purchased from Molecular Probes (Eugene, OR, USA). BMDC were incubated in 1 μg/ml AcLDL for 20 min at 37°C. Cells were then washed and analyzed by FACS for AcLDL uptake. Bacterial binding assays were performed with heat-killed, fluorescently labeled E. coli, which were heat-killed prior to labeling by incubation for 1 h at 90°C and fluorescently labeled as described previously [34, 35]. Binding assays were performed by incubating BMDC in the presence of labeled bacteria at 4°C for 30 min, washing cells three times with PBS, and assaying for fluorescence by FACS.
Bacterial strains
A DH5α E. coli strain transfected with a pTrcHis plasmid (Invitrogen, Carlsbad, CA, USA) to confer ampicillin resistance was used for all phagocytosis assays for Gram-negative bacteria uptake. The Staphylococcus aureus strain SMC4371 is an erythromycin-resistant variant of the laboratory strain RN4220 and was used for phagocytosis assays of Gram-positive bacteria and was obtained from Dr. George O’Toole (Dartmouth College, Lebanon, NH, USA).
Gentamycin protection assays
Phagocytosis of live E. coli or S. aureus was performed as a modified version of protocols described previously [35, 36]. Overnight cultures of E. coli or S. aureus were washed twice in 10 ml serum-free HBSS and centrifuged at 6000 rpm. The bacterial pellets were resuspended in HBSS, and the bacteria concentration was determined by spectrophotometric absorbance at 600 nM. BMDC (2.5×105) from the various mouse genotypes described were incubated with bacteria at the indicated multiplicity of infection (MOI) for 45 min at 37°C. BMDC were washed three times in serum-free HBSS and incubated in 100 μg/mL gentamycin for 30 min at 37°C. Cells were then washed and resuspended in 0.1% Triton X-100 in PBS. Lysates were plated on Luria-Bertani (LB)-agar plates containing ampicillin (E. coli) or tryptic soy agar plates containing erythromycin (S. aureus) and incubated overnight at 37°C. Subsequently, ampR or ermR colonies were counted, and CFUs were calculated based on the fraction of the total sample plated. For kinetic analysis, assays were performed as described above with gentamycin added at incremental time-points to permit analysis of bacterial uptake at early stages in the phagocytic process. For analysis of phagocytic killing, BMDC were incubated with bacteria as above and, after gentamycin addition, BMDC were harvested in 15-min increments to assess of death of internalized bacteria over the course of the assay.
FACS analyses and confocal microscopy
For FACS analyses, cells were analyzed by flow cytometry at the Norris Cotton Cancer Center (NCCC) Englert Cell Analysis Laboratory (Dartmouth College), using a FACSCaliber cytometer and subsequently analyzed using CellQuest software. For confocal microscopy, cells affixed to coverslips were mounted in buffered glycerol with 1 mg/ml phenylenediamine. Microscopy was performed on a Zeiss LSM510 Meta microscope taking single optical sections with a 63× lens, followed by viewing on an LSM5 Image Browser.
In vivo bacterial uptake assay
Live DH5α E. coli bacteria (5×106) were i.p.-injected into the indicated genotypes of mice, which were killed 1 h postinjection, and peritoneal lavages were collected. Peritoneal cells were pelleted, washed twice in serum-free HBSS, and resuspended in 1 ml HBSS. Each suspension (250 μl) was incubated in the presence of gentamycin for 30 min at 37°C to kill noninternalized bacteria. Cells were washed twice in serum-free HBSS and resuspended in 500 μl 0.1% Triton X-100 in PBS. Resuspended cells/bacteria (20 μl) were plated on LB-agar/ampicillin plates and incubated overnight at 37°C. Colonies were counted the following morning, and CFUs were calculated based on fraction of total sample plated.
Statistical analyses
All graphs and datasets were analyzed using unpaired, two-tailed t-test analyses to compare experimental data with control data. Ninety-five percent confidence intervals were used in these analyses. For all graphs, P ≤ 0.05, where statistical significance is indicated.
RESULTS
SR-A+/–TLR4+/– BMDC are impaired in the phagocytosis of E. coli
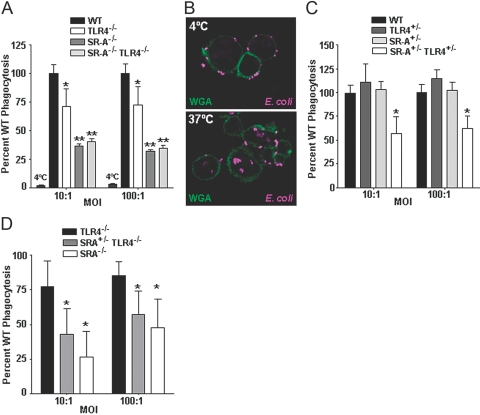
We and others [35, 37] have shown previously that SR-A is expressed on murine BMDC. In these studies, we used a gentamycin protection assay to test the relative capability of cells from various murine genotypes to phagocytose live DH5α bacteria. Unlike protocols using fluorescently labeled beads or apoptotic cells, the gentamycin protection assay rigorously distinguishes bacterial uptake from bacteria bound to the cell surface and enables quantitative analyses of the relative levels of bacteria actually internalized by phagocytic cells. The DH5α laboratory strain of E. coli was chosen specifically for these studies, as it has been shown previously to exhibit an intermediate SR-A dependency for its uptake by phagocytic cells compared with other E. coli strains tested [12]. We have shown previously that SR-A−/−, but not SR-A+/−, BMDC are impaired in their ability to phagocytose E. coli [35]. To test the hypothesis that TLR4 and SR-A are involved cooperatively in bacterial phagocytosis, we first assessed the role of TLR4 (Fig. 1A). Some previous reports indicate that TLR4−/− BMDC and aveolar macrophages are fully competent compared with WT cells at phagocytosis of E. coli [38, 39], and other reports suggest a role for TLR4 in peritoneal macrophage phagocytosis of E. coli during sepsis [40]. We observed that BMDC from TLR4−/− mice exhibit 25% impairment in the phagocytosis of live E. coli. The deficit in TLR4−/− BMDC remained constant over a range of MOIs tested (Fig. 1A). These findings demonstrate that loss of TLR4 negatively impacts phagocytosis of E. coli by BMDC. At 4°C, more than 97% of the phagocytic activity was abolished in WT cells compared with cells at 37°C (Fig. 1A); this demonstrates low levels of background associated with the gentamycin protection assay and specificity of the assay for internalized bacteria. To confirm that the gentamycin protection assay was indeed reflective of bacterial phagocytosis we performed confocal microscopy, which showed that BMDC incubated at 4°C are able to bind bacteria but not internalize them, and complete engulfment (phagocytosis) of E. coli occurs at 37°C (Fig. 1B). We then tested whether cells lacking TLR4 and SR-A exhibited an additive or synergistic defect. Although we observed the expected 70% deficit in SR-A−/− BMDC, we found that the TLR4−/−SR-A−/− BMDC were no further impaired than the SR-A−/− cells (Fig. 1A), indicating that SR-A deficiency is functionally dominant to TLR4 deficiency regarding the phagocytosis of live E. coli by BMDC.
Fig. 1.
SR-A+/−TLR4+/− BMDC are impaired in the phagocytosis of E. coli. (A) C57BL/10J (WT), TLR4−/−, SR-A−/−, and SR-A−/−TLR4−/− BMDC were incubated with live E. coli bacteria at 4°C (where noted) or 37°C at the indicated MOIs in a gentamycin protection assay. The mean CFUs of the experimental groups are expressed as a percentage of the mean WT CFU value. TLR4−/− cells were found to be impaired significantly (P<0.05) compared with WT cells. SR-A−/− and SR-A−/−TLR4−/− BMDC were impaired significantly (P<0.01) in the phagocytosis of E. coli compared with WT and TLR4−/− cells but were not significantly different from each other. At 4°C, WT BMDC exhibit >97% reduction in phagocytosis compared with 37°C conditions; n ≥ 12 for all genotypes. (B) C57BL/6 (WT) BMDC were incubated with Alexa647-labeled E. coli for 1 h at 4°C or 37°C. BMDC cell membranes were stained with FITC-labeled wheat germ agglutinin (WGA), and cells were imaged by confocal microscopy. Bacterial internalization was observed at 37°C but not 4°C, consistent with the gentamycin protection assay (A). (C) C57BL/10J, TLR4+/−, SR-A+/−, and SR-A+/−TLR4+/− BMDC were assayed for their quantitative ability to phagocytose live E. coli using a gentamycin protection assay as in (A). TLR4+/− and SR-A+/− BMDC exhibited a similar phagocytic ability as WT cells. However, SR-A+/−TLR4+/− BMDC were impaired significantly (*, P<0.01) in the phagocytosis of E. coli compared with WT BMDC at both MOIs tested. (D) TLR4−/−, SR-A+/−TLR4−/−, and SR-A−/− BMDC were tested for bacterial phagocytosis as in (A and C). BMDC from SR-A+/−TLR4−/− mice exhibited significantly lower levels of phagocytosis than BMDC from TLR4−/− cells; n ≥ 9 for all genotypes. For all graphs, the mean and sd are shown, and statistical significance (P≤0.05) from the WT values (A, C) or the TLR4−/− values (D) is indicated by *, and statistical significance from the WT and TLR4−/− values is indicated by **.
To address how subtle changes in expression of these receptors might affect the process of bacterial uptake, we next tested phagocytosis by BMDC heterozygous for TLR4 and SR-A. Although BMDC heterozygous for TLR4 or SR-A showed no deficit in phagocytosis compared with WT cells, double heterozygous SR-A+/−TLR4+/− BMDC were impaired significantly (P<0.01) in the phagocytosis of live E. coli, exhibiting a 50% deficit compared with WT cells (Fig. 1C). This is the first report of any functional deficit associated with heterozygosity for the SR-A gene and the first demonstration of TLR involvement in SR-A-mediated bacterial trafficking. Since the loss of a single copy of TLR4 renders cells functionally sensitive to the loss of a single copy of SR-A, we then tested whether cells completely deficient for TLR4 would likewise exhibit SR-A haplo-insufficiency. BMDC from mice that lacked TLR4 and were heterozygous for SR-A (SR-A+/−TLR4−/−) were impaired further in their ability to phagocytose E. coli compared with TLR4−/− cells (Fig. 1D). The phagocytosis levels of E. coli by SR-A+/−TLR4−/− BMDC trended toward a deficit intermediate between the SR-A+/−TLR4+/− and SR-A−/− BMDC, however the SR-A+/−TLR4−/− BMDC were not significantly (P>0.05) different from the SR-A+/−TLR4+/− cells. From these experiments, we conclude that partial or total loss of TLR4 expression sensitizes phagocytic cells to SR-A heterozygosity, genetically demonstrating that TLR4 contributes to SR-A-mediated phagocytic trafficking of E. coli.
The SR-A+/–TLR4+/– phagocytic deficit is specific for Gram-negative bacteria, and SR-A+/−TLR2+/− cells show a phagocytic deficit for the Gram-positive bacteria S. aureus
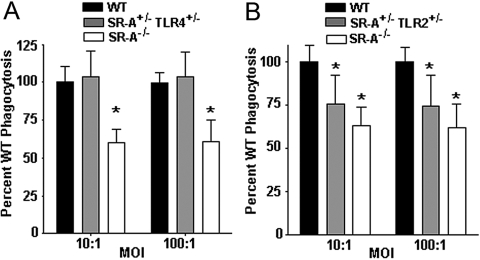
TLR4 signaling is well-documented to be activated by cell-wall products of Gram-negative bacteria [14], however an alternative explanation for the deficit observed in the SR-A+/−TLR4+/− BMDC is that for unknown reasons, loss of one or both copies of TLR4 causes a global, phagocytic defect that is not specific to interaction with a bacterial ligand. To test the specificity of the SR-A+/−TLR4+/− phenotype, we examined the phagocytosis of the Gram-positive bacteria S. aureus, which expresses the SR-A ligand LTA [10] but does not contain LPS in its cell membrane and thus does not engage the TLR4 signaling complex. Consistent with previous data using macrophages [17], SR-A−/− cells were impaired in their ability to phagocytose S. aureus compared with WT cells (Fig. 2A). Importantly, SR-A+/−TLR4+/− BMDC were not impaired in their phagocytosis of S. aureus (Fig. 2A). This result reveals novel and nuanced information about SR-A-mediated phagocytic trafficking. First, SR-A+/−TLR4+/− BMDC are fully competent at phagocytosis of S. aureus, while being grossly impaired for phagocytosis of E. coli, which demonstrates that the deficit for E. coli uptake is not the result of pleiotropic phagocytic defects in these cells. Second, these data show that it is ligand-specific engagement by TLR4 that leads to the deficit seen in SR-A+/−TLR4+/− BMDC. Furthermore, we found that SR-A+/−TLR2+/− BMDC are impaired for phagocytosis of S. aureus (Fig. 2B), which indicates that a more global relationship exists between TLRs and SR-A during phagocytosis. Together, these data show that TLR-specific signals, dependent on the type of bacteria engaged by the phagocytes, control SR-A-mediated phagocytic trafficking and that this mechanism is negatively affected in the context of concomitant heterozygosity in pathogen-specific TLRs and in SR-A. From these data, we conclude that the phagocytic deficit observed in SR-A+/−TLR4+/− BMDC is specific to altered TLR4 engagement by Gram-negative bacteria in these cells. Moreover, we show that loss of TLR4 does not impart a global deficit in phagocytosis and that multiple TLRs regulate SR-A-mediated phagocytosis of bacteria, depending on the specific type of bacteria involved. These results implicate a shared downstream molecule of TLR2 and TLR4 as mechanistically responsible for the phagocytic deficits observed in double- heterozygous BMDC.
Fig. 2.
SR-A+/−TLR2+/−, but not SR-A+/−TLR4+/−, cells are impaired at phagocytosis of the Gram-positive bacteria S. aureus. (A) Gentamycin protection assays were performed on C57BL/10J (WT), SR-A+/−TLR4+/−, and SR-A−/− BMDC, as in Figure 1, with the use of S. aureus bacteria. SR-A+/−TLR4+/− BMDC were comparable with WT cells at the phagocytosis of S. aureus, and SR-A−/− BMDC were impaired significantly (*, P<0.01) compared with WT cells. (B) Gentamycin protection assays were performed on WT, SR-A+/−TLR2+/−, and SR-A−/− BMDC using S. aureus bacteria. SR-A+/−TLR2+/− and SR-A−/− BMDC showed significant impairment in phagocytosis of S. aureus bacteria (*, P<0.001) compared with WT cells. For all graphs, n ≥ 18 for all genotypes, and the means and sd are shown.
TLR4 deficiency does not affect SR-A-mediated endocytic trafficking
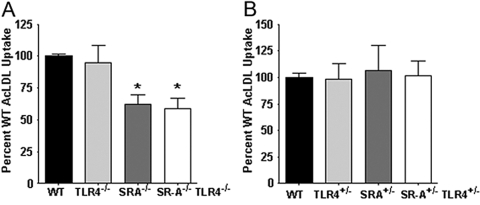
Scavenger receptors are able to traffic ligands through the mechanistically distinct phagocytic and endocytic internalization processes [41, 42], depending on the particulate nature of the ligand [43]. Since TLR4 heterozygosity affects SR-A-dependent phagocytic trafficking, we asked if this were also true for SR-A-mediated endocytic trafficking. To assess SR-A-mediated endocytic uptake by FACS analysis, we used Alexa488-labeled AcLDL. We observed the anticipated [18, 44] deficit for AcLDL uptake by SR-A−/− and SR-A−/−TLR4−/− BMDC (cells from both of these genotypes were functionally similar in this regard), and TLR4−/− BMDC were not impaired in comparison with WT cells (Fig. 3A). Endocytic uptake of AcLDL by SR-A+/−TLR4+/− BMDC was comparable with that by a single heterozygote and by WT BMDC (Fig. 3B). Thus, the trafficking deficit observed in SR-A+/−TLR4+/− BMDC appears to be specific to phagocytic events and is not recapitulated during SR-A-mediated endocytosis.
Fig. 3.
The trafficking deficit exhibited by SR-A+/−TLR4+/− BMDC is specific to SR-A-mediated phagocytosis and does not impair SR-A-mediated endocytic uptake. (A) WT, TLR4−/−, SR-A−/−, and SR-A−/−TLR4−/− BMDC were incubated with Alexa488-labeled AcLDL for 20 min at 37°C and analyzed quantitatively by FACS for endocytic uptake of AcLDL. SR-A−/− and SR-A−/−TLR4−/− BMDC exhibited significant (*, P<0.01) deficits in AcLDL uptake compared with WT cells, and TLR4−/− BMDC were similar to WT BMDC. (B) WT, TLR4+/−, SR-A+/−, and SR-A+/−TLR4+/− BMDC were treated as in A and analyzed for AcLDL uptake by FACS. Single heterozygous genotypes and the double heterozygous BMDC exhibited WT levels of AcLDL uptake.
SR-A+/–TLR4+/– BMDC exhibit WT levels of bacterial killing and bacterial binding
To identify the mechanism behind the phagocytic impairment in SR-A+/−TLR4+/− BMDC, we assessed multiple aspects of the phagocytic process. Since the gentamycin protection assay measures bacterial CFUs, one potential explanation for our results is that the relative decrease of CFUs in SR-A+/−TLR4+/− BMDC is a result of differential bacterial killing in these cells compared with WT controls. To test this, we performed gentamycin protection assays on WT, SR-A+/−TLR4+/−, and SR-A−/− BMDC and assessed phagocytic killing over the time period in which the previous assays (Figs. 1, A and C, and 2, A and B) occur. The gentamycin protection assay was performed as described previously with the exception that for each genotype, cells were harvested every 15 min following the addition of gentamycin to determine the rate of bacteria decrease (phagocytic killing) following internalization (Fig. 4A). By normalizing later time-points to the CFU levels of each genotype at 15 min after antibiotic addition, we were able to determine the relative rates of bacterial killing by BMDC. These experiments demonstrated that all of the genotypes exhibited the same rates of bacterial killing and that the level of killing observed over this duration (∼10% of the phagocytosed bacteria) does not account for the >50% impairment of bacterial uptake by the SR-A+/−TLR4+/− BMDC (Fig. 4A). We thereby eliminated differential bacteriocidal activity as an explanation for our data.
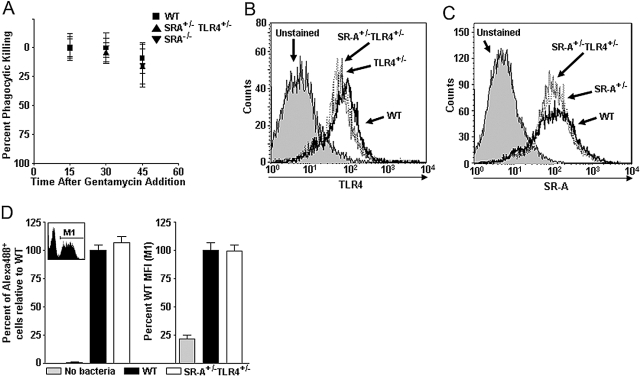
Fig. 4.
SR-A+/−TLR4+/− BMDC exhibit WT levels of bacterial killing and bacterial binding. (A) WT, SR-A+/−TLR4+/−, and SR-A−/− BMDC were incubated with E. coli in a gentamycin protection assay. After gentamycin addition, BMDC from each genotype were harvested every 15 min and lysed to assess the rate of killing of internalized bacteria over the course of the gentamycin protection assay. The graph is shown as percent phagocytic killing over time, and bacterial levels at the first time-point (15 min after gentamycin addition) were set as the baseline by which the later time-points are compared; n ≥ 8 for all genotypes, and mean and sd are shown. No statistical differences were observed among the genotypes. (B) TLR4 surface expression (assayed by FACS) of SR-A+/−TLR4+/− BMDC (dotted line) showed reduced TLR4 expression compared with WT (solid line) cells but identical expression to TLR4+/− cells (dashed line). (C) SR-A surface expression (assayed by FACS) of SR-A+/−TLR4+/− BMDC showed reduced expression of SR-A compared with WT (solid line) cells but identical expression to SR-A+/− cells (dashed line). (D) BMDC were incubated with Alex488-labeled E. coli in the linear range of binding. Gate M1 of the FACS histogram (inset) indicates BMDC that have bound Alexa488 E. coli. Neither the percent of the BMDC population that bound bacteria (number of cells within Gate M1 divided by the total number of cells; left panel) nor the relative number of bacteria bound to the cell surface per BMDC [mean fluorescence intensity (MFI) of Alex488+ BMDC within M1; right panel] was found to differ between WT and SR-A+/−TLR4+/− BMDC. For each parameter, the data are represented as a percentage of mean WT levels.
Another potential explanation for the phagocytic defect in SR-A+/−TLR4+/− BMDC is that heterozygosity of either gene affected the expression of the other gene. FACS analyses of TLR4 and SR-A expression revealed a modest decrease in protein expression in the respective single heterozygotes compared with WT cells (Fig. 4, B and C). However, importantly, expression levels of each receptor in the double heterozygous SR-A+/−TLR4+/− BMDC were identical to receptor expression levels observed in the single heterozygote BMDC (Fig. 4, B and C). Thus, loss of a single copy of either gene does not affect expression of the other gene. Another possible explanation related to receptor numbers is that fewer bacteria are able to be bound at the cell surface of SR-A+/−TLR4+/− BMDC. To discern whether the observed defect is a function of surface attachment of bacteria to the BMDC, fluorescently labeled, heat-killed E. coli were incubated with BMDC at 4°C, and the BMDC were subsequently assessed for bacterial binding by FACS analyses. We have described previously how this type of analysis allows assessment of bacterial binding along two parameters [35]. When the assay is performed in the linear range, only a portion of the BMDC binds bacteria, leaving a bimodal distribution of fluorescent cells that bound bacteria (Gate M1, Fig. 4D, inset histogram) and nonfluorescent cells that did not bind bacteria (ungated portion in Fig. 4D, inset histogram). The first parameter that was analyzed in this assay was the percent of the total BMDC population that bound E. coli bacteria during the assay (cells within Gate M1 as a percentage of the total number of cells analyzed). Our results show that SR-A+/−TLR4+/− BMDC do not differ significantly from WT cells in the percentage of the population that bound bacteria (Fig. 4D, left graph). The second parameter analyzed was the MFI of the cells that bound bacteria (within Gate M1 in Fig. 4D, inset histogram) among the different genotypes of BMDC, indicative of the relative amount of bacterial binding that occurred within those cells that did bind bacteria. By this analysis, we found that SR-A+/−TLR4+/− BMDC were not impaired in their average binding of fluorescently labeled E. coli (Fig. 4D, right graph). These data demonstrate that the defect in phagocytosis in SR-A+/−TLR4+/− BMDC is not a result of a defect in recognition or binding of the bacteria to the surface of BMDC. This indicated to us that the deficit was likely a result of impairment in the internalization of the bacteria.
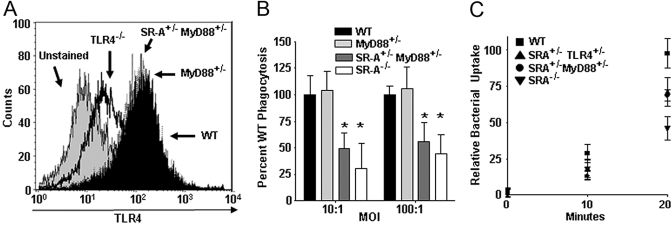
SR-A+/–MyD88+/– cells recapitulate the SR-A+/–TLR4+/– phagocytic deficit
Since SR-A+/−TLR4+/− BMDC showed a specific deficit for Gram-negative bacteria, and SR-A+/−TLR2+/− BMDC showed a similar deficit for Gram-positive bacteria, we hypothesized that a common downstream signaling molecule might be mechanistically responsible for the defective phenotypes we observed in double heterozygous cells. MyD88 is a shared signaling adaptor downstream of many of the TLRs (including TLR4 and TLR2), and loss of MyD88 (MyD88−/−) has been reported previously to impair phagocytic efficiency [3, 4]. Therefore, MyD88 represented a logical, functional pathway by which the TLR molecules were exerting their effects on SR-A-mediated phagocytosis. To test this hypothesis, we bred SR-A+/−MyD88+/− mice and assessed BMDC from these mice for their ability to recapitulate the defect seen in SR-A+/−TLR4+/− BMDC. First, we examined whether MyD88 heterozygosity affected surface expression of TLR4 in BMDC. Loss of MyD88−/− has been reported previously to have no effect on the cell-surface expression of TLR4 [45]. In accord with this, we found that MyD88+/− and SR-A+/−MyD88+/− BMDC exhibit WT levels of surface TLR4 expression, and TLR4−/− BMDC show no TLR4 expression by FACS analysis (Fig. 5A). Using a gentamycin protection assay to test the quantitative phagocytosis of E. coli by WT, MyD88+/−, SR-A+/−MyD88+/−, and SR-A−/− BMDC, we found that MyD88+/− BMDC display WT levels of E. coli phagocytosis, and SR-A+/−MyD88+/− and SR-A−/− show significant (P≤0.05) impairment in phagocytosis of E. coli compared with WT cells (Fig. 5B). Since MyD88+/− and SR-A+/−MyD88+/− BMDC express identical levels of surface TLR4, we can conclude definitively that the phagocytosis defect seen in the double heterozygous cells is not a result of aberrant distribution of receptor numbers at the cell surface or a cell surface-binding deficit reflective of total receptor numbers.
Fig. 5.
SR-A+/−TLR4+/− BMDC exhibit impaired kinetics of bacterial internalization and SR-A+/−MyD88+/− cells recapitulate this defect. (A) FACS histogram showing TLR4 cell-surface expression on BMDC from WT, TLR4−/−, MyD88+/−, and SR-A+/−MyD88+/− mice. MyD88 heterozygosity does not affect cell-surface expression of TLR4. (B) Gentamycin protection assays were performed on WT, MyD88+/−, SR-A+/−MyD88+/−, and SR-A−/− BMDC using E. coli bacteria. SR-A+/−MyD88+/− and SR-A−/− BMDC showed significant impairment in phagocytosis of E. coli (*, P<0.05) compared with WT cells. For all graphs, n ≥ 18 for all genotypes, and the means and sd are shown. (C) Kinetic analysis of bacterial uptake was performed on WT, SR-A+/−TLR4+/−, SR-A+/−MyD88+/−, and SR-A−/− BMDC. Quantitative uptake of bacteria by BMDC was assessed every 10 min using a gentamycin protection assay to determine the relative rates of phagocytosis for the indicated BMDC genotypes. SR-A+/−TLR4+/−, SR-A+/−MyD88+/−, and SR-A−/− BMDC show impairment for the internalization of bacteria as early as 10 min after BMDC-bacteria coincubation, and this deficit is fully apparent by 20 min after bacterial exposure. For all graphs, the means and sd are shown.
Having shown that TLR4 heterozygosity does not impair BMDC from binding to E. coli (Fig. 4D), we next tested the kinetics of phagocytosis in our genotypes of interest to see if differences in internalization kinetics were responsible for the deficit in bacterial uptake that we observed in SR-A+/−TLR4+/− and SR-A+/−MyD88+/− BMDC. To assess the rate of bacterial phagocytosis we modified the gentamycin protection assay described previously. The various genotypes of BMDC were incubated with bacteria at 37°C; then, at 10-min intervals, starting at 0 min, gentamycin was added to the assay. At Time 0 min, there were essentially no bacteria protected from the gentamycin, which demonstrates that there is little background associated with this assay (Fig. 5C). Within 10 min following BMDC-bacteria coincubation, measurable phagocytosis had occurred (bacteria protected from gentamycin), and the number of bacteria taken up by SR-A+/−TLR4+/−, SR-A+/−MyD88+/−, and SR-A−/− BMDC was lower than WT levels (Fig. 5C). By 20 min, three distinct phagocytic levels were observed, and the SR-A+/−TLR4+/− and SR-A+/−MyD88+/− BMDC had an intermediate level between WT and SR-A−/−. These findings are consistent with the defects observed in Figures 1C and 5B. From these results, we conclude that the phagocytic deficit in TLR4+/−SR-A+/− and SR-A+/−MyD88+/− BMDC is a result of an impaired rate of internalization of bacteria into these cells that is controlled mechanistically through the MyD88-dependent pathway.
SR-A+/−TLR4+/− mice are impaired in the in vivo phagocytosis of E. coli
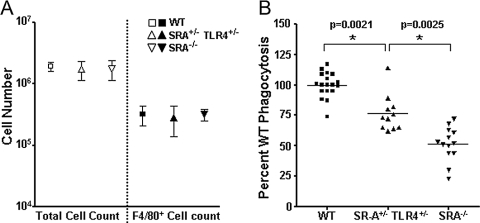
Since SR-A+/−TLR4+/− cells are defective in the phagocytosis of E. coli in vitro, we then asked if SR-A+/−TLR4+/− mice would be deficient for bacterial clearance in vivo. One hour following the injection of 5 × 106 live E. coli bacteria into their peritoneal cavity, the experimental groups of mice were killed and peritoneal cells were collected by lavage. Peritoneal cells were counted and stained with the macrophage marker F4/80 to determine total peritoneal cells and macrophage numbers per mouse. No differences were observed in total peritoneal cell numbers or in F4/80+ cell numbers among WT, SR-A+/−TLR4+/−, and SR-A−/− mice (Fig. 6A). Peritoneal cells were then treated in a gentamycin protection assay to determine the levels of in vivo phagocytosis. Relative CFUs of phagocytosed bacteria from each treatment group were assessed as described in Materials and Methods. Peritoneal phagocytes in SR-A+/−TLR4+/− mice were impaired significantly (P<0.005) at uptake of live E. coli bacteria compared with WT mice and consistent with the in vitro data, were intermediate between WT and SR-A−/− mice (Fig. 6B). These data demonstrate that peritoneal phagocytes from SR-A+/−TLR4+/− mice are impaired in their ability to take up E. coli. The in vivo data replicate our in vitro observations and confirm that concomitant heterozygosity for SR-A and TLR4 imparts a defect in the ability of murine phagocytes to clear E. coli bacteria.
Fig. 6.
SR-A+/−TLR4+/− mice are impaired at the in vivo phagocytosis of E. coli. WT, SR-A+/−TLR4+/−, and SR-A−/− mice were i.p.-injected with 5 × 106 live E. coli. After 1 h, mice were killed and cells harvested from the peritoneal lavage were treated in a gentamycin protection assay. Relative phagocytosis was determined as a percentage of WT CFU numbers. (A) WT, SR-A+/−TLR4+/−, and SR-A−/− mice showed similar numbers of total cells and F4/80+ cells in peritoneal exudates 1 h after peritoneal injection with live E. coli. Mean and sd are shown. (B) SR-A+/−TLR4+/− mice were impaired significantly (*, P<0.005) for the in vivo uptake of E. coli compared with WT mice, and SR-A−/− mice were impaired even further relative to SR-A+/−TLR4+/− mice (*, P<0.005). Values from individual mice in each group are represented on the graph along with the mean value for each group.
DISCUSSION
The innate immune system is an organism’s first line of defense against infection by pathogenic bacteria [46]. This initial immune response is largely dependent on a variety of PRR that respond to pathogens and initiate phagocytic clearance of microbes as well as the induction of inflammation at the site of infection [1, 2]. TLRs and SR-A are PRR that recognize major components of bacterial cell walls and are involved in the process of phagocytosis [3, 12, 35, 47]. In this report, we used mouse genetics to assess functional synergy between TLR4 and SR-A during the phagocytic uptake of the Gram-negative bacteria E. coli. We chose the DH5α strain based on previous findings that the DH5α strain of E. coli has an intermediate level of SR-A dependence for phagocytic uptake compared with other bacterial strains tested [12]. Thus, the avirulent DH5α strain provides a facile model for a number of virulent pathogenic microbes that are likewise cleared through SR-A-dependent mechanisms [16, 17].
Although SR-A−/− phagocytes are known to be deficient in bacterial uptake, the literature reports are inconsistent about the contribution of TLR4 to bacterial phagocytosis. BMDC and primary alveolar macrophages derived from the TLR4−/−C57BL/10ScNCr and C3H/HeJ strains of mice are reported to be fully competent at bacterial phagocytosis [38, 39]. However, TLR4 expression has also been reported to affect macrophage clearance of bacteria during peritoneal sepsis [39, 40]. Other studies have shown that TLR4 and its downstream signaling adaptor molecule MyD88 are critical for proper phagocytic efficiency [3, 4]. In our system, a single copy of the TLR4 gene is sufficient for normal phagocytic activity, and TLR4−/−BMDC show modest impairment in the phagocytosis of live E. coli bacteria in comparison with WT BMDC. Consistent with our previous findings [35], SR-A−/− BMDC exhibited substantial (>70% deficiency) impairment in their ability to phagocytose live bacteria. We hypothesized initially that deficiency in TLR4 and SR-A may exhibit an additive or synergistic, phagocytic deficiency. However, loss of TLR4 did not impair phagocytosis further in SR-A−/−cells. This demonstrates genetically and functionally that loss of SR-A is dominant to the role of TLR4 in the phagocytosis of E. coli.
The primary purpose of our investigation was to analyze how subtle changes in these cell-surface receptors impact the recognition and uptake of pathogenic bacteria. Although we found that heterozygosity for TLR4 or SR-A did not impact phagocytosis of E. coli, double heterozygous SR-A+/−TLR4+/− BMDC are impaired significantly and substantially in the phagocytosis of E. coli. To our knowledge, no functional defects associated with SR-A heterozygosity have been reported previously. TLR4 polymorphisms are well-documented in humans [48,49,50] and heterozygosity for these polymorphisms is associated with LPS hyporesponsiveness and increased susceptibility to bacterial infection [48, 51,52,53]. However, it is unclear whether these phenotypes reflect a phagocytic deficiency. The phagocytic deficiency that we have identified in SR-A+/−TLR4+/− and SR-A+/−TLR4−/− BMDC is greater than the defect seen in TLR4−/− BMDC, and single heterozygote cells for each gene display WT phagocytic activity. Together, these data demonstrate that loss of a single copy of TLR4 or complete loss of the TLR4 gene imparts sensitivity to SR-A heterozygosity with regard to phagocytic activity.
One simple explanation for the phagocytic defect observed in SR-A+/−TLR4+/− BMDC would be if heterozygosity of one of the genes down-regulated the expression of the other gene. However, SR-A+/−TLR4+/− cells exhibit the same TLR4 and SR-A expression levels as single heterozygote cells and thus we concluded that the observed, synthetic defect is not a result of differing protein expression on the double heterozygous genetic background. An alternative explanation for the observed phagocytic defect in SR-A+/−TLR4+/− BMDC is that reduction of the expression of two different PRR at the same time results in less-efficient cell-surface binding of the bacteria, thereby decreasing phagocytic uptake. However, SR-A+/−, TLR4+/−, and SR-A+/−TLR4+/− cells all bound bacteria with equal efficiency and were similar to WT BMDC. These observations led us to test if the defect in SR-A+/−TLR4+/− cells is a result of impairment of phagocytic trafficking as opposed to a defect in bacterial recognition. To test this we assayed for the rate of bacterial uptake with the gentamycin protection assay. Kinetic analysis of bacterial phagocytosis demonstrated that SR-A+/−TLR4+/− BMDC exhibit impaired rates of bacterial internalization compared with WT cells, and this deficit was apparent at early time-points, within 10 min, following induction of the phagocytic process. These data suggest that a rapid, TLR4-mediated process facilitates phagocytosis and that alteration of this signaling process (i.e., through TLR4 heterozygosity) makes phagocytes sensitive to SR-A heterozygosity. In support of this hypothesis, we observed that we were able to recapitulate the SR-A+/−TLR4+/− BMDC phagocytosis deficit in SR-A+/−MyD88+/− BMDC. This result strongly supports a model in which the phagocytic deficit observed in SR-A+/−TLR4+/− BMDC is functionally controlled by MyD88 signaling. Moreover, since MyD88 deficiency does not alter TLR4 expression at the cell surface, this eliminates the possibility that the SR-A+/−TLR4+/− defect is a function of reduced cell-surface binding of the bacteria.
Our identification of an analogous role for TLR2 in the phagocytosis of the Gram-positive bacteria S. aureus indicates that there is a global, but TLR-specific, regulation of SR-A-mediated phagocytic uptake by MyD88 signaling. SR-A functions in the recognition and uptake of Gram-positive and Gram-negative bacteria. To test the specificity of the SR-A+/−TLR4+/− phagocytic deficit, we tested the ability of SR-A+/−TLR4+/− cells to phagocytose S. aureus bacteria, which are Gram-positive and also phagocytosed via SR-A (ref. [10] and Fig. 2); however, their cell-wall products are predominantly ligands for TLR2 rather than TLR4 [54,55,56,57,58]. Although SR-A−/− BMDC exhibited an expected deficit for S. aureus uptake, SR-A+/−TLR4+/− cells were not defective for S. aureus uptake compared with WT cells. This specificity by the SR-A+/−TLR4+/− cells demonstrates that there is not a global phagocytic defect in these cells and that the deficit is restricted to TLR4 ligands. Importantly, we observed that TLR2 and SR-A exhibit a parallel relationship for the phagocytosis of Gram-positive bacteria, as SR-A+/−TLR2+/− BMDC were impaired significantly for the uptake of S. aureus. These data support a shared mechanism, whereby multiple TLRs signal through MyD88 to facilitate SR-A-mediated phagocytic trafficking in a TLR ligand-specific manner.
As an additional control for the specificity of the observed phenotypes, we used AcLDL, a well-established ligand for SR-A, which is internalized through an endocytic, rather than phagocytic, pathway [59]. SR-A+/−TLR4+/− BMDC exhibited WT levels of endocytic accumulation of AcLDL, which demonstrated that TLR4 modulation of SR-A-mediated uptake processes is restricted to phagocytic trafficking of bacteria and does not influence SR-A-dependent endocytosis.
With regard to the phagocytosis of bacteria, our in vivo data recapitulated the in vitro data, and SR-A+/−TLR4+/− mice are observed to be deficient in the clearance of live E. coli, despite having WT numbers of macrophages in the peritoneum within 1 h of bacterial challenge. These in vivo data provide some interesting implications about the role for TLR4 and SR-A synergy in host immunity to bacteria. In particular, we demonstrate that modest (twofold), but concomitant, alterations in TLR4 and SR-A expression levels can impact host bacterial clearance significantly. Two-fold changes in expression levels of these receptors are well within documented regulation of these genes. For example, IFN-γ, TNF-α, TGF-β, and IL-6 have all been shown to down-regulate SR-A expression at the mRNA and protein level [60,61,62,63,64,65], and TLR3 activation and cytokine stimuli have been demonstrated to down-regulate TLR4 expression [66,67,68]. Additionally, TLR4 and SR-A polymorphisms have been documented in humans, which could cause a predisposition to impaired bacterial clearance if the complementary receptor were down-regulated. Human TLR4 polymorphisms that lead to signaling defective receptors have been characterized and are associated with increased susceptibility to bacterial infection and LPS hyporesponsiveness [48,49,50,51, 53]. Although SR-A polymorphisms have been documented in humans [69, 70], no clear resultant phenotype has been identified yet; there are conflicting epidemiological reports about a connection between SR-A polymorphisms and increased risk for prostate cancer [69,70,71,72,73]. Our findings provide evidence that concomitant alterations in TLR4 and SR-A, through gene dosage or through gene regulation, may result in the impaired ability of innate immune cells to clear invading bacteria and subsequently would lead to increased susceptibility to pathogenic bacterial infection.
In summary, we report the novel finding that heterozygosity for two distinct PRR found on phagocytic innate immune cells, TLR4 and SR-A, results in impaired phagocytic function in vitro and in vivo for the clearance of the Gram-negative bacteria E. coli. We show in parallel that concomitant heterozygosity for TLR2 and SR-A results in a similar phagocytic defect for the clearance of the Gram-positive bacteria S. aureus. Finally, we demonstrate that heterozygosity for a common signaling adaptor molecule MyD88 recapitulates these phagocytic defects, identifying a novel, mechanistic role for MyD88 in the regulation of SR-A-dependent phagocytic trafficking. This is the first report of any phenotypic haplo-insufficiency for SR-A, and these findings have clear relevance to innate immune protection in humans where polymorphisms and regulation of expression in the TLR4 and SR-A genes have been well documented. These data provide new insight regarding functional synergy between TLRs and SR-A during bacterial phagocytosis and clearance.
Acknowledgments
This research was supported by Centers of Biomedical Research Excellence P20RR016437, National Institutes of Health (NIH) R01 AI067405, Dartmouth Clinical and Translational Science Award (B. B.), and NIH Training Grant T32 AI07363 (E. A.). We thank the NCCC Englert Cell Analysis Laboratory for help with FACS analysis and Rashalai Currington (University of Maryland-Eastern Shore); Drs. George O’Toole, Lloyd Kasper, and Deborah Hogan (Dartmouth College); Soman Abraham (Duke University, Durham, NC, USA); Kari Ann Shirey and Stefanie Vogel (University of Maryland, College Park, MD, USA); and Lester Kobzik (Harvard University, Cambridge, MA, USA) for help and advice.
References
- Janeway C A, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C A., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- Blander J M, Medzhitov R. Regulation of phagosome maturation by signals from Toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- Blander J M, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006;7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- Gough P J, Gordon S. The role of scavenger receptors in the innate immune system. Microbes Infect. 2000;2:305–311. doi: 10.1016/s1286-4579(00)00297-5. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Gordon S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology. 2004;209:39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14:123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- Gerold G, Zychlinsky A, de Diego J L. What is the role of Toll-like receptors in bacterial infections? Semin Immunol. 2007;19:41–47. doi: 10.1016/j.smim.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol. 2007;19:3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Dunne D W, Resnick D, Greenberg J, Krieger M, Joiner K A. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc Natl Acad Sci USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R Y, Golenbock D T, Penman M, Krieger M, Raetz C R. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature. 1991;352:342–344. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- Peiser L, Gough P J, Kodama T, Gordon S. Macrophage class A scavenger receptor-mediated phagocytosis of Escherichia coli: role of cell heterogeneity, microbial strain, and culture conditions in vitro. Infect Immun. 2000;68:1953–1963. doi: 10.1128/iai.68.4.1953-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K A, Rowe D C, Barnes B J, Caffrey D R, Visintin A, Latz E, Monks B, Pitha P M, Golenbock D T. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K A, Rowe D C, Golenbock D T. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. 2004;6:1361–1367. doi: 10.1016/j.micinf.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem Sci. 2002;27:474–482. doi: 10.1016/s0968-0004(02)02145-x. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Naito M, Yamamoto T, Hasegawa G, Gejyo F, Mitsuyama M, Suzuki H, Kodama T. Role of macrophage scavenger receptors in response to Listeria monocytogenes infection in mice. Am J Pathol. 2001;158:179–188. doi: 10.1016/S0002-9440(10)63956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C A, Li Y, Kodama T, Suzuki H, Silverstein S C, El Khoury J. Protection from lethal gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. J Exp Med. 2000;191:147–156. doi: 10.1084/jem.191.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth R, Platt N, Keshav S, Hughes D, Darley E, Suzuki H, Kurihara Y, Kodama T, Gordon S. The macrophage scavenger receptor type A is expressed by activated macrophages and protects the host against lethal endotoxic shock. J Exp Med. 1997;186:1431–1439. doi: 10.1084/jem.186.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Miyaji C, Watanabe H, Umezu H, Hasegawa G, Abo T, Arakawa M, Kamata N, Suzuki H, Kodama T, Naito M. Role of macrophage scavenger receptor in endotoxin shock. J Pathol. 2000;192:263–272. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH692>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Seimon T A, Obstfeld A, Moore K J, Golenbock D T, Tabas I. Combinatorial pattern recognition receptor signaling alters the balance of life and death in macrophages. Proc Natl Acad Sci USA. 2006;103:19794–19799. doi: 10.1073/pnas.0609671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Du W, McClellan S A, Barrett R P, Hazlett L D. TLR4 is required for host resistance in Pseudomonas aeruginosa keratitis. Invest Ophthalmol Vis Sci. 2006;47:4910–4916. doi: 10.1167/iovs.06-0537. [DOI] [PubMed] [Google Scholar]
- Malley R, Henneke P, Morse S C, Cieslewicz M J, Lipsitch M, Thompson C M, Kurt-Jones E, Paton J C, Wessels M R, Golenbock D T. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci USA. 2003;100:1966–1971. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjathoor V V, Febbraio M, Podrez E A, Moore K J, Andersson L, Koehn S, Rhee J S, Silverstein R, Hoff H F, Freeman M W. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277:49982–49988. doi: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Sakaguchi H, Kruijt J K, Higashi T, Suzuki T, van Berkel T J, Horiuchi S, Takahashi K, Yazaki Y, Kodama T. The multiple roles of macrophage scavenger receptors (MSR) in vivo: resistance to atherosclerosis and susceptibility to infection in MSR knockout mice. J Atheroscler Thromb. 1997;4:1–11. doi: 10.5551/jat1994.4.1. [DOI] [PubMed] [Google Scholar]
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Kim W S, Ordija C M, Freeman M W. Activation of signaling pathways by putative scavenger receptor class A (SR-A) ligands requires CD14 but not SR-A. Biochem Biophys Res Commun. 2003;310:542–549. doi: 10.1016/j.bbrc.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Cook D N, Whitehead G S, Burch L H, Berman K G, Kapadia Z, Wohlford-Lenane C, Schwartz D A. Spontaneous mutations in recombinant inbred mice: mutant Toll-like receptor 4 (Tlr4) in BXD29 mice. Genetics. 2006;172:1751–1755. doi: 10.1534/genetics.105.042820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A, Whitman S C, Block A E, Rateri D L. Polymorphism of class A scavenger receptors in C57BL/6 mice. J Lipid Res. 2000;41:1568–1577. [PubMed] [Google Scholar]
- Castellino F, Boucher P E, Eichelberg K, Mayhew M, Rothman J E, Houghton A N, Germain R N. Receptor-mediated uptake of antigen/heat shock protein complexes results in major histocompatibility complex class I antigen presentation via two distinct processing pathways. J Exp Med. 2000;191:1957–1964. doi: 10.1084/jem.191.11.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Deguchi M, Hagi K, Yasumizu R, Ikehara S, Muramatsu S, Steinman R M. Granulocytes, macrophages, and dendritic cells arise from a common major histocompatibility complex class II-negative progenitor in mouse bone marrow. Proc Natl Acad Sci USA. 1993;90:3038–3042. doi: 10.1073/pnas.90.7.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T, Volkmann A, Stockinger B. Mechanisms of tolerance induction in major histocompatibility complex class II-restricted T cells specific for a blood-borne self-antigen. J Exp Med. 1994;180:2089–2099. doi: 10.1084/jem.180.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwin B, Delneste Y, Lovingood R V, Post S R, Pizzo S V. SREC-I, a type F scavenger receptor, is an endocytic receptor for calreticulin. J Biol Chem. 2004;279:51250–51257. doi: 10.1074/jbc.M406202200. [DOI] [PubMed] [Google Scholar]
- Berwin B, Hart J P, Pizzo S V, Nicchitta C V. Cutting edge: CD91-independent cross-presentation of GRP94(gp96)-associated peptides. J Immunol. 2002;168:4282–4286. doi: 10.4049/jimmunol.168.9.4282. [DOI] [PubMed] [Google Scholar]
- Restrepo C I, Dong Q, Savov J, Mariencheck W I, Wright J R. Surfactant protein D stimulates phagocytosis of Pseudomonas aeruginosa by alveolar macrophages. Am J Respir Cell Mol Biol. 1999;21:576–585. doi: 10.1165/ajrcmb.21.5.3334. [DOI] [PubMed] [Google Scholar]
- Amiel E, Nicholson-Dykstra S, Walters J J, Higgs H, Berwin B. Scavenger receptor-A functions in phagocytosis of E. coli by bone marrow dendritic cells. Exp Cell Res. 2007;313:1438–1448. doi: 10.1016/j.yexcr.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M J, Li G, Shin J S, Carson J L, Abraham S N. Bacterial penetration of bladder epithelium through lipid rafts. J Biol Chem. 2004;279:18944–18951. doi: 10.1074/jbc.M400769200. [DOI] [PubMed] [Google Scholar]
- Becker M, Cotena A, Gordon S, Platt N. Expression of the class A macrophage scavenger receptor on specific subpopulations of murine dendritic cells limits their endotoxin response. Eur J Immunol. 2006;36:950–960. doi: 10.1002/eji.200535660. [DOI] [PubMed] [Google Scholar]
- Lee J S, Frevert C W, Matute-Bello G, Wurfel M M, Wong V A, Lin S M, Ruzinski J, Mongovin S, Goodman R B, Martin T R. TLR-4 pathway mediates the inflammatory response but not bacterial elimination in E. coli pneumonia. Am J Physiol Lung Cell Mol Physiol. 2005;289:L731–L738. doi: 10.1152/ajplung.00196.2005. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Rimoldi M, Valzasina B, Rotta G, Granucci F, Ricciardi-Castagnoli P. Toll-like receptor 4 is not required for the full maturation of dendritic cells or for the degradation of Gram-negative bacteria. Eur J Immunol. 2002;32:2800–2806. doi: 10.1002/1521-4141(2002010)32:10<2800::AID-IMMU2800>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Anand R J, Kohler J W, Cavallo J A, Li J, Dubowski T, Hackam D J. Toll-like receptor 4 plays a role in macrophage phagocytosis during peritoneal sepsis. J Pediatr Surg. 2007;42:927–932. doi: 10.1016/j.jpedsurg.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Shirai H, Murakami T, Yamada Y, Doi T, Hamakubo T, Kodama T. Structure and function of type I and II macrophage scavenger receptors. Mech Ageing Dev. 1999;111:107–121. doi: 10.1016/s0047-6374(99)00079-2. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Doi T, Hamakubo T, Kodama T. Scavenger receptor family proteins: roles for atherosclerosis, host defence and disorders of the central nervous system. Cell Mol Life Sci. 1998;54:628–640. doi: 10.1007/s000180050191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aderem A, Underhill D M. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Greaves D R, Gough P J, Gordon S. Recent progress in defining the role of scavenger receptors in lipid transport, atherosclerosis and host defence. Curr Opin Lipidol. 1998;9:425–432. doi: 10.1097/00041433-199810000-00006. [DOI] [PubMed] [Google Scholar]
- Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Steinman R M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Platt N, Suzuki H, Kurihara Y, Kodama T, Gordon S. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc Natl Acad Sci USA. 1996;93:12456–12460. doi: 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour N C, Lorenz E, Schutte B C, Zabner J, Kline J N, Jones M, Frees K, Watt J L, Schwartz D A. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- Kiechl S, Lorenz E, Reindl M, Wiedermann C J, Oberhollenzer F, Bonora E, Willeit J, Schwartz D A. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347:185–192. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- Rallabhandi P, Bell J, Boukhvalova M S, Medvedev A, Lorenz E, Arditi M, Hemming V G, Blanco J C, Segal D M, Vogel S N. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- Lorenz E, Mira J P, Frees K L, Schwartz D A. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- Rallabhandi P, Awomoyi A, Thomas K E, Phalipon A, Fujimoto Y, Fukase K, Kusumoto S, Qureshi N, Sztein M B, Vogel S N. Differential activation of human TLR4 by Escherichia coli and Shigella flexneri 2a lipopolysaccharide: combined effects of lipid A acylation state and TLR4 polymorphisms on signaling. J Immunol. 2008;180:1139–1147. doi: 10.4049/jimmunol.180.2.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F F, Marks K, Wong M, Watson S, de Leon E, McIntyre P B, Sullivan J S. Clinical relevance of TLR2, TLR4, CD14 and FcγRIIA gene polymorphisms in Streptococcus pneumoniae infection. Immunol Cell Biol. 2008;86:268–270. doi: 10.1038/sj.icb.7100155. [DOI] [PubMed] [Google Scholar]
- Lien E, Sellati T J, Yoshimura A, Flo T H, Rawadi G, Finberg R W, Carroll J D, Espevik T, Ingalls R R, Radolf J D, Golenbock D T. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- Schroder N W, Morath S, Alexander C, Hamann L, Hartung T, Zahringer U, Gobel U B, Weber J R, Schumann R R. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J Biol Chem. 2003;278:15587–15594. doi: 10.1074/jbc.M212829200. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP) Annu Rev Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- Bottalico L A, Wager R E, Agellon L B, Assoian R K, Tabas I. Transforming growth factor-β 1 inhibits scavenger receptor activity in THP-1 human macrophages. J Biol Chem. 1991;266:22866–22871. [PubMed] [Google Scholar]
- Draude G, Lorenz R L. TGF-β1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am J Physiol Heart Circ Physiol. 2000;278:H1042–H1048. doi: 10.1152/ajpheart.2000.278.4.H1042. [DOI] [PubMed] [Google Scholar]
- Geng Y J, Hansson G K. Interferon-γ inhibits scavenger receptor expression and foam cell formation in human monocyte-derived macrophages. J Clin Invest. 1992;89:1322–1330. doi: 10.1172/JCI115718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H Y, Nicholson A C, Hajjar D P. Inhibition of macrophage scavenger receptor activity by tumor necrosis factor-α is transcriptionally and post-transcriptionally regulated. J Biol Chem. 1996;271:7767–7773. doi: 10.1074/jbc.271.13.7767. [DOI] [PubMed] [Google Scholar]
- Liao H S, Matsumoto A, Itakura H, Doi T, Honda M, Kodama T, Geng Y J. Transcriptional inhibition by interleukin-6 of the class A macrophage scavenger receptor in macrophages derived from human peripheral monocytes and the THP-1 monocytic cell line. Arterioscler Thromb Vasc Biol. 1999;19:1872–1880. doi: 10.1161/01.atv.19.8.1872. [DOI] [PubMed] [Google Scholar]
- van Lenten B J, Fogelman A M. Lipopolysaccharide-induced inhibition of scavenger receptor expression in human monocyte-macrophages is mediated through tumor necrosis factor-α. J Immunol. 1992;148:112–116. [PubMed] [Google Scholar]
- Jiang W, Sun R, Wei H, Tian Z. Toll-like receptor 3 ligand attenuates LPS-induced liver injury by down-regulation of Toll-like receptor 4 expression on macrophages. Proc Natl Acad Sci USA. 2005;102:17077–17082. doi: 10.1073/pnas.0504570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller T, Terada T, Rosenberg I M, Shibolet O, Podolsky D K. Th2 cytokines down-regulate TLR expression and function in human intestinal epithelial cells. J Immunol. 2006;176:5805–5814. doi: 10.4049/jimmunol.176.10.5805. [DOI] [PubMed] [Google Scholar]
- Muzio M, Bosisio D, Polentarutti N, D'Amico G, Stoppacciaro A, Mancinelli R, van't Veer C, Penton-Rol G, Ruco L P, Allavena P, Mantovani A. Differential expression and regulation of Toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng S L, Komiya A, Mychaleckyj J C, Isaacs S D, Chang B, Turner A R, Ewing C M, Wiley K E, Hawkins G A, Bleecker E R, Walsh P C, Meyers D A, Isaacs W B. Common sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Am J Hum Genet. 2003;72:208–212. doi: 10.1086/345802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zheng S L, Komiya A, Mychaleckyj J C, Isaacs S D, Hu J J, Sterling D, Lange E M, Hawkins G A, Turner A, Ewing C M, Faith D A, Johnson J R, Suzuki H, Bujnovszky P, Wiley K E, DeMarzo A M, Bova G S, Chang B, Hall M C, McCullough D L, Partin A W, Kassabian V S, Carpten J D, Bailey-Wilson J E, Trent J M, Ohar J, Bleecker E R, Walsh P C, Isaacs W B, Meyers D A. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32:321–325. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- Chen Y C, Giovannucci E, Kraft P, Hunter D J. Association between genetic polymorphisms of macrophage scavenger receptor 1 gene and risk of prostate cancer in the health professionals follow-up study. Cancer Epidemiol Biomarkers Prev. 2008;17:1001–1003. doi: 10.1158/1055-9965.EPI-07-0744. [DOI] [PubMed] [Google Scholar]
- Hsing A W, Sakoda L C, Chen J, Chokkalingam A P, Sesterhenn I, Gao Y T, Xu J, Zheng S L. MSR1 variants and the risks of prostate cancer and benign prostatic hyperplasia: a population-based study in China. Carcinogenesis. 2007;28:2530–2536. doi: 10.1093/carcin/bgm196. [DOI] [PubMed] [Google Scholar]
- Seppala E H, Ikonen T, Autio V, Rokman A, Mononen N, Matikainen M P, Tammela T L, Schleutker J. Germ-line alterations in MSR1 gene and prostate cancer risk. Clin Cancer Res. 2003;9:5252–5256. [PubMed] [Google Scholar]