Summary
How size is controlled during animal development remains a fascinating problem despite decades of research. Here we review key concepts in size biology and develop our thesis that much can be learned by studying how different organ sizes are differentially scaled by homeotic selector genes. A common theme from initial studies using this approach is that morphogen pathways are modified in numerous ways by selector genes to effect size control. We integrate these results with other pathways known to regulate organ size in developing a comprehensive model for organ size control.
What can our toes teach us about size control?
The range of sizes found in the animal kingdom is extraordinary. Consider, for example, the mouse and elephant, two mammals with similar body plans but that differ nearly a million fold in size. Then compare the size of a mouse with that of a fruit fly. In considering how variations in size arise, we can expect that animals use roughly the same genes to regulate size and that their cells are about the same size. Recent reviews have summarized our current understanding of how environmental, nutritional and genetic factors can influence organism size and body part proportions.(1,2) In this review, we take a different perspective and argue that insight into these questions can also be gained by asking how the individual body parts of a single animal become different sizes. Our fingers, toes and ribs are sets of structures whose members are nearly identical in nature but differ in size. Uncovering the mechanisms by which these structures attain their different sizes will point to the genes and pathways used by nature to manipulate size and may shed light on other prominent questions in size biology such as how entire animals, either within or between species, attain their various sizes.
Selector gene control of tissue identity
What are the genes that control differences in body parts? The clearest answers come from studying mutations that cause one structure to develop in place of another — homeotic mutations. Homeotic genes are a class of selector genes, transcription factors that sit atop regulatory hierarchies to determine the gene activity status of the tissue in which they are expressed.(3) By manipulating selector genes, it is possible to completely transform one tissue into another, a transformation that includes differences in size. These transformations therefore allow us to ask questions about the molecular basis of differential tissue sizes.
Many selector genes belong to the Hox family of homeodomain transcription factors. Famous examples of Hox mutations, isolated in the fruit fly Drosophila melanogaster, include the four winged Ultrabithorax (Ubx) mutant (Fig. 1C,D), the antenna-to-leg Antennapedia (Antp) transformation and the mouthparts-to-leg proboscipedia (pb) transformation. In flies, as in vertebrates, the Hox genes are differentially expressed along the segments that comprise the anterior–posterior (AP) body axis. Hox genes (or combinations thereof) instruct the identities of the segments in which they are expressed. In the developing vertebrate limb and torso, Hox genes are expressed in such a way that each digit or rib expresses a unique combination of Hox genes — a Hox code that is thought to specify individual identity of serially iterated structures.(4,5)
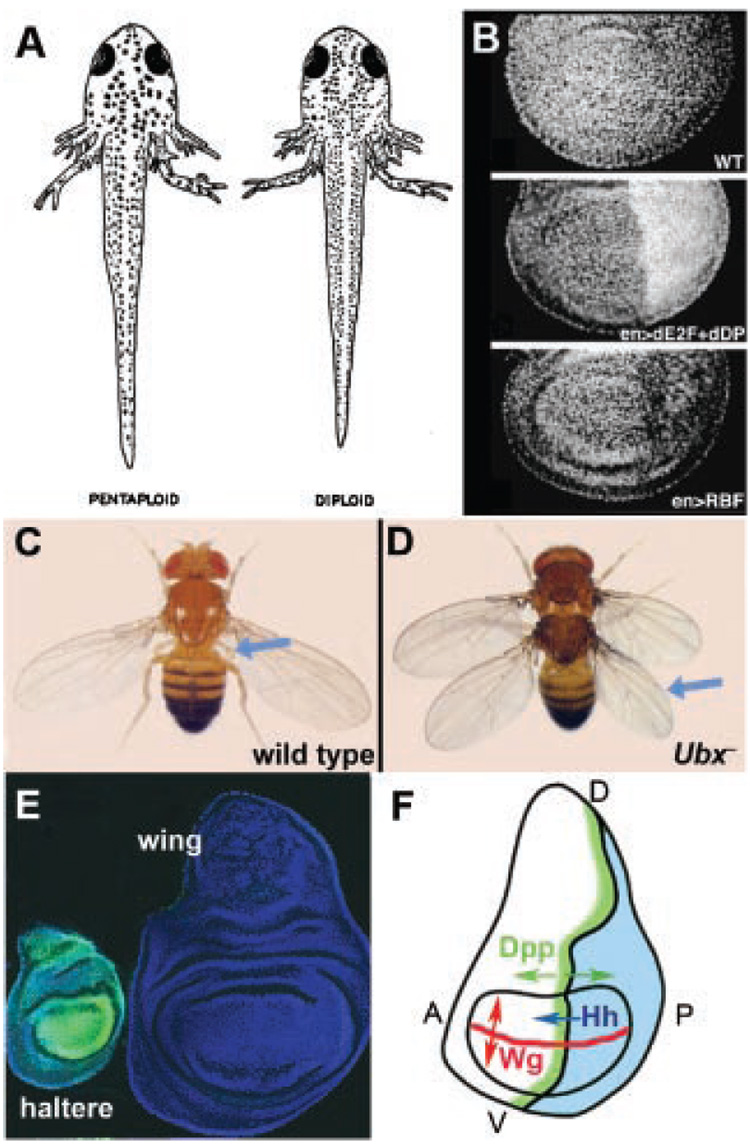
Figure 1. Tissue growth is independent of cell size and cell number but depends on selector genes.
A: Salamanders grow to the same size whether they have large, polyploid cells (left) or smaller, diploid cells (right). Reprinted from Fankhauser G 1940 Proc Natl Acad Sci USA 26:526–532. with permission.(77) B: Wing imaginal discs from Drosophila grow to the correct size even when ~half of the tissue is composed of an abnormally large number of smaller cells (middle panel) or an abnormally small number of larger cells (lower panel; wild type is shown in the top panel). Reprinted from Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA 1998 Cell 93:1183–1193 with permission. C,D:. Wild-type flies (C) have wings on T2 and halteres (blue arrow) on T3. Ubx mutant flies (D) exhibit a complete transformation of T3 to T2, including a transformation of haltere to wing (blue arrow), resulting in a four-winged fly.(6) E: Haltere imaginal discs (left) express the Ubx gene (visualized by green immunofluorescence) while wing imaginal discs (right) do not express Ubx. All cells are stained with a ubiquitous nuclear protein (blue). F: Wing and haltere discs have a very similar arrangement of morphogens. Hh is expressed by all cells in the P compartment (blue) and activates the expression of dpp in adjacent A compartment cells. Dpp (green) is secreted by these cells and diffuses in both directions to activate downstream genes in both the anterior (A; left) and posterior (P; right) directions. Wg (red) is expressed along the dorsal-ventral compartment boundary and is secreted in both the dorsal (D; up) and ventral (V; down) directions. In the haltere, Wg is not expressed in the P compartment due to repression by Ubx (not shown).
The clearest instances of Hox selector gene activity come from flies. Ubx, for example, is expressed in the third thoracic segment (T3) and mutations affecting the Ubx locus cause T3 to develop as T2(6) (Fig. 1C,D). This transformation is most obvious when examining the flight appendages of these segments. The fly’s wings are located on T2. At an equivalent position on T3, tiny balloon-shaped balancing organs called halteres are formed. When Ubx function is lost from T3, wings form on both T2 and T3 and halteres are lost (Fig. 1). These and other observations show that Ubx is responsible for modifying the wing-determining program of T2 into a haltere-determining program on T3. One of the key differences between the haltere and wing is size, which includes a fivefold difference in cell number. Thus, the control of haltere size by Ubx provides a simple and genetically dissectible system for understanding how selector genes modify organ size.
The phenomenology of size control
An important concept in size biology is the idea that size itself is regulated. A classic demonstration of this principle comes from polyploid salamanders (Fig. 1A).(7) The cells of polyploid salamanders can be twice as large as the cells of diploid salamanders, but the animals are the same size and proportion.(7) To remain normal size, a polyploid salamander compensates for its ~2-fold larger cells by halving its cell number. A modern equivalent of this experiment was performed by genetically manipulating the cell division rate of the cells in one half of the Drosophila wing imaginal disc, the developmental precursor to the adult wing and thorax(8) (Fig. 1B). When the cell cycle is sped up, cells achieve a smaller overall size before division, whereas when the cell cycle is slowed, cells become bigger. In both cases, the genetically manipulated half of the disc compensated for the differences in cell size by changing cell number, allowing final organ size to remain the same. Analogous results have also been obtained in vertebrates (see for example see Ref. 9). Observations such as these argue that animals and tissues “know” what their final size should be and grow to attain that size irrespective of the number or size of their constituent cells.
Final size has also been shown to be independent of cellular growth rate. When some cells of a tissue express higher levels of cell growth promoting genes (e.g. myc) than do other cells in the same tissue, a phenomenon called “cell competition” arises.(10–12) The faster growing cells “out compete” the slower growing cells in the sense that they come to occupy a larger fraction of the tissue than would be expected. In extreme cases, the final structures derived from tissues developing as mosaics of fast and slow growing cells will be completely composed of the fast-growing population. Though the mechanisms of cell competition are only beginning to be understood,(13) the phenomenon itself underscores the regulation of size at the level of the whole organ: tissue size is maintained whether it is composed of slow-growing cells, fast-growing cells, or some warring combination of the two. As such, genes that control the rates of biomass accumulation, by themselves, are unlikely candidates for size control targeting by selector genes (Fig. 2).
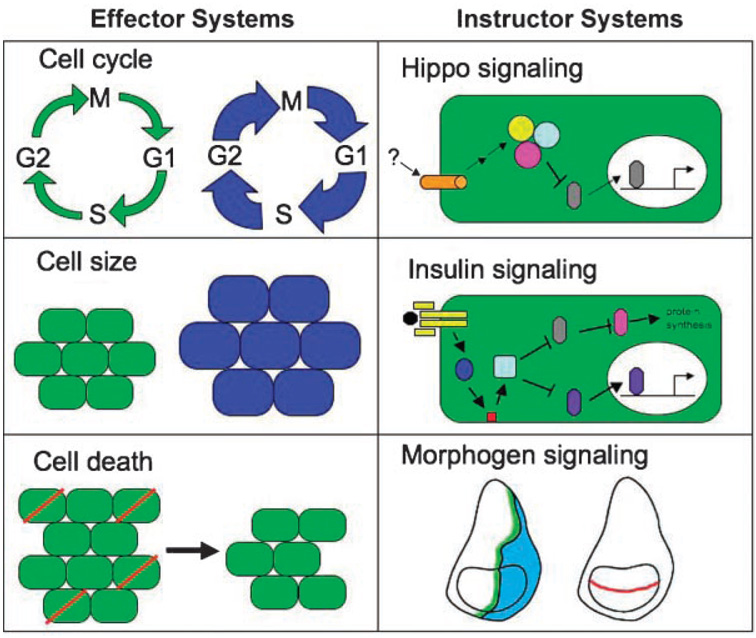
Figure 2. Effector versus instructor size regulating pathways.
We distinguish between ‘effector’ (left column; cell cycle, cell growth, and cell death) and ‘instructor’ (right column; nutritional/systemic inputs, Hippo signaling, morphogen signaling) size regulating systems. Manipulating individual effect or systems is compensated for by alterations in the other two, resulting in no net change in overall size. On the other hand, when Instructor systems are modified, changes in final size result. See text for details on the interactions between these systems.
In many cases, tissue size has also been shown to be independent of the environment in which the tissue is grown, that is, size information is intrinsic to the tissue itself. Infant rat hearts transplanted into mature rats attain the proper size, as do fetal mouse thymuses transplanted into mature mice.(14) These organs possess information about how big they should become, even in a different developmental context. This seems to be the rule in Drosophila as well. Wing discs dissected from developing larvae and placed into adult abdomens grow until their proper size is attained.(15) The regenerating liver, on the other hand, seems to get growth cues from circulating hormones and therefore depends on its environment for size information.(16) Still, the fact that the liver can regenerate to its proper size, even after surgical removal of two-thirds of its mass, demonstrates that it also contains a robust size-determination program, but one that also gets input from external cues. Although the liver is in some ways a special case because of its regenerative potential throughout the life of the animal, many tissues demonstrate a great deal of plasticity during developmental stages. For example, fruit flies can fully recover from severe irradiation damage during larval stages that causes the loss of up to 60% of their imaginal disc cells.(17) The massive cell death experienced by these tissues does not destroy the “knowledge” possessed by the fly’s tissues about what their proper size should be. Similar compensatory growth is seen when large portions of developing chicken limbs are removed.(18) Aside from demonstrating the remarkable plasticity of final organ size to developmental perturbations, these observations argue against simple “cell counting” or “amount of time spent growing” models of size regulation and instead argue for regulation at the level of final organ size.
Recently, a possible exception to size-based growth regulation described above has emerged. The size of the mouse pancreas was shown to be unable to compensate for experimentally induced decreases in progenitor cell number early in development.(19) These results led the authors to argue that the pancreas achieves its final size by counting cell divisions from a given start point, a mode of regulation that has not been previously observed and that is distinct from the observations discussed above. In light of these results, it will be interesting to see if the pancreas is able to compensate for cell size changes or if pancreas size depends on the size of its constituent cells. It will also be of interest to see if increasing the size of the pancreas progenitor population would cause an increase in final pancreas size, as would be predicted by a precursor-size-dependent mode of size regulation.
Size-determining pathways
Selector genes are transcription factors and are therefore likely to effect size control by regulating the activity of specific target genes. Although cell growth, proliferation and death are the ultimate determinants of tissue size, as described above, individually altering these processes does not lead to changes in size. In contrast, as discussed in more detail below and shown schematically in Figure 2, several pathways have been shown to be bona fide regulators of organ size.
The hippo pathway
While it is unlikely that selector genes work primarily at the level of cell growth, proliferation or apoptosis to control organ size, the fact remains that more cells accumulate in larger tissues. It may be that the coordinated regulation of all three of these factors together is necessary to alter final size. It is therefore of particular interest that, in recent years, the Hippo pathway has been shown to be a master regulator of cell growth, proliferation and death.(20) The cell cycle progression gene cyclinE, the anti-apoptotic protein dIAP and the growth-promoting microRNA bantam have all been shown to be regulated by the Hippo pathway.(21–23) The Hippo pathway has been delineated from a large number of genes that either positively or negatively regulate growth in a cell-autonomous manner (Fig. 2). Though this pathway has largely been worked out in flies, its function is conserved in vertebrates.(24) The upstream components of the Hippo pathway, the atypical cadherins Fat and Dachsous, are membrane proteins that engage in heterophilic interactions between cells.(25,26) Both fat and dachsous cause dramatic tissue overgrowth when mutated.(27,28) The Fat/Dachsous growth regulation signal is transduced through the kinase Warts,(25) itself a tumor suppressor.(29) Warts negatively regulates the activity of the transcription factor Yorkie, which acts in the nucleus to promote tissue growth.(21) The Hippo pathway therefore seems to convert information received from the cell membrane into a cell-autonomous growth profile. Thus, the Hippo pathway is clearly part of the mechanism by which organ sizes are determined and may, directly or indirectly, be targeted by selector genes.
Systemic and nutritional signals
The fact that individuals within a species can grow to different overall sizes, yet retain similar proportions implies systemic control of size. In humans, overall size is controlled by the levels of Human Growth Hormone (HGH) secreted from the pituitary gland. Too much HGH leads to gigantism while too little leads to dwarfism.(14) The levels of systemic size-determining factors like Insulin and HGH are influenced by genetics, nutrition and environment.
Tissue growth requires energy. It follows that the amount of energy available to an organism will influence its size. In many species, an animal that is malnourished during development will grow to a smaller overall size than a well-fed counterpart. Alterations in Insulin Receptor (InR) signaling or TOR signaling pathways phenocopy this starvation induced size decrease.(1,30–32) These pathways have therefore been implicated in the sensing and responding of tissues to the global energy state of the animal. Systemic size-determining pathways can also be altered at the genetic level. For example, small breeds of dogs have lower levels of circulating Insulin-like Growth Factor-1 (IGF-1) than do larger breeds, and this difference is associated with a polymorphism in the IGF-1 gene.(33) A similar molecular pathway underlies size differences mediated by HGH, which boosts the production of IGF-1 for secretion from the liver and other organs.(34) Interestingly, altering the activity of Inr or TOR pathways within a tissue can autonomously increase or decrease it size.(31,35) Although recent results in Drosophila argue against the idea that selector genes modulate the cell autonomous interpretation of systemic nutrient-sensing pathways (see below), their ability to alter organ sizes means that this information must somehow be integrated with selector gene information.
Morphogen signaling
A morphogen is a molecule that is produced in and secreted from a set of organizing cells to control growth and patterning of entire fields of cells.(36) Cells perceive extracellular morphogen levels through cell surface receptors. Morphogens instruct the patterning of tissues by activating different target genes at different thresholds of perceived morphogen concentration, that is, at different positions within the morphogen gradient. This results in multiple domains of gene expression that are translated into the morphological characteristics of the tissue.
The ability of organisms to scale up or down in size while retaining their patterning has led to the idea that pattern and growth are linked. Molecular support for this hypothesis comes from the fact that morphogens, in addition to controlling pattern, also have a strong influence on organ size. For example, three morphogens are active in the Drosophila wing disc: Hedgehog (Hh), Wingless (Wg) and Decapentaplegic (Dpp) (Fig. 1). When any of these molecules (or downstream signaling components) are impaired, wing growth is strongly decreased, while increased morphogen production in the wing leads to tissue overgrowth. However, the mitogenic potential of morphogens is context-dependent. There are several instances, even in wing development, where activation of morphogen signaling pathways causes decreased growth.(37–40) Our overall picture of how exactly morphogens regulate growth is far from complete, and several recent papers and reviews address this issue.(39,41–44) However, as described below, it has recently been shown that morphogen signaling is altered between differently sized, but otherwise similar tissues.(45–48) Thus, it seems that selector genes indeed manipulate morphogen pathways to alter tissue sizes.
Lessons on size control from the fly wing and haltere
We and others have studied the haltere-to-wing transformation seen in Drosophila Ubx mutants as a model for understanding how size is encoded in tissues and how selector genes control size differences between tissues.(45–48) Progenitor cells (called imaginal discs) of both the wing and haltere are specified about mid-way through embryogenesis. At the embryo/larval transition, the wing imaginal disc consists of 30–50 cells and the haltere disc has 15–25 cells.(49) After 4 days of larval growth, the mature wing disc contains ~50,000 cells while the haltere consists of ~10,000 cells(50) (Fig. 1). At this point in development, both discs have the same shape, and there are few obvious differences between them other than size. During the metamorphosis that follows larval development, the cells of the haltere and wing take on their adult characteristics. During this time, wing cells flatten and become ~eight times larger in surface area than haltere cells.(51) As cell sizes are generally similar throughout the animal kingdom,(52) the main interest for size biology lies in the mechanisms by which Ubx limits the number of cells in the haltere relative to the wing, a process that is completed at the end of larval development.
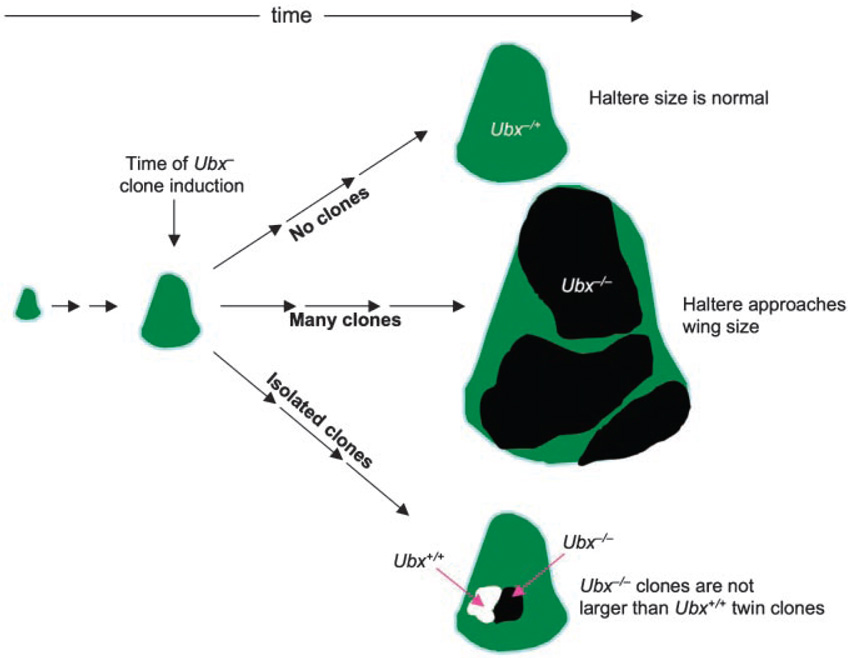
When Ubx function is removed from large groups of cells in the developing haltere, even mid-way through development, transformations toward a wing identity are observed.(6,53) As expected, part of this transformation includes an increase in cell number. However, a crucial and surprising finding is that when isolated patches of Ubx− tissue are generated at the same time, the mutant cells do not overgrow relative to their neighboring wild-type (Ubx+) cells(45,47) (Fig. 3). Thus, Ubx is not dictating the growth or proliferation rate of each individual haltere cell. Instead, this observation argues that Ubx works through cell non-autonomous mechanisms to control the size of the haltere.
Figure 3. Ubx does not control haltere size cell-autonomously.
The loss of Ubx function from large patches (clones, black regions) of cells in the developing haltere imaginal disc (middle scenario) causes the disc to overgrow. The overgrowth seen in these discs is not restricted to the Ubx mutant tissue. In contrast, when isolated Ubx− clones are generated at the same time, they grow to a similar size as simultaneously generated Ubx+ twin spots (white patch; bottom scenario). Therefore Ubx must work through cell-nonautonomous mechanisms to reduce haltere size relative to the wing. See Ref. 45 for details.
The lack of an autonomous control of size seen in the Ubx− clone experiment argues against this selector gene controlling proliferation by modifying the intracellular components of most of the pathways discussed above, including the apoptotic, cell cycle, cell growth, Hippo and insulin/nutrition pathways. If Ubx were altering the way haltere cells execute any of these pathways, their growth and proliferation should strictly depend on whether they expressed Ubx or not. Instead, that Ubx controls growth and proliferation non-cell autonomously points to a potential role for morphogen signaling in mediating the size differences between the haltere and wing. Indeed, several groups have identified multiple alterations in morphogen signaling between the wing and haltere(45–48) and we have found these alterations to underlie differences in size and proportioning of the two tissues.(45,46)
Selector gene regulation of morphogen production and mobility
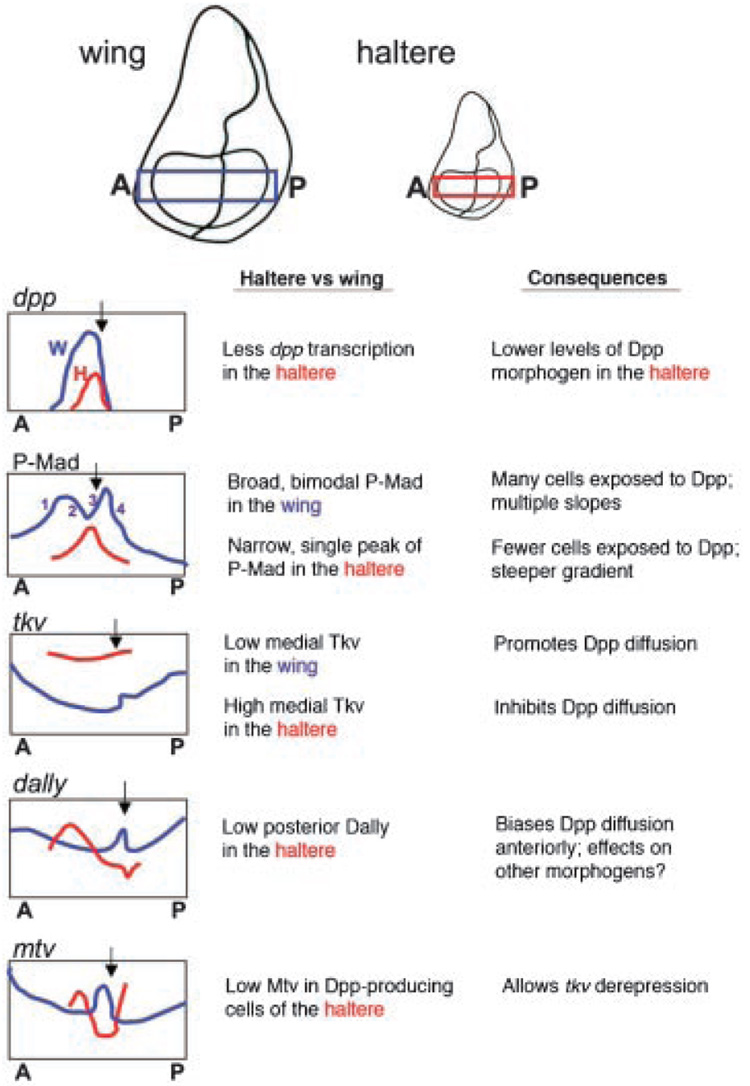
The way in which morphogens are produced in the haltere and wing are qualitatively similar but quantitatively different(45,47,48,54) (Fig. 4). In both tissues, the posteriorly produced morphogen Hedgehog (Hh) travels into the A compartment to activate Dpp production in adjacent A cells (the AP organizer), from which Dpp is secreted. In the haltere, less Dpp is produced in the AP organizer relative to that seen in the wing (Fig. 4). Dpp production in the haltere is decreased in both the number of cells that express dpp, and the levels of dpp transcribed per cell. This finding is very exciting because we have long known that mutations that decrease Dpp production in many Drosophila tissues, including the wing, result in reduced tissue growth.(55,56) Thus the reduction in Dpp production in the haltere strongly implicates regulation of Dpp signaling in the control of haltere size by Ubx. Ubx also changes the amount of Hh and Wg produced in the haltere.(46,54) The effects of decreased Wg and Hh production in the haltere have never been explicitly tested but, based on growth phenotypes seen in wg and hh mutants, are also expected to also contribute to its small size relative to the wing.
Figure 4. Alterations in Dpp signaling between the wing and haltere.
Some of the many differences in Dpp signaling between the wing and haltere are described here as plots of gene expression patterns (dpp, tkv, dally and mtv) or Dpp signaling activity (P-Mad). Plots are representative of intensity traces taken from the boxed region of wing (blue) and haltere (red) discs stained for the indicated markers. For each readout, the haltere and wing traces are positioned such that their AP boundaries (black arrows) are in the same position.
Because morphogens are typically produced by a small number of cells but act broadly within tissues, their distributions and consequent activity profiles within tissues are critical to how they instruct morphological outputs. Morphogen activity profiles are determined by the parameters of morphogen mobility through tissues. Two classes of extracellular proteins, receptors and heparin sulphate proteoglycans (HSPGs), interact with morphogens and control their movement. Both receptors and HSPGs are differentially regulated between the haltere and wing and are relevant to size control (Fig. 4). Upon morphogen binding to its receptor, in addition to activating the signal transduction cascade, it ceases moving through the tissue. Conversely, HSPGs generally promote the movement of morphogens. Morphogens interact with the heparin side chains of HSPGs and this binding facilitates morphogen mobility. Dpp, for example, cannot traverse wing cells from which HSPGs (or their heparin side chains) are missing.(57) Interestingly, both receptors and HSPGs control morphogen mobility in a concentration dependent manner. The amount of morphogen trapped by a cell correlates with the level of receptor expression(45,58,59) and morphogens have a higher tendency to associate with cells that have higher levels of HSPGs.(60–62) Therefore, the levels and expression patterns of receptors and HSPGs are critical in shaping morphogen activity profiles in developing tissues.
Work on the Dpp pathway has produced reagents that enable monitoring of Dpp diffusion through tissues (Dpp-GFP(63,64)), as well as intracellular Dpp pathway activation (anti-P-Mad(65)). Using these reagents, Dpp signaling in the the haltere was found to differ from the wing in several respects,(45,47,48,54) and these differences have been shown to contribute significantly to their different sizes.(45,46) Most strikingly, the Dpp pathway activity profile (measured by anti-P-Mad staining) is very broad along the AP axis in the wing disc, and much sharper and narrower along the AP axis of the haltere disc (Fig. 4). These observations are explained as follows: when Dpp is secreted from the AP organizer of the haltere, it encounters different levels of receptor and HSPG than it does in the wing. In the wing, the cells in and around the AP organizer express low levels of the Dpp receptor, thickveins (tkv)(65,66) (Fig. 4). The relatively few receptor molecules in wing AP organizer cells are quickly saturated, allowing the remaining Dpp to travel many cell diameters. The result accounts for the broad Dpp activity profile that is observed in the wild-type wing imaginal disc (Fig. 4). This broad profile gradually decays on either side of the AP organizer. In the haltere, tkv expression is high in all cells (Fig. 4). High Tkv levels create a restrictive environment for Dpp diffusion in the haltere as these cells are capable of binding and internalizing high amounts of Dpp. Therefore, Dpp molecules have an increased tendency to bind receptor at or near the site of production in the haltere, resulting in a highly compact activity gradient compared to the wing (Fig. 4). Importantly, compacting the Dpp activity gradient is sufficient to cause reduced wing size, suggesting that it is a critical determinate of the small haltere size.(45)
In addition to a general mobility restriction, Dpp mobility is directionally biased in the haltere. In the wing, expression levels of dally, which encodes a glypican that is modified to become an HSPG, are roughly symmetrical with respect to the AP organizer(67) (Fig. 4). In the haltere, dally is repressed in the P compartment, creating an AP asymmetry in dally expression (Fig. 4). As a consequence, Dpp mobility and pathway activation is biased away from the P compartment expressing low dally levels and into the A compartment of the haltere, where higher dally levels are present.(46) This restriction of Dpp mobility causes a reduction in size of the haltere in general and in the P compartment specifically.(46)
Receptors and HSPGs are known to influence the mobility of secreted proteins other than Dpp,(68) so their levels and expression patterns are likely to play prominent roles in the trafficking of morphogens in addition to Dpp. A second glypican, dally-like (dlp), is encoded in the Drosophila genome and also controls morphogen mobility. In the wing, dlp levels seem to be most relevant for Wg signaling. It has been shown that dlp repression from the DV boundary of the wing is important for the distribution of extracellular Wg and the regulation of wg target genes.(61,62,69) It is of interest that, in addition to a generally lower level of Dlp, the DV domain of dlp repression is absent from the haltere.(46) Finally, the enzyme Notum has been proposed to modify the morphogen binding capabilities of both Dally and Dally-like.(61,62,69) notum expression is also altered between the wing and haltere,(46) but the consequences of altered notum regulation have not been investigated. Thus HSPGs and their modifiers seem to be commonly targeted by selector genes as a means of sculpting morphogen activity profiles, and thus organ shapes and sizes.
It is important to include in this discussion the targets of the Dpp pathway that also play a critical role in tissue growth. In the Drosophila wing, for example, two Dpp targets, vestigial (vg) and spalt (sal), are both required for the correct patterning and growth of this appendage.(70,71) Importantly, both of these targets are repressed by Ubx in the haltere.(54) vg, like the morphogens Dpp and Wg, is essential for the growth of the wing disc. However, vg activity is not sufficient to drive cell proliferation in the absence of morphogen signaling, and the mechanism by which vg drives growth is not known.(71,72) Regardless, these observations underscore the idea that selector genes like Ubx regulate many steps in the Dpp pathway to modify organ size—from expression of the ligand itself, to molecules that shape its extracellular gradient (tkv, dally), to its downstream targets (vg, sal). It is likely that the sum of all of these Ubx-mediated regulatory events is necessary to fully explain the size differences between these two flight appendages.
How selector genes modulate complex patterns of gene expression
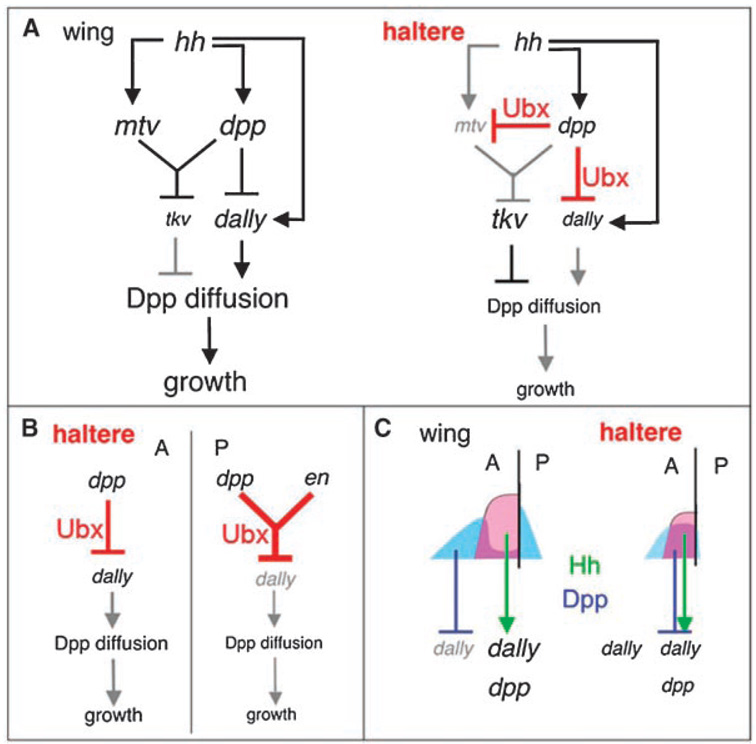
In this review, we have discussed how selector genes alter gene expression patterns to sculpt morphogen activity profiles. But how do selector genes, which are typically expressed uniformly within a field of cells, create the various patterns of gene expression that we see, for example, in the Dpp pathway components in the haltere? Studies of Ubx control of Dpp pathway components in the fly have provided insight into the mechanisms by which a uniformly expressed selector gene can give rise to diverse gene expression patterns. One mechanism is collaborative control of gene expression by selector genes and the other patterning genes in the tissue. A simple example involves the transcription factor engrailed (en), which is expressed in all posterior cells of the fly. While neither Ubx nor En are individually sufficient to repress dally (one of the genes that differ in expression between the wing and haltere), the combination of these two factors results in the repression of dally in the posterior compartment of the haltere(46) (Fig. 5).
Figure 5. Gene regulatory networks in the wing and haltere controlling size.
A: The wing and haltere have different versions of a core regulatory network designed to produce different amounts of Dpp production, Dpp diffusion and growth. Gene regulatory steps in the haltere that use Ubx are shown in red. See text for further details. B: In Ubx-expressing cells of the haltere, dpp is a stronger repressor of dally than it is in the wing. In the P compartment of the haltere, repression is augmented further by the posterior selector gene, engrailed (en), thus biasing the diffusion of Dpp in the anterior direction. C: In both the wing and haltere, peak Hh signaling (pink shading) occurs in cells immediately anterior to the AP boundary and activates dpp and dally transcription (green arrows). In contrast, due to differences in Tkv levels, peak Dpp signaling (blue shading, an indication of P-Mad levels), which represses dpp and dally transcription, occurs at different positions in the wing and haltere (blue bars).
A more-complicated example of combinatorial control by Ubx comes from the uniform expression of the Dpp receptor, tkv, found in the haltere. Since Ubx is expressed throughout the haltere, a naïve prediction would be that Ubx simply activates transcription at the tkv locus. However, reality is more complicated (Fig. 5). Repression of tkv in the wing by Hh and Dpp requires master of thickveins (mtv), a zinc-finger-containing nuclear protein.(73) In the haltere, Ubx represses mtv, but only in the presence of Dpp signaling(45) (Fig. 5). Therefore, in the presence of Ubx (i.e. in the haltere), one tkv repressor (Dpp signaling) represses mtv, which is also required for tkv repression. This relationship between Dpp signaling and mtv in the haltere ensures that no haltere cell contains high levels of both of these repressors, resulting in tkv derepression throughout the haltere. Ubx has been shown to work closely with the Dpp pathway transcription factor, Mad, to alter the expression of the spalt gene in the haltere,(74) arguing that Hox intervention into the readouts of Dpp signaling is likely a commonly used mechanism of pattern generation.
Another mechanism used by Ubx to modulate gene expression patterns is by changing the spatial domains of morphogen signaling. For example, through tkv derepression, Ubx changes the relative positions of peak Dpp and Hh signaling in the haltere(45) (Fig. 5). In both the wing and haltere, peak Hh signaling occurs in the AP organizer—those cells that lie immediately anterior to the AP compartment boundary. In contrast, Dpp signaling is different in these two appendages, peaking on either side of the AP organizer in the wing, but within the AP organizer in the haltere. This alteration in relative position for these two signaling pathways has consequences for the expression of any target gene that is regulated by both pathways (Fig. 5). In the wing, these two conflicting inputs are non-overlapping and contribute to the dpp and dally expression patterns. In the haltere, these two inputs coincide, buffering their opposing effects. The overlap in Dpp and Hh signaling inputs in the AP organizer of the haltere contributes to lower dpp expression levels in the haltere, which in turn contributes to its smaller size.(45)
A unified model for organ size regulation
As described above, several non-morphogen signaling pathways can also influence tissue growth and overall organ sizes. For example, starvation or the disruption of Insulin signaling results in the development of smaller animals, with smaller, though still properly proportioned, organs. How can these observations be meshed with a primary role for morphogen signaling in organ size determination? Interestingly, when Insulin signaling is decreased in the posterior (P) half of the wing, the corresponding portion of the Dpp gradient compacts, and that portion of the organ undergrows.(64) Conversely, when Insulin signaling is hyperactivated in P cells, the Dpp gradient expands in these cells and overgrowth is seen. Thus, Insulin signaling appears to have command over morphogen signaling in a manner reminiscent of that which we have described for selector genes. The difference is that, as a systemic signal, Insulin signaling provides the entire tissue with information about the overall size and nutritional status of the animal, while selector genes modify morphogen signaling in a tissue- and organ-specific manner. It is not known how Insulin signaling modifies the shape of the Dpp gradient, but based on how selector genes modify the gradient, the regulation of extracellular surface molecules such as tkv and dally is an attractive hypothesis.
The Hippo pathway controls growth but is not known to involve secreted molecules and so does not constitute a morphogen pathway. How can we reconcile the dramatic effects of Hippo pathway components with a morphogen-centric model for size control? Interestingly, a recent study by Rogulja et al.44 has shown that morphogens control growth through modulation of Hippo pathway activity. This paper shows that the levels and cellular localizations of upstream Hippo pathway components are influenced by morphogen signaling. Differences in morphogen signaling between cells are proposed to be perceived via intercellular interactions between Hippo pathway cadherins and transduced through the Hippo pathway into a cellular growth profile. These data provide a first look at the mechanisms used by morphogens in the control of tissue growth and fit well with the long-held but contentious idea that the slope of the morphogen gradient dictates the growth of the tissue.(75) Though there is strong evidence that morphogens do not always work through their gradient slopes to control size,(39,40,76) the potent growth-regulating capabilities of the Hippo pathway cast it as a likely means through which morphogens generally dictate organ size.
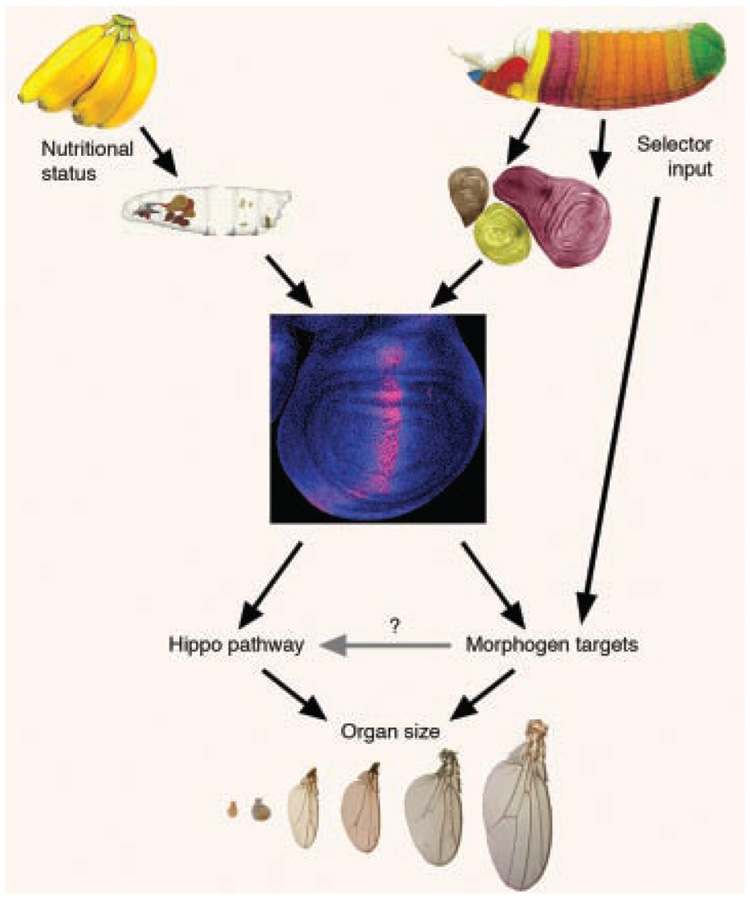
Putting these ideas together, we suggest that nutrient signals provide information to tissues about the overall size of the animal in which it is developing, whereas selector genes provide organ-specific information (Fig. 6). The cells of the tissue respond to both of these inputs by altering the expression of morphogen pathway components and target genes. Morphogen activity gradients and target genes, in turn, alter the activation status of the Hippo pathway. The result is a dynamic cellular growth profile that integrates information obtained from genetic background, nutritional status, general location within the animal, and the status of surrounding cells. In this model, this integration of constantly streaming, multi-source information, translated into patterns of morphogen signaling within a tissue and read out by individual cells through the Hippo pathway, endows animals with the remarkable plasticity and reproducibility of size control observed in nature and in the lab.
Figure 6. Model for control of organ size.
Nutritional and hormonal inputs control the overall size of the organism and selector genes control the relative sizes of the organs within an animal. Both inputs work, at least in part, by altering morphogen signaling that has been proposed to control size through the Hippo pathway. See text for details.
Open questions and future outlook
In this review, we have argued that the tissue transformations orchestrated by selector genes can be used to probe the mechanisms that nature uses to determine organ size. Initial experiments using the ability of the Hox gene Ubx to transform the large wing into the smaller haltere have shown that regulation of morphogen production and mobility are central components of organ-size-determining mechanisms. But exactly how these changes in morphogen gradients are transduced into different sizes is just beginning to be understood. The identification of two differently shaped morphogen gradients in these two differently sized tissues provides an excellent opportunity for addressing the long-standing question of how morphogens control size.
What about size differences between animals? For example, how is the 25% difference in size between male and female fruit flies specified by the selector gene-like sex-determination pathway? Does this size difference between animals also involve morphogen signaling? What about instances of size differences as vast as those found between mice and elephants? Can differences in morphogen production and mobility possibly explain million fold differences in size? It seems likely that size-controlling outputs downstream of morphogen signaling must also be altered in such enormous size discrepancies, but at what level? Is the Hippo pathway altered in the elephant so that a given morphogen stimulus causes much more growth it does in the mouse? We envision that, now that we have an initial grasp on the genes used to control animal sizes, studies comparing sizes of whole animals and the structures structures within them will provide the clearest path for understanding the mechanisms that control the amazing array of animal sizes on display in nature.
Acknowledgments
We thank Barry Honig and Dragana Rogulja for comments on the manuscript, and members of the Mann and Johnston labs for discussions.
Funding agencies: this work was supported by an NIH grant to R.S.M. M.A.C. was supported by NIH training grants DK07328 and GM008798.
References
- 1.Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays. 2007;29:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- 2.Shingleton AW, Frankino WA, Flatt T, Nijhout HF, Emlen DJ. Size and shape: the developmental regulation of static allometry in insects. Bioessays. 2007;29:536–548. doi: 10.1002/bies.20584. [DOI] [PubMed] [Google Scholar]
- 3.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 4.Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- 5.Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 6.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 7.Fankhauser G. The effect of changes in chromosome numbers on amphibian development. Q Rev Biol. 1945;20:20–78. [Google Scholar]
- 8.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 9.Henery CC, Bard JB, Kaufman MH. Tetraploidy in mice, embryonic cell number, and the grain of the developmental map. Dev Biol. 1992;152:233–241. doi: 10.1016/0012-1606(92)90131-y. [DOI] [PubMed] [Google Scholar]
- 10.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 11.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 12.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129:1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 14.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 15.Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- 16.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 17.Haynie JL, Bryant PJ. The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc of Drosophila melanogaster. Develop Genes Evol. 1977;183:85–100. doi: 10.1007/BF00848779. [DOI] [PubMed] [Google Scholar]
- 18.Kalthoff K. Analysis of Biological Development. New York: McGraw-Hill; 1996. [Google Scholar]
- 19.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 20.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 23.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Dong J, Feldmann G, Huang J, Wu S, Zhang N, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 26.Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- 27.Clark HF, Brentrup D, Schneitz K, Bieber A, Goodman C, Noll M. Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila. Genes Dev. 1995;9:1530–1542. doi: 10.1101/gad.9.12.1530. [DOI] [PubMed] [Google Scholar]
- 28.Bryant PJ, Huettner B, Held LI, Jr, Ryerse J, Szidonya J. Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev Biol. 1988;129:541–554. doi: 10.1016/0012-1606(88)90399-5. [DOI] [PubMed] [Google Scholar]
- 29.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 30.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 31.Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158–166. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Rother KI, Accili D. Role of insulin receptors and IGF receptors in growth and development. Pediatr Nephrol. 2000;14:558–561. doi: 10.1007/s004670000351. [DOI] [PubMed] [Google Scholar]
- 33.Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heyner S, Garside WT. Biological actions of IGFs in mammalian development. Bioessays. 1994;16:55–57. doi: 10.1002/bies.950160109. [DOI] [PubMed] [Google Scholar]
- 35.Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- 36.Tabata T, Takei Y. Morphogens, their identification and regulation. Development. 2004;131:703–712. doi: 10.1242/dev.01043. [DOI] [PubMed] [Google Scholar]
- 37.Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- 38.Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development. 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- 39.Rogulja D, Irvine KD. Regulation of cell proliferation by a morphogen gradient. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- 41.Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124:318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 43.Hufnagel L, Teleman AA, Rouault H, Cohen SM, Shraiman BI. On the mechanism of wing size determination in fly development. Proc Natl Acad Sci USA. 2007;104:3835–3840. doi: 10.1073/pnas.0607134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008 doi: 10.1016/j.devcel.2008.06.003. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crickmore MA, Mann RS. Hox Control of Organ Size by Regulation of Morphogen Production and Mobility. Science. 2006;313:63–68. doi: 10.1126/science.1128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crickmore MA, Mann RS. Hox control of morphogen mobility and organ development through regulation of glypican expression. Development. 2007;134:327–334. doi: 10.1242/dev.02737. [DOI] [PubMed] [Google Scholar]
- 47.de Navas LF, Garaulet DL, Sanchez-Herrero E. The Ultrabithorax Hox gene of Drosophila controls haltere size by regulating the Dpp pathway. Development. 2006;133:4495–4506. doi: 10.1242/dev.02609. [DOI] [PubMed] [Google Scholar]
- 48.Makhijani K, Kalyani C, Srividya T, Shashidhara LS. Modulation of Decapentaplegic gradient during haltere specification in Drosophila. Dev Biol. 2006;302:243–255. doi: 10.1016/j.ydbio.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 49.Cohen SM. Imaginal Disc Development. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Volume II. Cold Spring Harbor, NY: CSH Lab Press; 1993. pp. 747–842. [Google Scholar]
- 50.Martin P. Direct determination of the growth rate of Drosophila imaginal discs. J Experi Zool. 1982;222:97–102. [Google Scholar]
- 51.Roch F, Akam M. Ultrabithorax and the control of cell morphology in Drosophila halteres. Development. 2000;127:97–107. doi: 10.1242/dev.127.1.97. [DOI] [PubMed] [Google Scholar]
- 52.Raff MC. Size control: the regulation of cell numbers in animal development. Cell. 1996;86:173–175. doi: 10.1016/s0092-8674(00)80087-2. [DOI] [PubMed] [Google Scholar]
- 53.Morata G, Garcia-Bellido A. Developmental Analysis of Some Mutants of the Bithorax System of Drosophila. Roux’s Arch Develop Biol. 1976;179:125–143. doi: 10.1007/BF00848298. [DOI] [PubMed] [Google Scholar]
- 54.Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spencer FA, Hoffmann FM, Gelbart WM. Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell. 1982;28:451–461. doi: 10.1016/0092-8674(82)90199-4. [DOI] [PubMed] [Google Scholar]
- 56.Zecca M, Basler K, Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- 57.Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, et al. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 59.Lecuit T, Cohen SM. Dpp receptor levels contribute to shaping the Dpp morphogen gradient in the Drosophila wing imaginal disc. Development. 1998;125:4901–4907. doi: 10.1242/dev.125.24.4901. [DOI] [PubMed] [Google Scholar]
- 60.Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, et al. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- 61.Kirkpatrick CA, Dimitroff BD, Rawson JM, Selleck SB. Spatial Regulation of Wingless morphogen distribution and signaling by Dally-like protein. Developmental Cell. 2004;7:513–523. doi: 10.1016/j.devcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Developmental Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Entchev EV, Schwabedissen A, Gonzalez-Gaitan M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 64.Teleman AA, Cohen SM. Dpp gradient formation in the Drosophila wing imaginal disc. Cell. 2000;103:971–980. doi: 10.1016/s0092-8674(00)00199-9. [DOI] [PubMed] [Google Scholar]
- 65.Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- 66.Brummel TJ, Twombly V, Marques G, Wrana JL, Newfeld SJ, et al. Characterization and relationship of Dpp receptors encoded by the saxophone and thick veins genes in Drosophila. Cell. 1994;78:251–261. doi: 10.1016/0092-8674(94)90295-x. [DOI] [PubMed] [Google Scholar]
- 67.Fujise M, Izumi S, Selleck SB, Nakato H. Regulation of dally, an integral membrane proteoglycan, and its function during adult sensory organ formation of Drosophila. Dev Biol. 2001;235:433–448. doi: 10.1006/dbio.2001.0290. [DOI] [PubMed] [Google Scholar]
- 68.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 69.Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc 10.1242/dev.01636. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- 70.de Celis JF, Barrio R, Kafatos FC. A gene complex acting down-stream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- 71.Zecca M, Struhl G. Control of Drosophila wing growth by the vestigial quadrant enhancer. Development. 2007;134:3011–3020. doi: 10.1242/dev.006445. [DOI] [PubMed] [Google Scholar]
- 72.Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development. 2007;134:3001–3010. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- 73.Funakoshi Y, Minami M, Tabata T. mtv shapes the activity gradient of the Dpp morphogen through regulation of thick veins. Development. 2001;128:67–74. doi: 10.1242/dev.128.1.67. [DOI] [PubMed] [Google Scholar]
- 74.Walsh CM, Carroll SB. Collaboration between Smads and a Hox protein in target gene repression. Development. 2007;134:3585–3592. doi: 10.1242/dev.009522. [DOI] [PubMed] [Google Scholar]
- 75.Day SJ, Lawrence PA. Measuring dimensions: the regulation of size and shape. Development. 2000;127:2977–2987. doi: 10.1242/dev.127.14.2977. [DOI] [PubMed] [Google Scholar]
- 76.Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 77.Fankhauser G. A pentaploid larva of the newt, Triturus viridescens. Proc Natl Acad Sci USA. 1940;26:526–532. doi: 10.1073/pnas.26.8.526. [DOI] [PMC free article] [PubMed] [Google Scholar]