Abstract
We describe a detailed study of the RhoA-binding epitope of the GAP domain of Graf, including the determination of the thermodynamic and kinetic parameters of the interaction of wild-type domain, and of its fifteen single-site mutants, with cognate GTPases. We show that residues important for the structural integrity of the Arg-finger loop are critical for binding Rho and for the catalytic activity of GAP, but GTPase selectivity appears to be modulated by a much more subtle interplay of electrostatic and hydrophobic interactions involving residues on the periphery of the main interface. The eight residues targeted in this study are involved in three distinct patches on the surface, two of which appear to interact with highly conserved regions of the GTPase, while the third plays a role in GTPase selectivity.
Keywords: GNBP, GTPase activating protein, BH domain, GAP, titration calorimetry
Small GTPases of the Rho (Ras-homology) family control a multitude of biological functions, ranging from cytoskeletal rearrangements, cell cycle regulation and membrane trafficking, to transcription regulation and apoptosis (Etienne-Manneville and Hall, 2002; Hall, 2005; Jaffe and Hall, 2005; Raftopoulou and Hall, 2004). There are currently twenty two known members of the Rho family in the human genome, and all exert their biological function as binary switches by binding to and activating downstream targets when bound to the GTP (guanosine triphosphate) nucleotide. To ensure its biological potency, this complex must have a long half-life, and so Rho GTPases are poor enzymes, with kcat values for the hydrolysis of GTP to GDP as low as ∼0.01min-1, or, in exceptional cases (Rnd proteins and RhoH) no detectable GTPase activity is observed. To initiate and to terminate the biological signal, i.e. to load GTP for GDP, and then to hydrolyze GTP to GDP, Rho GTPases utilize two distinct families of accessory proteins: GEFs (guanine nucleotide exchange factors) (Rossman et al., 2005) and GAPs (GTPase activating proteins) (Moon and Zheng, 2003). Both GAPs and GEFs are typically large, multidomain proteins, which are themselves regulated in a spatial and temporal manner by a broad variety of mechanisms, in response to extra- and intracellular stimuli. The actual catalytic activities responsible for nucleotide exchange (GEFs) and hydrolysis (GAPs) reside in specific domains within these large proteins, which are responsible for the selectivity and specificity towards the Rho family targets.
Current human genome analyses indicate that there are ∼80 proteins containing GAP domains for Rho GTPases (Moon and Zheng, 2003). A number of them show selectivity for a specific GTPase, but many others act on two or more different targets. The molecular basis for GTPase selectivity of GAP domains is not well understood. One of the complicating factors is the discrepancy between the observed specificity of isolated GAP domains in vitro and their apparent in vivo selectivity as judged by corresponding phenotypes. For example, some GAP domains stimulate in vitro GTP hydrolysis of two or even three of the major Rho GTPases, i.e. Cdc42, RhoA and Rac1, while the in vivo experiments suggest activation of only one specific GTPase (Tcherkezian and Lamarche-Vane, 2007).
A typical RhoGAP domain encompasses ∼150 amino acids, and on average there is 20% sequence conservation within the family. Crystal structure of the GAP domain from the p50RhoGAP protein revealed an all α-helical architecture, similar to that observed for the Ras-specific GAP (Barrett et al., 1997; Rittinger et al., 1998). Subsequently reported complexes of this domain with GDP-bound and transition-state mimic complexes of RhoA and Cdc42 GTPases, (Nassar et al., 1998; Rittinger et al., 1997a; Rittinger et al., 1997b) showed how this domain assists in GTP hydrolysis by inserting a critical Arg side chain into the active site on the GTPase, resulting in stabilization of the transition-state. The so-called ‘arginine-finger’ mechanism was originally proposed for the Ras GTPase activating proteins, and clearly shares common evolutionary ancestry (Ahmadian et al., 1997b; Scheffzek et al., 1997; Scheffzek et al., 1998).
Graf is a GTPase activating protein associated with focal adhesion kinase (Hildebrand et al., 1996). Its GAP domain is one of only three with known crystal structure (Longenecker et al., 2000). Unlike p50RhoGAP, which acts on all principal RhoGTPases, GAPGraf acts on RhoA and Cdc42, but not Rac1 (Hildebrand et al., 1996). In this paper, we describe a detailed study of the RhoA-binding epitope of the GAP domain of GAPGraf, including the determination of the thermodynamic and kinetic parameters of the interaction of wild-type domain, and of its fifteen single-site mutants, with cognate GTPases. We specifically investigate the role of select surface GAP residues involved in the interaction with Rho GTPases, as well as the role of the catalytic Arg220 in the stabilization of the transition state complex. We show that residues important for the structural integrity of the Arg finger loop are critical for binding Rho and for the catalytic activity of GAP, but GTPase selectivity appears to be defined by complex interplay of diverse elements, with both electrostatic and hydrophobic interactions at play.
Materials and Methods
Modeling of complexes of the GAP domain of GAPGraf with GTPases and MD simulations
Starting models of the complexes of the GAP domain of Graf (residues 156-391) with RhoA and Cdc42 were generated by superposition of the crystallographic coordinates (PDB code 1F7C) onto the p50RhoGAP molecule in the crystal structures of the complexes with RhoA (PDB code 1TX4) or Cdc42 (PDB code 1GRN), respectively. All hydrogen atoms were added to the models and the structures were solvated in a cubic box. Additionally, AlF4- present in the nucleotide binding site of the RhoA and AlF3 in the Cdc42 were replaced with PO4-. A 1.0 ns molecular dynamics simulation was carried out for both ternary models (GAPGraf-RhoA–GTP and GAPGraf-Cdc42–GTP) using NWCHEM (Group, 2000) and AMBER99 force field (Cornell et al., 1995). For the Mg2+ ion in the active site we used a non-bonded model with a charge +2.0 and Lenard-Jones parameters as defined elsewhere (Åqvist, 1990).
Production and Purification of Recombinant Proteins
The GAP domain (residues 156-391) from avian GTPase Regulator Associated with Focal Adhesion Kinase (Graf), human Cdc42 (residues 1-191), human RhoA (residues 1-193) and human Rac1 (residues 1-192), were cloned in pET Uni1 vectors containing an N-terminal polyHis tag followed by rTEV cleavage site (Sheffield et al., 1999). QuikChange™ kit (Stratagene) was used for all mutagenesis. All recombinant proteins were produced in E.coli BL21(DE3)-RIL cells. Bacterial pellets were sonicated in an ice-cold 50 mM Tris/HCl, 1 mM PMSF, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 8.5 buffer. GTPases were purified using Ni-NTA agarose chromatography in 50 mM Tris/HCl, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 8.5 followed by gel filtration on Superdex-75 in 25 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 7.8. The GAP domain and its mutants were purified with an additional step of salting out with 55% ammonium sulfate followed by gel filtration on Superdex-75 in 25 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 7.8. At different stages, proteins were concentrated using CentriPrep (Millipore) and Vivaspin (Sartorious) filtration systems. All of the proteins were shock frozen and stored at -80°C. The protein purity was > 95% as judged by SDS-PAGE and Coomassie Blue staining. Final protein products were confirmed by ESI-MS analysis. The homogeneous, monomeric state of protein samples at high concentration required by the isothermal titration calorimetry (ITC) experiments (see below) was confirmed by gel filtration using analytical Superdex-75 column. Additionally, in order to confirm that the introduced mutations had no impact on the structure of the GAP domain, we recorded their circular dichroism (CD) spectra in the 200 - 250 nm range. We also determined thermal denaturation curves of all the proteins used in the study. All of the GAP domain mutants were properly folded and exhibited virtually the same CD spectra as the wild-type protein and denatured cooperatively but irreversibly.
Nucleotide Exchange
The nucleotide (GDP or GMPPNP) exchange reaction was carried out in 50 mM Tris/HCl or 25 mM HEPES, 50 mM NaCl, 1 mM DTT, pH 7.5. To start the exchange reaction, appropriate GTPase solution at 2 mg/ml was incubated on ice with 20 molar excess of the nucleotide and 15 mM EDTA (Hoffman et al., 1998). After 2 h the reaction was stopped by addition of MgCl2 to 30 mM concentration and the buffer was changed to 25 mM HEPES, 5 mM MgCl2, 1 mM DTT, pH 7.8 by gel filtration on HiPrep desalting column (Amersham Biosciences).
Calorimetric (ITC) and Spectroscopic Measurements
Isothermal titration calorimetry (ITC) experiments were performed using a CSC 4200 and 5300 calorimeters. The concentration of the GTPase (loaded with GDP or GMPPNP) in the sample cell (1.25 or 1.05 mL) was in the range of 0.05 to 0.15 mM. The GAPGraf proteins concentration was in the range of 0.6 to 1.7 mM. Except for the ΔCp determinations, which were carried out at 10, 15 and 20°C, all other experiments were carried out at 20 °C. Prior to experiment, the protein solution was extensively dialyzed at 4 °C against 25 mM HEPES, 5 mM MgCl2, 1 mM DTT, pH 7.8. If not stated otherwise, the buffer was supplemented with 25 mM NaF and 1 mM AlCl3. The titration thermograms were analyzed with BindWorks®Applied Thermodynamics and Origin software to obtain n, Ka and ΔHa values. The free energy of binding (ΔGa) and the entropy of association (ΔSa) were calculated from the Gibbs-Helmholtz equation. The experimental error in ΔHa arises mainly from the concentration inaccuracies of the active protein and is estimated to be 5-10%, whereas the experimental error on Ka of 20% is estimated from the iterative ITC runs.
Concentration of RhoA, Rac1, Cdc42, GAPGraf and its mutants was estimated using the ε280 molar absorbance coefficient based on the predicted number of Trp and Tyr residues (Pace et al., 1995). Additionally, for GTPases loaded with GDP or GMPPNP nucleotides, a molar absorbance coefficient ε280= 8000 M1-cm-1 for the nucleotide moiety was determined and taken into account.
GTPase Activity Assay
For kinetic measurements of GAP-stimulated GTP hydrolysis, the MESG/PNP spectroscopic assay protocol (Merck) was used, as described elsewhere (Zhang and Zheng, 1998). The initial rates of GTP-hydrolysis monitored by γPi release were determined in the presence of a constant amount of GAP (treated as an enzyme) and increasing concentrations of GTP-loaded GTPases (treated as substrate). The GAP-catalyzed GTP hydrolysis rates were fitted using GraFit 3, Erithacus Software Ltd. to the modified Michaelis-Menten equation to yield the KM and Vmax values of the reactions (Zhang and Zheng, 1998). The kcat values were calculated as Vmax/[GAPGraf], allowing determination of overall catalytic efficiencies kcat/KM. All experiments were carried out in 50 mM Tris/HCl, 5 mM MgCl2, pH 7.8 at 20 °C using Cary 300 Varian spectrophotometer with Peltier accessory.
Results and Discussion
Modeling of complexes of GAPGraf with cognate GTPases
Efforts to crystallize the GAP domain of Graf, i.e. GAPGraf, in complex with either RhoA or Cdc42 were unsuccessful. In order to gain insights into the structural features of the GAPGraf-GTPase interactions, we used molecular modeling. As a starting point, we used the p50RhoGAP-RhoA and p50RhoGAP-Cdc42 crystal structures (1TX4 and 1GRN), in which we replaced p50RhoGAP with the crystallographic model of GAPGraf (1F7C). We then applied molecular dynamics to optimize the structure of the complexes, as described in the Methods section.
As expected, the simulated complexes retain most of the key stereochemical features of the p50RhoGAP complexes, especially within the switch regions of the GTPases. GAPGraf does not undergo any significant conformational changes during the simulation on going from the isolated structure to one bound to either of the two GTPases. This was also expected based on the previous studies of p50RhoGAP in isolation and in complexes with cognate GTPases (Nassar et al., 1998; Rittinger et al., 1997b).
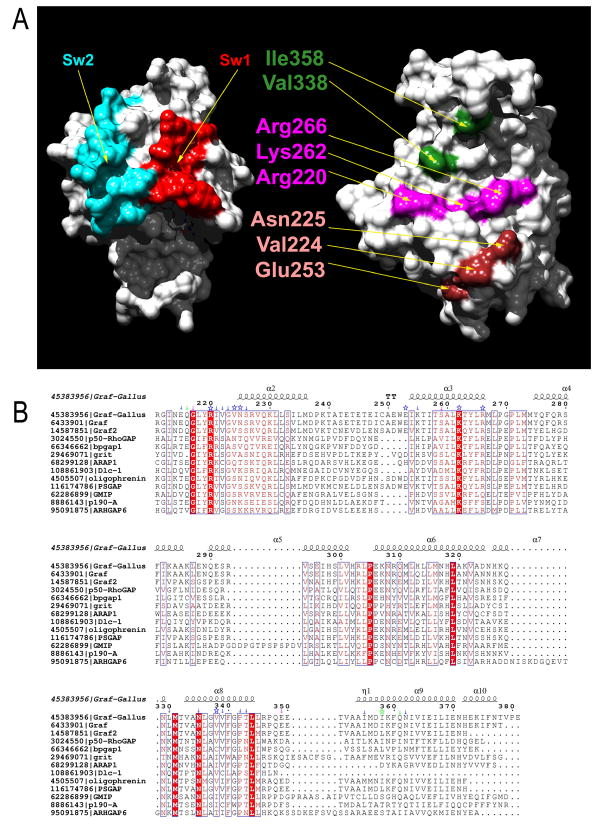
Figure 1 illustrates the nature of the contacts between the GAPGraf domain and the two GTPases in the respective models. In general terms, GAPGraf interacts with the GTPase via a concave large surface patch (997 Å2 and 1019 Å2 for RhoA and Cdc42 complexes, respectively) generated by the solvent exposed faces of helices α3, α7 and α8. The arginine finger, which includes a loop connecting α1 and α2, is located at the edge of this patch which binds an extensive surface on the nucleotide-binding face of the GTPase. The arginines-finger loop of GAPGraf, including the catalytic Arg220, is inserted in a canonical fashion allowing for a direct interaction of the latter with the catalytic Gln63 of RhoA (or Gln61 in Cdc42).
Figure 1.
(A) The binding interface of RhoA- GAPGraf complex. The switch regions I (residues 34-42) and II (residues 62-79) are colored in red and blue, respectively. Mutation sites in the GAPGraf are marked in magenta for patch I, brown for patch II and dark green for patch III. (B) Alignment of amino acid sequence of GAP domains from several RhoGAPs. P50-RhoGAP, bpgap1 and grit show in-vitro specificity towards Cdc42 over RhoA, whereas ARAP1, Dlc-1, PSGAP, GMIP, p190-A and ARHGAP6 prefer RhoA over Cdc42. Oligophrenin shows no preference towards neither Cdc42 nor RhoA. ARHGAP15 and chimaerin1 are GAPs specific for Rac1 both in vitro and in vivo (Tcherkezian and Lamarche-Vane, 2007). Fully conserved residues are red highlighted. Mutated GAPGraf amino acids are marked with asterisks and other residues in contact with GTPases are marked with arrows and colored as follows: green indicates contact with RhoA, pink – Cdc42 and blue – both RhoA and Cdc42. Residues in contact were defined based on modeled complex structures. Residue numbering as in the full-length Graf protein. Alignment was visualized using ESPript (Gouet et al., 1999).
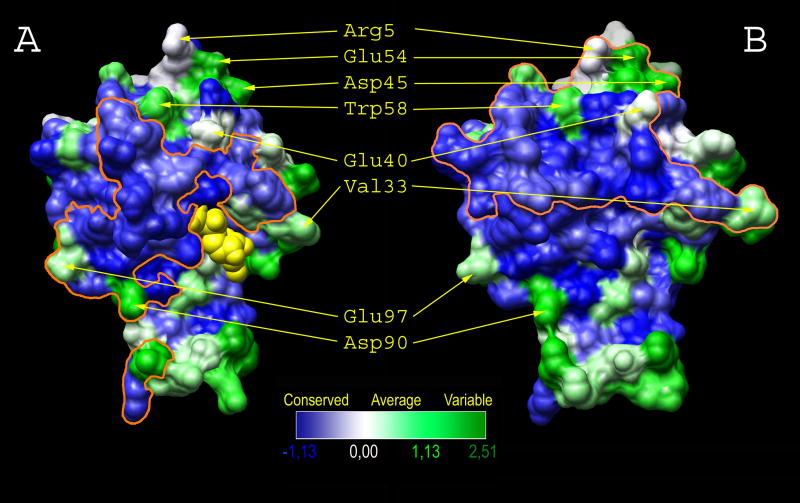
It is instructive to compare the surface that is engaged by GAPGraf on RhoA, to that involved in the interaction with the DH (Dbl-homology) domain of a RhoA-specific nucleotide exchange factor (GEF), as seen, for example, in the complex with PDZRhoGEF (Derewenda et al., 2004) (Figure 2). The interface buried between RhoA and the DH domain is significantly larger (1514 Å2) that the one engaged by GAPGraf (1165 Å2). Moreover, the DH domain, which is representative of the way that DH domains bind GTPases in other complexes (Kristelly et al., 2004; Rossman et al., 2005; Snyder et al., 2002) binds to a surface which is mostly (∼70%) conserved between RhoA, Cdc42 and Rac1, but which also contains variable and highly variable residues accounting for selectivity. In contrast, ∼90% of the surface engaged by the GAP domain is made up of highly conserved residues, with only ∼10% moderately variable amino acids. This is consistent with, and to some degree rationalizes the observed tendency of GAP domains to exhibit a higher level of promiscuity in their in vitro interactions with GTPases. Thus, in vivo selectivity is probably primarily controlled by spatial targeting through other domains.
Figure 2.
Conservation of residues within Rho family GTPases and involvement in the interaction with GAP and GEF regulators based on RhoA (GI:20379114), Rac1 (GI:8574038) and Cdc42 (GI:20379098). Binding interface of (A) RhoA with p50-RhoGAP (pdb:1TX4) (Rittinger et al., 1997b) and (B) RhoA with RhoGEF11 (pdb:1XCG) (Derewenda et al., 2004) is depicted by orange line. Conservation range is indicated by color intensity from deep blue through white and light green to dark green showing residues from strictly conserved to most variable, respectively. The color bar shows normalized conservation scores as obtained from ConSurf server (Landau et al., 2005). Several RhoA residues distinguishing GAP and GEF interacting epitopes are shown to help in navigation. Nucleotide is presented as yellow spheres. Figure was prepared using LIGPLOT (Wallace et al., 1995), ConSurf (http://consurf.tau.ac.il) and Chimera (Pettersen et al., 2004).
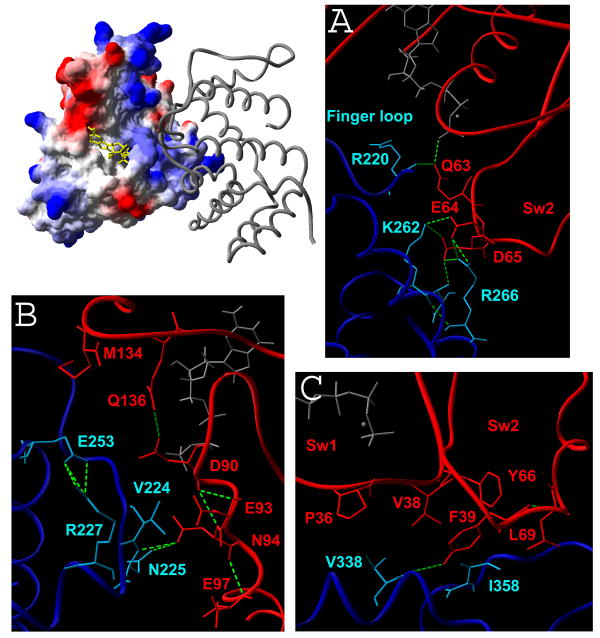
Within the complex-forming surface on GAPGraf, we identified three specific patches which are important for binding and/or catalysis, as judged by the models of the complexes with RhoA and Cdc42. The first such epitope, i.e. patch I, includes the catalytic Arg220 located within the arginines-finger and two residues invariant among RhoGAPs which appear to be essential for the structural integrity of the arginines-finger, i.e. Lys262 and Arg266. The catalytic Arg220 is suitably positioned to interact with Gln63 (RhoA numbering), while the positively charged Lys262 and Arg266, located on the solvent exposed face of helix α3, interact with a negatively charged fragment of Switch II containing Glu64 and Asp65, which is highly conserved among RhoA, Rac1 and Cdc42.
Patch II includes several residues including Val224, Asn225 and the adjacent Glu253. These amino acids interact with a mostly negatively charged region of RhoA including Asp90, Glu93 and Glu97. Of these, only Glu93 is conserved in Rac1, while Glu93 and Glu97 (but not Asp90) are conserved in Cdc42. We previously showed that the cohesive interaction between Asp225 of GAPGraf and Glu97 of Cdc42 provides a significant contribution to the observed RhoA/Cdc42 selectivity of GAPGraf (Longenecker et al., 2000), and so the entire patch II may play a role in selectivity in contrast to patch I.
Finally, a hydrophobic cradle (Patch III) formed by several solvent-exposed residues from helices 7 and 8, and in particular Val338 and Ile358, interacts with several conserved residues within the switch I region of RhoA/Cdc42, and specifically with the conserved Pro34 and Val36. Like patch I, this contact involves highly conserved amino acids and is less likely to be involved in specificity control.
Binding and enhancement of GTP hydrolysis on cognate GTPases by wild-type GAPGraf
It is well established that GAP domains bind target RhoGTPases only weakly when the latter are in the biologically inactive, GDP-bound state, and that the affinity increases dramatically for the transition state in which GTP transiently contains pentavalent phosphorus (Graham et al., 1999). This transition state can be effectively mimicked by adding fluoride ions in the presence of Al3+ or Mg2+ to a GDP-bound GTPase (Ahmadian et al., 1997a; Hoffman et al., 1998). In order to evaluate how wild type GAPGraf binds to RhoA and Cdc42 in both the GTP-bound state of the GTPase and to the transition complex, we used GTPases loaded with a slowly hydrolysable GTP analogue – GMP-PNP, and GDP with AlFx, respectively. The thermodynamics of binding were monitored using isothermal titration calorimetry (ITC) (Jelesarov and Bosshard, 1999). The results of ITC experiments are shown in Table 1. The association constants (Ka) for both GMP-PNP-Mg2+-bound RhoA and Cdc42 GTPases were 33.8 × 103 M and 26.1 × 103 M respectively. For the transition-state mimic these values increased 90 and 16 fold, respectively, suggesting preference for RhoA. This is in contrast to p50RhoGAP, where there is no preference in binding to two states of GTPase (Graham et al., 1999). These results reaffirm that many GAP-GTPase interactions responsible for the specificity and strength of the functional complex are formed in the transition state (Fersht, 1990).
Table 1.
The thermodynamic and kinetic parameters of GAPGraf WT interaction with RhoA and Cdc42. The ITC measurements were performed using GMP-PNP-Mg2+ and GDP·Mg2+-loaded Cdc42 and RhoA in the presence of AlFx. Kinetic parameters of GAPGraf stimulated GTP hydrolysis were obtained using GTP·Mg2+ loaded GTPases. For experimental condition see text. Ka is given in units of 104 M-1, ΔHa [kJ mol-1], ΔG is given in kJ mol-1, ΔSa in J·mol-1·K-1 and kcat/KM in μM-1·min-1.
| Thermodynamics (ITC) | ||||
|---|---|---|---|---|
| GAPGraf WT | Ka (×104) [M-1] |
ΔGa [kJ mol-1] |
ΔHa [kJ mol-1] |
ΔSa [J mol-1 K-1] |
| Cdc42·GDP-AlFx·Mg2+ | 242.5±25.0 | -35.8 | 62.8±0.4 | 336 |
| RhoA·GDP-AlFx·Mg2+ | 55.4±5.1 | -32.2 | 54.3±0.5 | 295 |
| Cdc42·GMPPNP·Mg2+ | 2.61±0.3 | -24.8 | 22.9±1.2 | 163 |
| RhoA·GMPPNP·Mg2+ | 3.38±1.0 | -25.4 | 6.7±0.5 | 110 |
| Kinetics (MESG/PNP assay) | ||||
| GAPGraf WT | KM [μM] |
kcat [min-1] |
kcat / KM [μM-1 min-1] |
|
| Cdc42·GTP·Mg2+ | 2.7±0.3 | 219.1±32 | 80.8 | |
| RhoA·GTP·Mg2+ | 6.8±0.8 | 87.8±17 | 13.0 | |
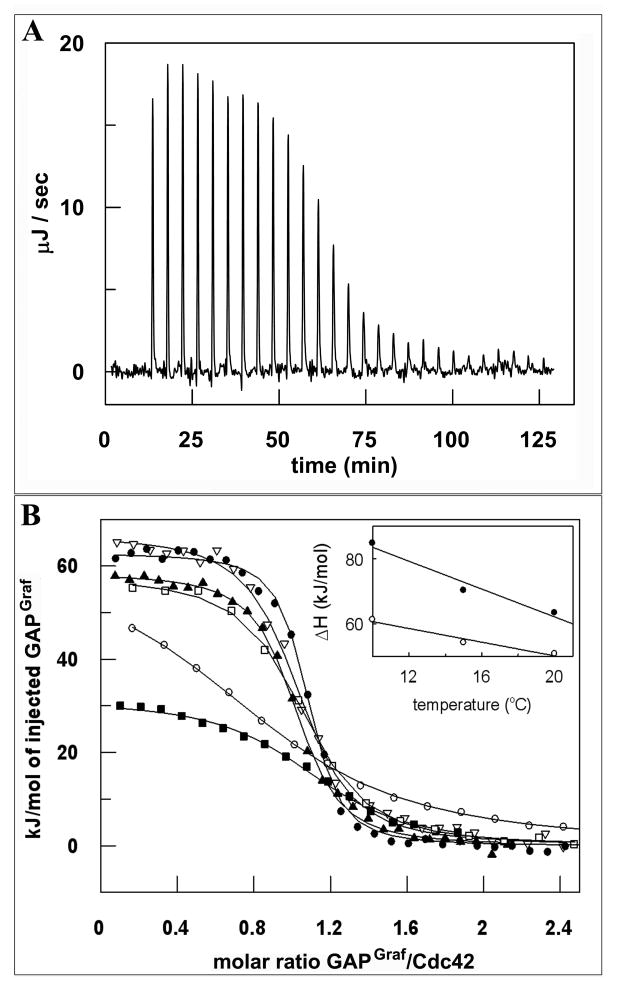
The interactions of GAPGraf with both Cdc42 and RhoA are strongly endothermic and the data could be fitted to a single-site binding model with calculated stoichiometry of 1.0 ± 0.15%. Large, unfavorable positive enthalpy changes (ΔHa) are in both cases compensated by positive (favorable) entropy changes (ΔSa) which effectively drive the reactions. Unfavorable values of ΔHa typically reflect the dehydration effect of polar groups associated with hydrogen bonding accompanying formation of a complex (Loladze et al., 2002). The observed positive change in entropy suggests that the reaction is driven by the release of water molecules from the proteins' surfaces. To further elucidate the nature of the driving forces during the formation of the GAPGraf -GTPase complex, we determined the change in heat capacity (ΔCp) of association, which directly characterizes the nature of protein-protein interaction; a negative ΔCp value indicates that a complex buries substantial hydrophobic surfaces. Figure 3 shows the temperature dependence of calorimetric ΔHa plot for Cdc42- GAPGraf and RhoA- GAPGraf. The linear dependence of ΔHa over the studied temperature range yields ΔCp values of -2.14 and -1.05 kJ/mol·K for Cdc42 and RhoA, respectively. To assess, if the GTPase- GAPGraf models are consistent with experimental results, we used an empirical relationship between the heat capacity and the change in the accessible surface area (ΔASA) (Murphy and Freire, 1992), and we used the models to calculate theoretical ΔCp values. We obtained -1.7 and -1.8 kJ/mol·K for Cdc42 and RhoA, respectively. These values are close to experimental data. The discrepancy between the measured and calculated ΔCp values probably reflects unaccounted water molecules in the RhoA- GAPGraf and Cdc42- GAPGraf binding interface, as noted in similar studies Ras interactions with effectors (Rudolph et al., 2001).
Figure 3.
Representative calorimetric titrations of Cdc42 with GAPGraf. (A) panel shows raw heat data corrected for baseline drift obtained from 27 consecutive 7 μl injections of 1.72 mM I358A mutant at 260 second intervals into the sample cell (1.05 mL) containing 0.0792 mM Cdc42 in 25mM HEPES, 5mM MgCl2, 1mM DTT, 25mM NaF, 1mM AlCl3 pH 7.8 at 20°C. (B) panel shows the representative binding isotherms for I358A(▲), V338L(▽), E253A(■), R266A(□), V224K(○) and WT (●), created by plotting the heat peak areas against the molar ratio of GAP proteins added to Cdc42 present in the cell. The line represents the best fit to the model of n independent binding sites. The heats of mixing (dilution) were subtracted. Insert. Temperature dependence of GAPGraf WT interaction with RhoA(○) and Cdc42(●). The slopes of the linear regressions provide the ΔCp values of -2.14 and -1.05 kJ/mol K for Cdc42 and RhoA, respectively.
In parallel to calorimetric titrations, we investigated the catalytic activity of wild-type GAPGraf on Cdc42 and RhoA using the MESG/PNP assay. Reactions were carried out under single turnover conditions and the release of free phosphate group was monitored as a function of time (Zhang and Zheng, 1998). This assay allowed us to determine both the KM and kcat/KM parameters for the hydrolysis reaction (Table 1, 2). The KM and kcat values of wild-type GAPGraf -stimulated GTP hydrolysis for Cdc42 were 2.5 times lower and higher, respectively, than for RhoA, resulting in the overall catalytic efficiency six times higher on Cdc42 than on RhoA. This is consistent with the calorimetric studies which showed that the Cdc42 transition-state mimic is favored over the RhoA mimic as the binding partner.
Table 2.
Table 2a. The thermodynamic and kinetic parameters of GAPGraf mutants interaction with Cdc42. The ITC measurements were performed in 25 mM HEPES, 1mM DTT, 5 mM MgCl2, 25 mM NaF, 1 mM AlCl3, pH 7.8 at 20°C using GMP-PNP-Mg2+ and GDP·Mg2+-loaded Cdc42 and RhoA. Kinetic parameters were measured using MESG/PNP assay in 50 mM Tris/HCl, 5 mM MgCl2, pH 7.8 using GTP·Mg2+ loaded GTPases. The fitting error is given. n/d – not determined due to very weak binding. Ka is given in units of 104 M-1, ΔHa [kJ·mol-1], ΔG and ΔΔGa are given in kJ·mol-1 and ΔSa in J·mol-1·K-1. ΔΔGa=ΔGWT-ΔGmutant [kJ·mol-1]; KM [μM], kcat/KM [μM-1·min1].
Table 2b. The thermodynamic and kinetic parameters of GAPGraf mutants interaction with RhoA. For description see Table2a.
| a | |||||||
|---|---|---|---|---|---|---|---|
| Thermodynamics (ITC) |
Kinetics (MESG/PNP assay) |
||||||
| GAPGraf | Ka (×104) [M-1] |
ΔHa [kJ mol-1] |
ΔSa [J mol-1 K-1] |
KM [μM] |
kcat / KM [μM-1 min-1] |
kin-WT/kin-MUT | Ka WT/Ka MUT |
| WT | 242.5±25.0 | 62.8±0.4 | 336 | 2.7±0.3 | 80.8 | - | - |
| K262A | <1 | n/d | n/d | n/d | n/d | - | - |
| R266E | <1 | n/d | n/d | n/d | n/d | - | - |
| R266A | 77.3±4.8 | 57.9±0.7 | 310 | 11.0±1.4 | 16.7 | 4.8 | 3.1 |
| V338L | 118.2±1.3 | 66.7±0.7 | 344 | 2.4±0.3 | 31.3 | 2.6 | 2.1 |
| V338A | 20.3±1.9 | 70.6±0.2 | 343 | 5.9±0.8 | 14.6 | 5.5 | 11.9 |
| I358E | 9.7±1.8 | 19.4±1.0 | 162 | 12.4±2.1 | 7.4 | 10.9 | 24.9 |
| I358A | 94.6±7.6 | 58.5±0.4 | 314 | 15.0±0.5 | 20.0 | 4.0 | 2.6 |
| E253A | 29.8±2.9 | 31.3±0.5 | 212 | 19.9±1.3 | 31.9 | 2.5 | 8.2 |
| E253K | 44.5±5.6 | 43.8±0.7 | 258 | 10.2±1.4 | 27.4 | 2.9 | 5.4 |
| N225A | 155.8+13.2 | 41.2±0.3 | 259 | 8.0±1.2 | 12.3 | 6.5 | 1.5 |
| V224K | 9.2±0.4 | 43.8±0.7 | 258 | 10.2±0.8 | 27.4 | 2.9 | 5.4 |
| b | |||||||
|---|---|---|---|---|---|---|---|
| Thermodynamics (ITC) |
Kinetics (MESG/PNP assay) |
||||||
| GAPGraf | Ka (×104) [M-1] |
ΔHa [kJ mol-1] |
ΔSa [J mol-1 K-1] |
KM [μM] |
kcat / KM [μM-1 min-1] |
kin-WT/kin-MUT | Ka WT/Ka MUT |
| WT | 55.4±5.1 | 54.3±0.5 | 295 | 6.8±0.8 | 13.0 | - | - |
| K262A | <1 | n/d | n/d | n/d | n/d | - | - |
| R266E | <1 | n/d | n/d | n/d | n/d | - | - |
| R266A | 9.3±0.7 | 42.9±0.1 | 242 | 25.0±4.2 | 2.9 | 4.5 | 6.0 |
| V338L | 22.0±4.7 | 41.0±2.2 | 242 | 8.7±1.1 | 2.0 | 6.4 | 2.5 |
| V338A | 9.3+1.1 | 60.7±2.1 | 302 | 31.3±3.3 | 1.6 | 8.2 | 6.0 |
| I358E | 3.3±0.7 | 16.4±2.1 | 143 | n/d | n/d | - | 16.7 |
| I358A | 25.5±2.6 | 60.4±2.4 | 309 | 18.2±2.6 | 2.5 | 5.2 | 2.2 |
| E253A | 45.6±5.0 | 42.2±0.9 | 252 | 11.3±1.2 | 19.8 | 1.5 | 1.2 |
| E253K | 64.3±5.4 | 50.1±0.4 | 282 | 8.8±2.0 | 24.8 | 0.5 | 0.9 |
| N225A | 30.3±0.2 | 44.9±2.5 | 258 | 24.4±3.2 | 12.3 | 1.1 | 1.8 |
| V224K | 33.8±1.4 | 70.5±0.3 | 346 | 10.3±2.2 | 5.9 | 2.2 | 1.6 |
We have also re-evaluated the activity of GAPGraf against Rac1. As expected, the association constant for Rac1·GDP·Mg2+·AlFx was over twenty times lower than for RhoA and a hundred times lower than for Cdc42. The MESG/PNP assay revealed only marginal activity on Rac1 (data not shown)
Design of mutants probing structure/function relationships in GAPGraf
Based on the model of the RhoA(Cdc42)/GAPGraf complex, we designed fifteen point mutants to probe how the different epitopes on GAPGraf contribute to the interaction with the cognate GTPase and to catalysis.
Within patch I (i.e. the arginine finger and its adjacent loop) we targeted the strictly conserved Arg220, Lys262 and Arg266. Arg220 was mutated to Ala, Ser, Lys and Glu, to assess the effect of loss of side chain, retention of positive charge and charge reversal. Lys262 was mutated to Ala, while Arg266 was mutated to both Ala and Glu to assess the impact of loss of side chain and charge reversal. Within patch II we targeted Val224, Asn225 and the adjacent Glu253: Val224 was mutated to Glu, Asn225 to Ala and Glu253 to both Ala and Lys. Finally, within patch III we targeted both Val338, and Ile358 and we mutated Val338 to Leu and Ala, while Ile358 was mutated to Ala and to Glu.
Binding of cognate GTPases by mutant GAPGraf proteins and their catalytic properties
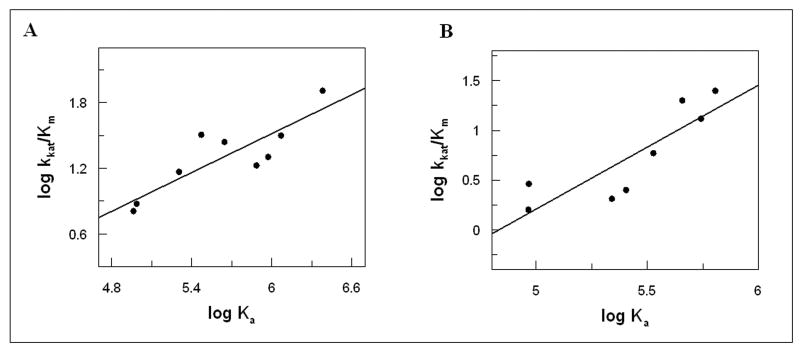
All mutants were assayed for their ability to bind the transition-state mimic of RhoA and Cdc42, and for their catalytic properties. In both series of experiments, using Cdc42 or RhoA as targets, we observe strong correlation between Ka, determined by calorimetry, and kcat/KM parameters, determined by MESG/PNP assay (Figure 5). The mutants that conform to this paradigm exclude all those at the Arg220 position, as well as mutations of Lys262 and the R266E mutant. These data confirm that, in general terms, residues outside the arginine finger play a purely structural and non-catalytic function.
Figure 5.
The correlation plot of Ka and kcat/KM parameters. A. Cdc42 B. RhoA. The Ka measurements were done in 25mM HEPES, 5mM MgCl2, 1mM DTT, 25mM NaF, 1mM AlCl3, pH 7.8 at 20°C, and kcat/KM determinations in 50mM Tris/HCl, GTP, 0,1mM MESG, 1U PNP, 5mM MgCl2 pH 7.8 at 20°C. The Pearson moment correlation coefficients are 0.898 and 0.894, respectively.
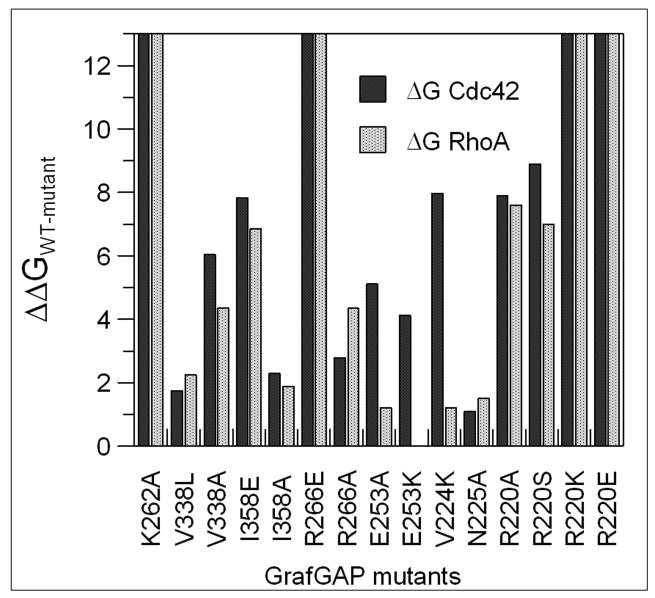
Figure 3 shows representative calorimetric data for the interactions of GAPGraf mutants with Cdc42. Figure 4 shows all determined ΔΔGa, defined as the ΔGa of the wild-type minus ΔGa of the GAPGraf mutant, for the interactions of all assayed GAPGraf mutants with both RhoA and Cdc42. For the majority of mutations, a decrease in the Gibbs association energy (ΔΔGa) is observed, with a maximum change of 8.9 kJ/mol. For some GAPGraf mutants, the decrease in association constant was so large, that accurate measurements of Ka required exceedingly high concentrations of the protein. In such cases, only the lower limit of Ka (below 1 × 104 M-1) could be estimated, corresponding to the reduction of Gibbs energy by at least 13 kJ/mol.
Figure 4.
Comparison of the ΔΔGa values determined for the interaction of GAPGraf mutants with RhoA and Cdc42. Measurements were done in 25mM HEPES, 5mM MgCl2, 1mM DTT, 25mM NaF, 1mM AlCl3, pH 7.8 at 20°C.
Table 2 lists all the numerical data of the calorimetric and catalytic assays. With one exception, all mutants showed a decrease in kcat/KM in comparison with wild-type GAPGraf. The results were similar for both RhoA and Cdc42 GTPases.
Arginine finger substitutions
Mutations of the catalytic Arg220 to Ser and Ala significantly reduced the Ka values for the interaction with both RhoA and Cdc42, while mutants containing Lys and Glu at this position showed no detectable binding to either RhoA or Cdc42. In all above cases the catalytic activity was completely abolished. Thus, Arg220 in GAPGraf plays a significant role in both binding the GTPase and in the protein's ability to stimulate the catalytic activity of Cdc42 and RhoA GTPases. While these data are consistent with experimental results reported for some RhoGAPs (Ahmed et al., 1994; Hoffman et al., 1998; Leonard et al., 1998; Muller et al., 1997), they are different from those reported for p50RhoGAP (Graham et al., 1999). In the latter case the mutations of the catalytic Arg85 to Lys or Ala cause only moderate, 3-fold, decrease in the association constant with RhoA in the presence of AlFx but result in a dramatic decrease in the catalytic activity on RhoA (Graham et al., 1999).
The two conserved residues critical to the structural integrity of the arginine finger are Lys262 and Arg266. The crystal structure of the p50RhoGAP complex and our models suggest that Lys262 and Arg266 are involved in direct hydrogen bonds to Glu64 and Asp65 of RhoA, which is present in the G3 loop within the switch II region (Figure 6A). These residues stabilize the transition state conformation of G3 loop that bears the catalytic Gln63 (Gln61 in Cdc42). The Ka values for the interaction of K262A/Q and R266E GAPGraf mutants with Cdc42/RhoA were not measurable and, as expected, no activity was observed in kinetic MESG/PNP experiments (Table 2). In contrast, the R266A mutant causes only a 3- and 6-fold decrease of association constant for Cdc42 and RhoA, respectively. The effects on kcat/KM are of similar magnitude. These results suggest that the Lys262 side chain and its H-bonds to Glu64 and Asp65 of RhoA, as well as an H-bond to the main chain carbonyl of Tyr219, play a critical role in the formation of the catalytically competent interaction between the arginine finger and Switch II. Lys262 is equivalent to Arg903 in RasGAP334 and Lys122 in p50RhoGAP, and has been previously identified as the secondary arginine finger (Graham et al., 1999). Loss of the side chain is as deleterious for Lys262 as charge reversal. In contrast, Arg266 appears to be less critical. The charge reversal mutant R266E is clearly disruptive, and must lead to severe electrostatic repulsion with Glu64 and Asp65, but the R266A mutant shows a milder effect, indicating that the side chain of Arg266 is electrostatically favorable, but not absolutely required for function.
Figure 6.
Modeled complex of RhoA (electrostatic potential representation) and GAPGraf (main chain). A, B and C - structural details of Patch I, II and III, respectively. GAPGraf is shown in blue, RhoA in red and hydrogen bonds as green broken lines.
The role of residues within Patch II
The distinct, negatively charged cluster of Asp90, Glu93 and Glu97 is located on the solvent exposed face of helix 3 in RhoA. Interestingly, the distribution of charge on this surface is different in all three GTPases. In Rac1, Asp90 and Glu97 are replaced by alanines, and in Cdc42 Asp90 is replaced by a Ser. Our models show that this region comes into an intimate contact with several GAPGraf residues clustered on two adjacent loops: the loop containing the catalytic Arg220, i.e. between the N-terminal α-helix and the second α-helix, and the loop leading to helix 3. These residues are Val224, Asn225 and Glu253.
The V224K mutation was engineered to probe if a positively charged amino acid could confer a higher selectivity for RhoA. However, this mutant binds RhoA with 1.6-fold lower affinity, suggesting that unfavorable interactions are also created. A much larger drop in affinity, ∼26-fold, was observed for Cdc42. This could be a result of the absence in Cdc42 of the negatively charged residue in the position analogous to Asp90, resulting in no new cohesive interactions. The binding affinities correlate well with the difference in kinetic parameters for both GTPases. The kcat/KM values for the GTP hydrolysis by RhoA in the presence of V224K mutant is only 2.3-fold lower, while for Cdc42, the effect is again more profound with kcat/KM about 12 times lower than determined for the wild-type.
The E253A/K mutants were tested to assess the consequences of side chain removal or charge reversal. Interestingly, both mutations affect the interaction with Cdc42 with 5.5- and 8-fold weaker binding, respectively, but with virtually no effect on the binding of RhoA. The effect on catalysis on Cdc42 is less significant with only about 2.7-fold decrease of kcat/KM.
The N225A was designed to assess the effect of loss of side chain. This mutation causes approximately 2-fold decrease in Ka for both RhoA and Cdc42. The effect on kcat/KM is negligible for RhoA but it produces much larger, 6.6-fold, decrease of catalytic efficiency in case of Cdc42.
The impact of mutations within Patch III
The V338A/L and I358A mutations have a moderate impact on GAPGraf ability to bind RhoA and Cdc42. Val338 is highly conserved among many RhoGAPs, although the position is often occupied by Ile. On the other hand, Ile358 is found in the variable C-terminal portion of the sequence which shows poor similarity to other GAPs, precluding credible sequence alignment. Both residues are in the center of a hydrophobic surface patch which packs between the Switch I and Switch II fragments of RhoA, and specifically Tyr66, Leu69, Val38 and Phe39 in RhoA (all these residues are conserved in Cdc42) The truncation of Ile358 to Ala resulted in 2.5-fold decrease in association constant and 4 to 5-fold reduced values of kcat/KM for the interaction with Cdc42 and RhoA. Insertion of negatively charged Glu in position of Ile358 is significantly more unfavorable, leading to about ∼20-fold decrease in the Ka values for both GTPases and lack of catalytic activity. Substitution of Val338 with Ala results in a 6-fold decrease of the association constant for the interaction with both GTPases, and a moderate decrease in kcat/KM. A similar effect is observed for the V338L mutant.
Conclusions
In general terms, we note that the surfaces on the Rho GTPase involved in the interaction with GAP domains, appear to be significantly more conserved between RhoA, Rac and Cdc42, than the surfaces responsible for the interaction of these GTPases with the DH domain of guanine nucleotide exchange factors (Derewenda et al., 2004; Snyder et al., 2002). This explains why most of GAPs show significant promiscuity in their substrate selectivity, while a larger proportion of GEFs show high selectivity. Most of the amino acids in the active surface of GAP are important for the formation of the catalytically productive complex with the GTPase, and some, notably the residues in the arginine finger, are essential for catalysis. Selectivity is modulated by a much more subtle interplay of electrostatic and hydrophobic interactions involving residues on the periphery of the main interface.
In an effort to better understand the structure-function relationships in the RhoA/Cdc42 specific GAP domain of Graf, we generated fifteen mutants of this domain and measured their ability to bind the two cognate GTPases and to accelerate the hydrolysis of GTP bound to the GTPase to GDP. The eight residues targeted in this study are involved in three distinct patches on the surface, two of which (including the catalytic arginine finger loop) appear to interact with highly conserved regions of the GTPase, while the third plays a role in GTPase selectivity.
As expected, we find that Arg220 and the adjacent residues within the arginine finger play a critical role. However, in contrast to previously published data, we find that both binding and enhancement of GTP hydrolysis are equally dependent on the structural integrity of the arginine finger. These residues appear to have a ‘generic’ role, in that the corresponding GTPase epitope is highly conserved and does not offer any means of selectivity. The contact mediated by the second patch, i.e. with Switch I of the GTPase, is of a similar nature, although the impact of mutations in this contact is significantly smaller and the mutants retain a significant fraction of catalytic potential. Mutants within the third patch, which interacts with a variable sequence on the GTPase, show noticeable differences with respect to their activities on RhoA and Cdc42.
Acknowledgments
Filip Jelen was supported by a young scientist fellowship from the Foundation for Polish Science. This work was supported in part by National Institutes of Health Grant PO1 HL48807.
BRIEFS
- GAP
GTPase activating protein
- GEF
guanidine nucleotide exchange factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian MR, Mittal R, Hall A, Wittinghofer A. Aluminum fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett. 1997a;408:315–318. doi: 10.1016/s0014-5793(97)00422-5. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997b;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Lee J, Wen LP, Zhao Z, Ho J, Best A, Kozma R, Lim L. Breakpoint cluster region gene product-related domain of n-chimaerin. Discrimination between Rac-binding and GTPase-activating residues by mutational analysis. J Biol Chem. 1994;269:17642–17648. [PubMed] [Google Scholar]
- Åqvist J. Ion-Water Interaction Potentials Derived from Free Energy Perturbation Simulations. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- Barrett T, Xiao B, Dodson EJ, Dodson G, Ludbrook SB, Nurmahomed K, Gamblin SJ, Musacchio A, Smerdon SJ, Eccleston JF. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458–461. doi: 10.1038/385458a0. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. Journal of American Chemical Society. 1995;117:5179–5197. [Google Scholar]
- Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS. The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure. 2004;12:1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. 1990. [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Graham DL, Eccleston JF, Lowe PN. The conserved arginine in rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with Rho.GDP and aluminum fluoride. Biochemistry. 1999;38:985–991. doi: 10.1021/bi9821770. [DOI] [PubMed] [Google Scholar]
- Group, HPCC. Pacific Northwest National Labolatory. 2000. NWChem, A Computational Chemistry Package for Parallel Computers, Version 4.0. [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Taylor JM, Parsons JT. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Oswald RE, Cerione RA. Fluoride activation of the Rho family GTP-binding protein Cdc42Hs. J Biol Chem. 1998;273:4392–4399. doi: 10.1074/jbc.273.8.4392. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J Mol Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard DA, Lin R, Cerione RA, Manor D. Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J Biol Chem. 1998;273:16210–16215. doi: 10.1074/jbc.273.26.16210. [DOI] [PubMed] [Google Scholar]
- Loladze VV, Ermolenko DN, Makhatadze GI. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J Mol Biol. 2002;320:343–357. doi: 10.1016/S0022-2836(02)00465-5. [DOI] [PubMed] [Google Scholar]
- Longenecker KL, Zhang B, Derewenda U, Sheffield PJ, Dauter Z, Parsons JT, Zheng Y, Derewenda ZS. Structure of the BH domain from graf and its implications for Rho GTPase recognition. J Biol Chem. 2000;275:38605–38610. doi: 10.1074/jbc.M007574200. [DOI] [PubMed] [Google Scholar]
- Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- Muller RT, Honnert U, Reinhard J, Bahler M. The rat myosin myr 5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell. 1997;8:2039–2053. doi: 10.1091/mbc.8.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Taylor WR, Smerdon SJ, Gamblin SJ. Support for shared ancestry of GAPs. Nature. 1998;392:448–449. doi: 10.1038/33043. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997a;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997b;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Linnemann T, Grunewald P, Wittinghofer A, Vetter IR, Herrmann C. Thermodynamics of Ras/effector and Cdc42/effector interactions probed by isothermal titration calorimetry. J Biol Chem. 2001;276:23914–23921. doi: 10.1074/jbc.M011600200. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zheng Y. Regulation of RhoA GTP hydrolysis by the GTPase-activating proteins p190, p50RhoGAP, Bcr, and 3BP-1. Biochemistry. 1998;37:5249–5257. doi: 10.1021/bi9718447. [DOI] [PubMed] [Google Scholar]