Abstract
Investigations using Drosophila melanogaster have shown that the circadian clock gene period can influence behavioral responses to cocaine, and the mouse homologues, mPer1 and mPer2, modulate cocaine sensitization and reward. In the present study, we applied DNAzyme targeting mPer1 to interfere the expression of mPer1 in CNS in mice and studied the role of mPer1 on morphine dependence. We found that the DNAzyme could attenuate the expression of mPer1 in CNS in mice. Mice treated with DNAzyme and morphine synchronously did not show preference to the morphine-trained side, whereas the control group did. In contrast, mice treated with DNAzyme after morphine showed preference to the morphine-trained side as well as the control group did. These results indicate that drug dependence seems to be influenced at least partially by mPer1, but mPer1 cannot affect morphine dependence that has been formed.
Keywords: drug dependence, DNAzyme, learning and memory, circadian, i.c.v.
Circadian clocks are molecular time-keeping mechanisms that reside in a diverse range of cell types in a variety of organisms. The primary role of these cell-autonomous clocks is to maintain their own approximately 24 h molecular rhythm and to drive the rhythmic expression of genes involved in physiology, metabolism and behavior. Components of the endogenous master clock were first identified in the fruit fly Drosophila melanogaster. The Period (Per) encodes one of the essential elements involved in the transcription/translation-based auto-regular loop of the endogenous master clock (Reppert and Weaver, 2001). Three homologues of Drosophila Per genes were subsequently identified in mice (mPer1, mPer2, and mPer3) (Albrecht, 2002), leading to great progress in elucidation of the molecular mechanism underlying circadian rhythm in the CNS.
It has been shown that repeated administration of methamphetamine caused behavioral sensitization as well as sensitized expression of mPer1 (Nikaido et al., 2001). Some studies implicate a role for Per genes in drug-induced behavioral sensitization processes. This suggestion is supported by investigations using Drosophila flies. Flies mutant in the Per gene did not sensitize after repeated exposure to volatilized free-base cocaine (Andretic et al., 1999; Hirsh, 2001). In mice, the mPer1 and mPer2 genes influence cocaine-induced sensitization and reward in an opposite manner. The lack or dysfunction of the mPer1 gene abolishes cocaine sensitization and reward whereas the dysfunction of the mPer2 gene induces a hypersensitized response to cocaine (Abarca et al., 2002).
DNAzyme is a suitable tool for studying gene function. The typical DNAzyme, known as the “10–23” model, is capable of cleaving single-stranded RNA at specific sites. The “10–23” model of DNAzymes has a catalytic domain of 15 highly conserved deoxyribonucleotides, flanked by two substrate-recognition domains, which can cleave effectively between any unpaired purine and pyrimidine of mRNA transcripts (Santoro and Joyce, 1997).
To further understand the role and the mechanism of mPer1 gene in morphine dependence, we first studied morphine-induced reward to be involved in morphine dependence by blocking the expression of mPer1 gene with the “10–23” DNAzyme.
EXPERIMENTAL PROCEDURES
Animals
In all experiments, 4- to 6-week-old male BALB/C mice were used. Mice were housed in groups of five and provided with food and water ad libitum. Artificial light was provided daily from 8:00 a.m. to 8:00 p.m. with room temperature and humidity kept constant (temperature: 22–24 °C; humidity: 55–65%). All procedures were performed in compliance with the local, international and institutional guidelines. All efforts were made to minimize the number of animals used and their suffering.
Conditioned place preference (CPP)
Place conditioning was conducted as described previously according to Suzuki et al. (1993). The apparatus consisted of a shuttle box (30×15×15 cm: length×width×height) made of an acryl-resin board. The box was divided into two compartments of equal size by means of a sliding partition. One compartment was white with a textured floor, and the other was black with a smooth one. When the CPP was measured, the partition separating the two compartments was raised to 7 cm above the floor. Preference for a particular place was assessed. The time spent in black compartment during a 900-s session was measured automatically. To avoid the introduction of systematic errors, the CPP experiment was carried out in a light and sound-controlled environment.
I.c.v. injection
The i.c.v. injection procedure was adapted from the method described earlier (Mistry et al., 1997). Briefly, the i.c.v. injections were given as follows: under light ether anesthesia, bregma was exposed. An injection volume of 20 µl was delivered over a 60-s period, 2 mm lateral and caudal to bregma at a depth of 2 mm by using a syringe. Proper placement was verified in the experiments by injection and localization of Methylene Blue dye.
DNAzyme
We designed a “10–23” DNAzyme, as described previously by Santoro and Joyce (1997), targeting mPer1 gene in mice (Fig. 1). The 15-nt catalytic domain is flanked by two eight-nt arms that recognized the mPer1 mRNA substrate from 287 to 303 nt except 296 nt (GenBank accession number: U49930). The 5′ and 3′ termini of the molecule are protected from exonucleases by a phosphorothioate linkage and a CPG-C3 cap respectively. To inactivate the DNAzyme and to generate a control oligonucleotide (ODN), two nts were changed in the catalytic domain of the DNAzyme (Fig. 1C). Transversion of two nts in the disabled ODN is sufficient to inactivate the catalytic activity (Santoro and Joyce, 1997; Wu et al., 1999; Sriram and Banerjea, 2000; Unwalla and Banerjea, 2001). The DNAzyme and the disabled DNAzyme ODN (control ODN) were synthesized by Invitrogen (Invitrogen, USA).
Fig. 1. DNAzyme and its mRNA substrate of mPer1.
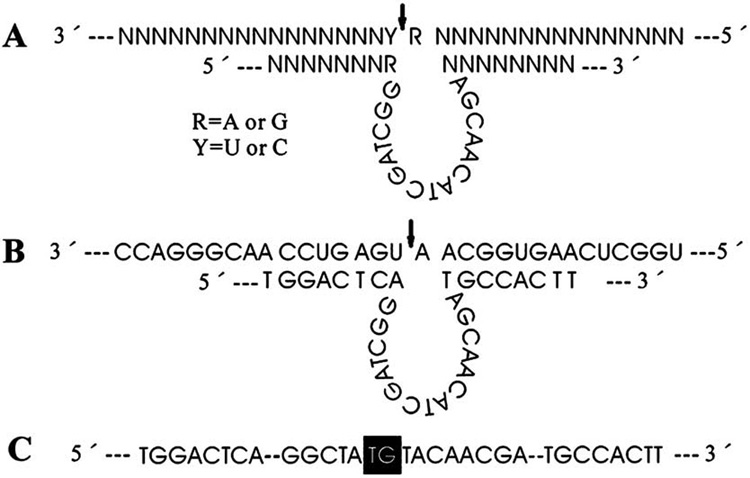
(A) A 10–23 DNAzyme structure with substrate cleavage occurring at the position indicated by the arrow. (B) Sequence and structure of mPer1 DNAzyme annealed to the mPer1 substrate. The arrow indicates the cleavage site. (C) Sequence of the control ODN. To inactivate enzymatic activity of the DNAzyme, two nts (black) in the intervening 15-nt catalytic domain were altered.
In vitro transcript of sequence of mPer1 mRNA for cleavage
A double stranded ODN containing the sequence of the mPer1 cDNA 140–845 nts (GenBank accession number: U49930) plus appropriate cloning sites was synthesized and introduced into the HindIII and BamH I sites of plasmid pBluescript II SK(+). This recombinant was prepared for cleavage experiments.
Cleavage experiments
DNAzyme cleavage experiments were performed as described previously (Santiago et al., 1999). The oligoribonucleotide substrate of the DNAzyme was labeled at the 5′ end with [33P] ATP (2500 Ci/mmol; Amersham Pharmacia Biotech) by using T4 polynucleotide kinase (New England Biolabs). DNAzyme (50 pmol) was added to 4 pmol of the in vitro transcript substrate. The reaction was then stopped at several time points. In these cleavage experiments, the cut and uncut substrates were separated by electrophoresis on a 5% urea denaturing polyacrylamide gel and detected by autoradiography at 4 °C. Signals were then scanned by Storm 840 instrument and analyzed by Image-Quant 5.0 software (Molecular Dynamics).
Treated with DNAzyme and morphine synchronously
DNAzyme and control ODN were enclosed with Lipofectamine (Invitrogen) according to the description respectively. After mice’s acclimatization, the basic CPP of mice was measured. The animals were divided into two groups (n=30 per group), including DNAzyme group (DMS) and control ODN group (CMS). The animals were given saline (the same volume as morphine, s.c.) at zeitgeber time (ZT) 2 (10:00 a.m.) before placed into black section of the shuttle boxes for 30 min. On the next day, the animals were given morphine (10 mg/kg, s.c.) at ZT2 (10:00 a.m.) and then placed into white section of the shuttle boxes for 30 min. These procedures were repeated four times in 8 days. At ZT12 (8:00 p.m.) mice of different group were injected intracerebroventricularly with DNAzyme and control ODN enclosed with Lipofectamine respectively once a day from the 1st to the 7th day of the experiment according to different group. At ZT8 (4:00 p.m.), the CPP of the mice was measured on the 8th day of the experiment. Thus four mice of the each group were killed at ZT12 (8:00 p.m.) and ZT16 (12:00 p.m.) on the 6th day and at ZT20 (4:00 a.m.), ZT0 (8:00 a.m.), ZT4 (12:00 a.m.) and ZT8 (4:00 p.m.) on the 9th day respectively. The brains of the killed mice were prepared for Western blot.
Treated with DNAzyme after morphine
Mice were also divided into two groups (n=30 per group), including DNAzyme group and (DMA) control ODN group (CMA). The basic CPP of mice was measured. The animals were given saline (the same volume as morphine, s.c.) at ZT2 (10:00 a.m.) before placed into black section of the shuttle boxes for 30 min. On the next day, the animals were given morphine (10 mg/kg, s.c.) at ZT2 (10:00 a.m.) and then placed into white section of the shuttle boxes for 30 min. These procedures were repeated four times in 8 days, and then the mice were injected intracerebroventricularly with DNAzyme and control ODN once a day respectively according to different group in the next 7 days at ZT12 (8:00 p.m.). The remained procedures were the same as the abovementioned.
Western blot
Mice were deeply anesthetized with ether. Whole brain homogenates were obtained as described (Hastings et al., 1999; Lee et al., 1998). Briefly, tissues were homogenized at 4 °C in buffer 1 (0.4 M NaCl, 20 mM HEPES [pH 7.5], 1 mM EDTA, 5 mM NaF, 1 mM dithiothreitol, 0.3% Triton X-100, 5% glycerol, 0.25 mM phenylmethylsulfonyl fluoride, 10 mg of aprotinin per ml, 5 mg of leupeptin per ml, 1 mg of pepstatin A per ml). Homogenates were cleared by centrifugation (twice, 12 min each, 12,000×g). Proteins were separated by electrophoresis through sodium dodecyl sulfate–6% polyacrylamide gels and then transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20 and then incubated with affinity-purified antisera to mPER1 (Alpha Diagnostics International). Immunoreactive bands were visualized using antigoat immunoglobulin G secondary antisera and enhanced chemiluminescence detection. Signals were then scanned by Storm 840 instrument and analyzed by Image-Quant 5.0 software (Molecular Dynamics).
Data analysis
Data were analyzed by one-way ANOVA for group differences and time difference respectively, and by two-way ANOVA for time and group differences (SPSS 11.5).
RESULTS
In vitro cleavage of mPer1 mRNA
We tested the efficacy of the DNAzyme targeting mPer1 to cleave the substrate of mPer1 mRNA prepared an in vitro transcript. The DNAzyme had the effect on cleaving mPer1 mRNA in vitro. Two expected cleavage products, 255-nt and 451-nt, were produced (Fig. 2A). With 4 pmol of the RNA transcript as the target, the DNAzyme at 0 min, 30 min, 60 min, 90 min and 120 min digested 0, 18.1, 38.9, 46.5 and 51.9%, respectively, of the substrate (Fig. 2A). Cleavage products were progressively increased until the last time point at 120 min. In contrast, the disabled DNAzyme ODN (control ODN) did not show any enzymatic activity (Fig. 2B).
Fig. 2. In vitro cleavage of mPer1 RNA.

(A) For DNAzyme cleavage of an in vitro transcript, 4 pmol of a 706-nt mPer1 mRNA transcript were digested with 50 pmol of DNAzyme. The expected 255-nt and 451-nt cleavage products were generated. (B) The control ODN did not show any enzymatic activity. The substrate and cleavage products were separated and analyzed on a urea denaturing polyacrylamide gel.
In vivo interfering expression of mPER1 protein in CNS
In this study, the results showed that the DNAzyme could attenuate the expression of mPer1 in whole brain in mice. Fig. 3 displayed examples of mPER1 protein with Western blotting in CNS. In mice treated with control ODN, the rhyme of the density of mPER1 immunoreactivity was display on Fig. 3A, whereas the density of mPER1 immunoreactivity of mice treated with DNAzyme was arrhythmic and lower than that of mice treated with control ODN (Fig. 3B). One-way ANOVA displayed significant difference of mPER1 immunoreactivity in the whole brain in DMS, CMS, DMA and CMA, and revealed significant difference in DMS to CMS, DMS to CMA, DMA to CMS and DMA to CMA, and non-significant difference in DMS to DMA and CMS to CMA (Fig. 4).
Fig. 3. Examples of mPER1 immunoreactivity in whole brain in mice.

(A) Treated with control ODN (top: mPER1; bottom: actin). mPER1 immunoreactivity of mice treated with control ODN started to rise in the late morning, reached its maximum around the usual time of ZT8, and then declined. (B) Treated with DNAzyme (top: mPER1; bottom: actin). The mPER1 immunoreactivity of mice treated with DNAzyme was arrhythmic and lower than that of mice treated with control ODN.
Fig. 4. mPER1 immunoreactivity in whole brain in mice of different group (mean±S.E.M.).
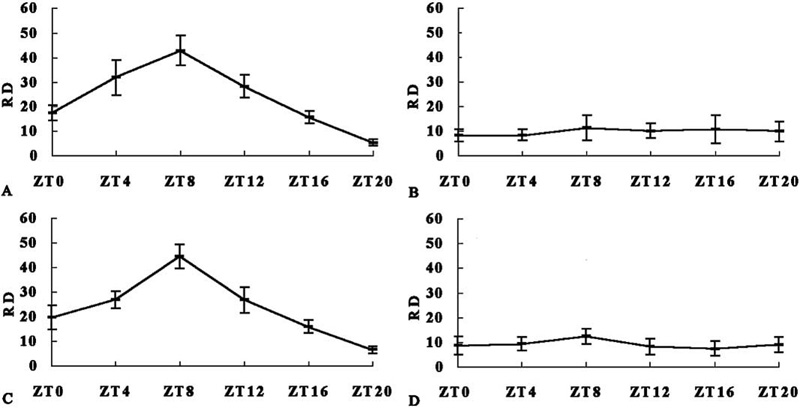
The relative density of mPER1 immunoreactivity (RD) was normalized against that of actin. Two-way ANOVA displayed that RD was different in groups and time points (intercept, F(1,87)=557.279, P<0.001; group, F(3,87)=32.775, P<0.001; time, F(5,87)=15.394, P<0.001). (A) CMS. One-way ANOVA revealed a significant daily rhythm (F(5,18)=34.516, P<0.001). The maximum of mPER1 immunoreactivity was at ZT8 (ZT8–ZT4: P=0.031; ZT8–ZT12: P=0.003; ZT8–ZT16: P<0.001; ZT8–ZT20: P<0.001; ZT8–ZT0: P<0.001), and the minimum was at ZT20 (ZT20–ZT4: P<0.001; ZT20–ZT8: P<0.001; ZT20–ZT12: P<0.001; ZT20–ZT16: P=0.05; ZT20–ZT0: P=0.001). (B) DMS. One-way ANOVA revealed a non-significant daily rhythm (F(5,18)=0.421, P=0.828). (C) CMA. One-way ANOVA revealed a significant daily rhythm (F(5,18)=33.109, P<0.001). The maximum of mPER1 immunoreactivity was at ZT8 (ZT8–ZT4: P=0.002; ZT8–ZT12: P=0.002; ZT8–ZT16: P<0.001; ZT8–ZT20: P<0.001; ZT8–ZT0: P<0.001), and the minimum was at ZT20 (ZT20–ZT4: P<0.001; ZT20–ZT8: P<0.001; ZT20–ZT12: P<0.001; ZT20–ZT16: P=0.037; ZT20–ZT0: P=0.003). (D) DMA. One-way ANOVA revealed a non-significant daily rhythm (F(5,18)=1.157, P=0.368).
Significant difference of mPER1 immunoreactivity in the whole brain in groups and time points was revealed (two-way ANOVA). One-way ANOVA revealed a significant daily rhythm of mPER1 immunoreactivity in the whole brain in mice treated with control ODN and morphine synchronously (CMS) and treated with control ODN after morphine (CMA). In contrast, there were non-significant daily rhythms of mPER1 in the whole brain in mice treated with DNAzyme and morphine synchronously (DMS) and treated with DNAzyme after morphine (DMA; Fig. 4).
Morphine-induced reward under treatment with DNAzyme
CPP has been widely used to measure the rewarding properties of drug dependence (Tzschentke, 1998). Therefore, we studied the response of mice treated with DNAzyme in the CPP paradigm to detect differences in morphine-induced reward. As expected, one-way ANOVA displayed DMS, unlike CMS, did not show preference to morphine-trained side (Fig. 5).
Fig. 5. CPP in mice.
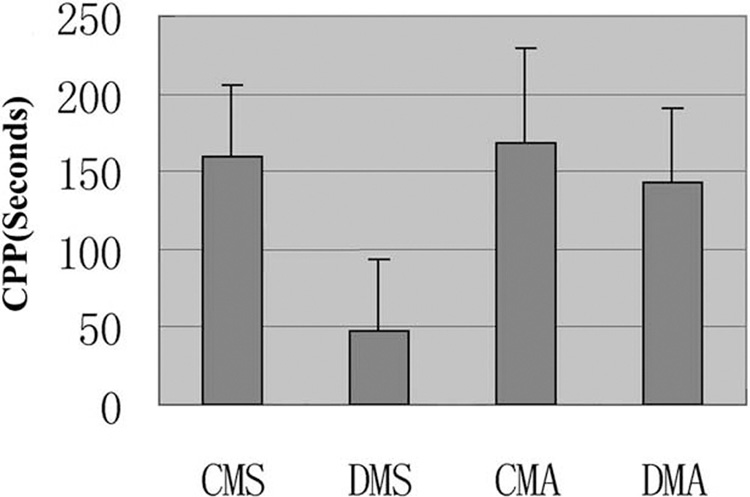
Data are given as mean (±S.E.M.) of difference between the basic CPP and CPP after treatment. One-way ANOVA revealed significant difference (F(3,116)=30.436, P<0.001). DMS was significant to other groups (DMS to CMS: P<0.001; DMS to CMA: P<0.001; DMS to DMA: P<0.001), but others were non-significant difference.
Treatment with DNAzyme when morphine addiction had been formed
When animals were given morphine for three to four times, the morphine dependence was formed, and the CPP would last for 3 weeks (Schecter, 1998). We studied the response of mice treated with the DNAzyme after given morphine in the CPP paradigm. DMA showed preference to the morphine-trained side as well as CMA group did (Fig. 5). One-way ANOVA showed non-significant difference.
DISCUSSION
The mechanism of circadian oscillation based on the molecular feedback loops consists of several circadian genes and their protein products. Several clock genes including the Per genes have been cloned in the last few years. Drosophila and mice were the models in these performed experiments (Wager-Smith and Kay, 2000; Forger and Peskin, 2003), which gave us viewpoints that circadian clock components affect many complex neuron activities like drug reward (Andretic et al., 1999). Clock, cycle, period, and timeless, doubletime were the object to be manipulate in experiment performed on Drosophila. Clock, cycle, period, and doubletime mutant phenotype showed no sensitization to cocaine. Mice with mPer1 gene mutation did not sensitize to cocaine and CPP test also indicated the block of reward to cocaine (Abarca et al., 2002). Those results showed that regulation of circadian clock gene expression might influence the process of drug dependence significantly.
DNAzymes are short DNA molecules that have the potential to cleave any target RNA in a sequence-specific manner, and two major catalytic motifs (10–23 and 8–17) have been described (Santoro and Joyce, 1997). DNAzymes are suitable tools for studying gene function, since they can down-regulate endogenous gene expression. This approach was used to interfere specifically with the intracellular function of the target genes (Famhe and Khachigian, 2004; Ordoukhanian and Jouce, 2002; Khachigian et al., 2002). Our study reveals that DNAzyme is an effective tool of researching gene function, which can cleave the targeting mRNA and attenuates its protein product in vivo and in vitro.
There is strong expression and significant circadian rhythm of mPER1 in SCN in mice, and furthermore Per1 mRNA and proteins are expressed in other brain areas (Hastings et al., 1999; Yan et al., 1999; Takumi et al., 1998; Field et al., 2000). The mPER1 in SCN starts to rise in the late morning, reaches its maximum around the usual time of the afternoon, and then declines, whereas the mPer1 mRNA cycle precedes the mPER1 protein cycle by 4–6 h, consistent with a role for the mPER protein in the autoregulation of its transcription. The rhythm of mPER1 in the whole brain in mice was demonstrated in the present results, and the pattern of circadian expression of mPER1 is similar to that in SCN. Masashi et al. (1999) used antisense ODN to inhibit mPer1 expression in vivo and in vitro, which showed that mPer1 mRNA in the SCN could was reduced by ODNs injected intracerebroventricularly (Masashi et al., 1999), but mPER1 protein was not detected in that report. In our research, the expression of mPER1 could be attenuated by i.c.v. injection of DNAzyme, and the pattern of circadian expression was interfered also.
The degree of response to cocaine-induced behavioral sensitization and reward is under the influence of the diurnal state of the animal (Abarca et al., 2002; Tei et al., 1997; Sun et al., 1997). A similar time-dependent profile was reported in methylphenidate-sensitized animals (Gaytan et al., 1999, 2000). The lack or dysfunction of the mPer1 gene abolishes cocaine sensitization and reward whereas the dysfunction of the mPer2 gene induces a hypersensitized response to cocaine (Abarca et al., 2002). Consistent with those results, animals attenuated mPer1 expression with the DNAzyme when treated with morphine synchronously did not show a preference to the morphine-paired side in this study. These results support that drug addiction seems to be influenced at least partially by the expression of mPer1.
It is known that different components of the midbrain dopamine system and the glutamatergic system play a critical role in drug-induced sensitization and reward (Spanagel and Wess, 1999; Vanderschuren and Kalivas, 2000; Cornish and Kalivas, 2001), and mesolimbic dopaminergic neurons are involved in strengthening formation of associations between salient contextual stimuli and internal rewarding or aversive events (Spanagel and Wess, 1999). The common long-term adaptations produced by opioids and other drugs of abuse in this system could enhance these processes and thereby play a major role in initiation and maintenance of compulsive drug use. Drugs of abuse cause long-lasting changes in the brain that underlie the behavioral abnormalities associated with drug addiction. Similarly, experience can induce memory formation by causing stable changes in the brain. Learning and memory and drug addiction are modulated by the same neurotrophic factors, share certain intracellular signaling cascades, and depend on activation of the transcription factor CREB. They are associated with similar adaptations in neuronal morphology, and both are accompanied by alterations in synaptic plasticity at particular glutamatergic synapses in the brain (Eric, 2002). In this study, animals interfered mPer1 expression with DNAzyme when treated with morphine synchronously did not show preference to the morphine-paired side in the CPP paradigm in comparison with the controlled. In contrast, animals with interfered-with mPer1 expression with DNAzyme after treatment with morphine showed the same preference to the morphine-trained side in the CPP paradigm in comparison with the controlled. These experiments indicate that mPer1 and its product do not influence morphine dependence when morphine dependence has been memorized. In fact, mPer1 is not related to learning and memory processes. The mPer1 and mPer2 mutants did not differ from wildtype animals in a fear conditioning paradigm, demonstrating that Per mutants do not differ in their learning abilities from wildtype animals (Abarca et al., 2002). This may be interpreted that the morphine dependence was not interfered with when mice were treated with DNAzyme after morphine.
In summary, the expression of the circadian clock gene, mPer1, can be attenuated by i.c.v. injection of DNAzyme targeting mPer1 in CNS in mice. Our data also show that animals with interfered-with expression with DNAzyme when treated with morphine synchronously did not show morphine dependence, but in contrast, animals with interfered-with mPer1 expression with DNAzyme after treatment with morphine display morphine dependence. These results indicate that drug dependence seems to be influenced at least partially by mPer1, but mPer1 cannot affect morphine dependence that has been formed.
Acknowledgments
This study was supported by the National Nature Science Foundation of China (No. 39970275; No. 30070288 to Z. Wang; No. 30070198 to C. Wan).
Abbreviations
- CPP
conditioned place preference
- ODN
oligonucleotide
- Per
Period
- ZT
zeitgeber time.
REFERENCES
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc Natl Acad Sci USA. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U. Invited review: regulation of mammalian circadian clock genes. J Appl Physiol. 2002;92:1348–1355. doi: 10.1152/japplphysiol.00759.2001. [DOI] [PubMed] [Google Scholar]
- Andretic R, Chaney S, Hirsh J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science. 1999;285:1066–1068. doi: 10.1126/science.285.5430.1066. [DOI] [PubMed] [Google Scholar]
- Cornish JL, Kalivas PW. Cocaine sensitization and craving: differing roles for dopamine and glutamate in the nucleus accumbens. J Addict Dis. 2001;20:43–54. doi: 10.1300/J069v20n03_05. [DOI] [PubMed] [Google Scholar]
- Eric JN. Common molecular and cellular substrates of addiction and memory. Neurobiol Learn Mem. 2002;78:637–647. doi: 10.1006/nlme.2002.4084. [DOI] [PubMed] [Google Scholar]
- Famhe RG, Khachigian LM. Locked nucleic acid modified DNA enzymes targeting early growth response-1 inhibit human vascular smooth muscle cell growth. Nucl Acids Res. 2004;32:2281–2285. doi: 10.1093/nar/gkh543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci USA. 2003;100:14806–14811. doi: 10.1073/pnas.2036281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaytan O, Lewis C, Swann A, Dafny N. Diurnal differences in amphetamine sensitization. Eur J Pharmacol. 1999;374:1–9. doi: 10.1016/s0014-2999(99)00243-5. [DOI] [PubMed] [Google Scholar]
- Gaytan O, Yang P, Swann A, Dafny N. Diurnal differences in sensitization to methylphenidate. Brain Res. 2000;864:24–39. doi: 10.1016/s0006-8993(00)02117-x. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci (Online) 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh J. Time flies like an arrow: fruit flies like crack? Pharmacogenom J. 2001;1:97–100. doi: 10.1038/sj.tpj.6500020. [DOI] [PubMed] [Google Scholar]
- Khachigian LM, Fahmy RG, Zhang G, Bobryshev YV, Kaniaros A. c-Jun regulates vascular smooth muscle cell growth and neointima formation after arterial injury: inhibition by a novel DNA enzyme targeting c-Jun. J Biol Chem. 2002;277:22985–22991. doi: 10.1074/jbc.M200977200. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Masashi A, Yasuko K, Satomi T, Hisanori W, Takahiro M, Miyuki M, et al. T inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry AM, Swick AG, Romsos DR. Leptin rapidly lowers food intake and elevates metabolic rates in lean and ob/ob mice. J Nutr. 1997;127:2065–2072. doi: 10.1093/jn/127.10.2065. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Akiyama M, Moriya T, Shibata S. Sensitized increase of Period gene expression in the mouse caudate/putamen caused by repeated injection of methamphetamine. Mol Pharmacol. 2001;59:894–900. doi: 10.1124/mol.59.4.894. [DOI] [PubMed] [Google Scholar]
- Ordoukhanian P, Jouce GF. RNA-cleaving DNA enzymes with altered regio- or enantioselectivity. J Am Chem Soc. 2002;124:12499–12506. doi: 10.1021/ja027467p. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Santiago FS, Lowe HC, Kavurma MM, Chesterman CN, Baker A, Atkins DG. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nat Med. 1999;5:1264–1269. doi: 10.1038/15215. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schecter MD. Continued trends in the conditioned place preference literature from 1992 to 1996, inclusive with site indexed. Neurosci Biobehav. 1998;22:827–846. doi: 10.1016/s0149-7634(98)00012-8. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Wess F. The dopamine hypotheses of reward: past and current status. Trends Neurosci. 1999;22:521–527. doi: 10.1016/s0166-2236(99)01447-2. [DOI] [PubMed] [Google Scholar]
- Sriram B, Banerjea AC. In vitro-selected RNA cleaving DNA enzymes from a combinatorial library are potent inhibitors of HIV-1 gene expression. Biochem J. 2000;352:667–673. [PMC free article] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Cheng CL. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Funada M, Narita M, Misawa M, Nagase, Suzuki H. Morphine-induced place preference in the CXBK mouse: characteristics of µopioid receptor subtypes. Brain Res. 1993;602:45–52. doi: 10.1016/0006-8993(93)90239-j. [DOI] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi H, Fukuhara C, Ozawa R, Hirose M. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM. Measuring reward with the conditioned place preference paradigm: a comprehensive review of drug effects, recent progress and new issues. Prog Neurobiol. 1998;56:613–672. doi: 10.1016/s0301-0082(98)00060-4. [DOI] [PubMed] [Google Scholar]
- Unwalla H, Banerjea AC. Inhibition of HIV-1 gene expression by novel macrophage-tropic DNA enzymes targeted to cleave HIV-1 TAT/Rev RNA. Biochem J. 2001;357:147–155. doi: 10.1042/0264-6021:3570147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren LJ, Kalivas PW. Alterations in dopaminergic and glutamatergic transmission in the induction and expression of behavioral sensitization: a critical review of preclinical studies. Psychopharmacology. 2000;151:99–120. doi: 10.1007/s002130000493. [DOI] [PubMed] [Google Scholar]
- Wager-Smith K, Kay SA. Circadian rhythm genetics: from flies to mice to humans. Nat Genet. 2000;26:23–27. doi: 10.1038/79134. [DOI] [PubMed] [Google Scholar]
- Wu Y, Yu L, McMahon R, Rossi JJ, Forman SJ, Snyder DS. Inhibition of bcr-abl oncogene expression by novel deoxyribozymes (DNAzymes) Hum Gene Ther. 1999;10:2847–2857. doi: 10.1089/10430349950016573. [DOI] [PubMed] [Google Scholar]
- Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]


