Abstract
Membrane lipids act as important regulators of a litany of important physiological and pathophysiological events. Many of them act as site-specific ligands for cytosolic proteins in binding events that recruit receptors to the cell surface and control both protein function and subcellular localization. Phosphatidylinositol phosphates (PIPns) are a family of signaling lipids that regulate numerous cellular processes by interacting with a myriad of protein binding modules. Characterization of PIPn-binding proteins has been hampered by the lack of a rapid and convenient quantitative assay. Herein, microplate-based detection is presented as an effective approach to characterizing protein-PIPn binding interactions at the molecular level. With this assay, the binding of proteins to isolated PIPn headgroups is detected with high sensitivity using a platform that is amenable to high-throughput screening.
In the studies described herein, biotinylated PI-(4,5)-P2 headgroup analogue 1 was designed, synthesized and immobilized onto 96-well streptavidin-coated microplates to study receptor binding. This assay was used to characterize the binding of the PH domain of β-spectrin to this headgroup. The high affinity interaction that was detected for surface association (Kd, surf = 6 nM ±3), demonstrates that receptor binding modules can form high affinity interactions with lipid headgroups outside of a membrane environment. The results also indicate the feasibility of the assay for rapid characterization of PIPn-binding proteins as well as the promise for high-throughput analysis of protein-PIPn binding interactions. Finally, this assay was also employed to characterize the inhibition of the binding of receptors to the PIPn-derivatized microplates using solution phase competitors. This showcases the viability of this assay for rapid screening of inhibitors of PIPn-binding proteins.
INTRODUCTION
Lipids present in cellular membranes act as key regulators of important biological processes. A primary role of these species is to act as site-specific ligands in molecular recognition events that result in the recruitment of cytosolic effector proteins to the membrane surface, a process that generally regulates both protein function and subcellular localization (1–3). Protein–lipid binding interactions lie at the heart of many of the most important cellular pathways, which is likely due to the ability to transmit information from the membrane into cellular signaling cascades.
One family of phospholipids involved in many crucial biological events consists of phosphate derivatives of phosphatidylinositol, collectively known as phosphoinositides (PIPns) (3–7). The study of PIPns is vital as these lipids have been heavily implicated in several high-profile pathophysiological events including the onset of cancer, leukemia and diabetes (8). A prominent example involves the conversion of phosphatidylinositol-4,5-bisphosphate (PI-(4,5)-P2) to phosphatidylinositol-3,4,5-trisphosphate, with the forward and reverse reactions catalyzed by phosphoinositide 3-kinase (PI-3K) (9) and phosphatase PTEN (10), respectively. Both PI-3K (11) and the tumor suppressor PTEN (12) are among the most frequently mutated enzymes in tumorigenesis.
The PIPn family consists of seven isomeric phosphodiester-linked myo-inositol headgroups containing every possible combination of phosphorylation at the 3-, 4- and 5-positions. Due to the structural similarity and yet functional diversity of these compounds, their study generally requires a synthetic approach to obtain homogeneous samples of the natural lipid or analogues thereof. However, the synthesis of PIPns represents a significant barrier to these efforts. In addition to this synthetic difficulty, the study of PIPn-effector protein binding events generally faces additional challenges. For example, the complexity of the membrane environment complicates binding characterization due to its inherent heterogeneity.
An important question pertaining to protein–PIPn binding is what structural features are required for membrane binding to occur, as some proteins show strong binding to the lipid headgroup alone, while others require the full membrane environment for interaction (4, 6). In particular, crystal structures of certain pleckstrin homology (PH) domains indicate a shallow binding pocket, which, combined with other evidence, indicates predominantly headgroup-driven association (3). However, instances of PH domain membrane-insertion are documented (13, 14), and other domains (phox homology (PX) and Fab1, YOTB, Vac1, EEA1 (FYVE)) require the membrane for lipid headgroup binding. This disparity makes it necessary to determine the headgroup-binding affinities of PIPn-binding proteins to understand how PIPns drive the specific membrane recruitment of diverse effectors for cell regulation.
A rapidly growing number of cellular proteins are known to bind PIPns. Many of these proteins contain specialized PIPn-binding modules, such as the PH, PX, FYVE, and the more recently discovered β-propellers that bind PIPn (PROPPIN) domains (3). Among the large number of reported PIPn-binding modules, PIPn affinities and specificities are known to vary widely, requiring the careful investigation of each distinct receptor with each PIPn isomer. Such studies are hampered by the diversity of domain families, exemplified by the PH domains, for which recent sequence homology searches have implicated over 250 family members in humans (3), many of which remain uncharacterized. Finally, proteins exist that interact with PIPns but lack a well-defined PIPn-binding site. Characterization of these important groups of proteins would be advanced by a rapid and reliable quantitative assay for determining the affinity and specificity for each PIPn isomer. Herein, we present a new microplate-based quantitative assay for PIPn binding which can be applied to large scale high-throughput screening.
EXPERIMENTAL PROCEDURES
General Experimental Details
Dry solvents were obtained from a Pure Solv solvent delivery system purchased from Innovative Technology, Inc. Column chromatography was performed using 230 – 400 mesh silica gel purchased from Sorbent Technologies. NMR spectra were obtained using a Bruker AC250 spectrometer updated with a TecMag data collection system, a Varian Mercury 300 spectrometer, and a Bruker Avance 400 spectrometer. Mass spectra were obtained with JEOL DART-AccuTOF spectrometer with high resolution capabilities and an ABI Voyager DE Pro MALDI spectrometer. HPLC data was obtained using an HP series 1100 HPLC with an Alltech Lichrosphere SI 60 50U column with HPLC grade solvents purchased from Fisher. Optical rotation values were obtained using a Perkin-Elmer 241 polarimeter. Microplate-based chemiluminescence measurements were performed using a BioTek Synergy 2 multi-detection microplate reader. White reacti-bind streptavidin high binding capacity (HBC) coated 96-well microplates for chemiluminescence studies were purchased from Pierce Biotechnology (Rockford, IL). Goat polyclonal anti-GST tag HRP-conjugate was purchased from Novus Biologicals (Littleton, CO). Supersignal ELISA femto maximum sensitivity substrate was purchased from Pierce Biotechnology (Rockford, IL). The GST-tagged PH domain of β-spectrin was expressed and purified as previously described.(15)
1D-4,5-O-Bisbenzyl-1-O-(tert-butyldiphenylsilyl-2,3,6-O-tris-(benzyloxymethyl)-myo-inositol (3)
To a solution of 2 (0.586 g, 0.96 mmol) in 1,2-dichloroethane (21 mL) were added diisopropylethylamine (1.7 mL, 9.83 mmol) and benzyloxychloromethyl ether (1.4 mL, 10.1 mmol). The reaction mixture was stirred at reflux for 24 h. Diisopropylethylamine (1.7 mL, 9.83 mmol) and benzyloxychloromethyl ether (1.4 mL, 10.1 mmol) were added to the solution, and the mixture was stirred at reflux for another 24 h. After the reaction was finished, water was added and the crude reaction mixture was extracted with methylene chloride (2 × 50 mL). The organic phase was dried with magnesium sulfate, filtered and then concentrated. The crude was then purified by chromatography on silica gel (acetone/hexane 1:5) giving 3 (0.631 g, 68%) as a syrup.
[α]D296K 25.8 (c = 0.95, CHCl3); 1H NMR (400 MHz, CDCl3) δ 7.88 (dd, J = 13.6, 7.3 Hz, 4H), 7.75 (t, J = 7.6 Hz, 4H), 7.42-7.14 (m, 23H), 6.94-6.89 (m, 4H), 5.92 (t, J = 10.1 Hz, 1H), 5.50 (t, J = 9.7 Hz, 1H), 5.05 (d, J = 6.8 Hz, 1H), 4.86-4.80 (m, 3H), 4.73 (d, J = 6.8 Hz, 1H), 4.55-4.45 (m, 2H), 4.33-4.02 (m, 7H), 3.67 (dd, J = 10.3, 2.0 Hz, 1H), 3.58 (m, 1H), 1.11 (s, 9H); 13C NMR (100.5 MHz, CDCl3) δ 166.0, 165.6, 138.2, 137.7, 137.4, 136.0, 135.9, 133.8, 132.8, 130.2, 130.0, 129.7, 129.6, 128.2, 128.1, 127.9, 127.7, 127.4, 127.3, 96.6, 95.5, 91.8, 77.7, 74.9, 74.5, 73.0, 72.8, 72.2, 69.9, 69.4, 68.9, 27.3, 27.2, 19.3. MALDI-HRMS [M + Na]+ calcd for C60H62O11SiNa 1009.3959, found 1009.3936.
1D-1-O-(tert-butyldiphenylsilyl-2,3,6-O-tris-(benzyloxymethyl)-myo-inositol (4)
A solution of 3 (0.631 g, 0.650 mmol) in methanol (40 mL) was treated with sodium methoxide (1.6 g, 29.6 mmol) at room temperature for 18 h. Then sodium methoxide (1.0 g, 18.5 mmol) was added to the reaction solution and the mixture was stirred at room temperature for 3 h, after which the contents were filtered. The resulting residue was dissolved in water (50 mL), and the aqueous solution was extracted with methylene chloride (2 × 50 mL). In addition, the previous filtrate was concentrated in vacuum and then extracted with methylene chloride (2 × 50 mL) from water (50 mL). The combined organic layer was then dried with magnesium sulfate, filtered, concentrated and purified by chromatography on silica gel (hexanes/acetone, 3:1) to provide pure diol 4 (0.393 g, 80%) as a syrup.
[α]D296K 86.1 (c = 1.36, CHCl3); 1H NMR (250 MHz, CDCl3) δ 7.68 (t, J = 7.3 Hz, 4H), 7.41-7.19 (m, 21H), 5.02 (d, J = 6.8 Hz, 1H), 4.90-4.83 (m, 2H), 4.73-4.45 (m, 8H), 4.34-4.25 (m, 2H), 3.99-3.91 (m, 2H), 3.77-3.75 (m, 2H), 3.40 (dd, J = 9.6, 1.8 Hz, 1H), 3.13-3.10 (m, 1H), 2.78 (m, 1H), 1.03 (s, 9H);13C NMR (62.9 MHz, CDCl3) δ 138.2, 135.9, 135.6, 134.1, 133.4, 129.9, 129.5, 128.5, 128.4, 128.3, 128.02, 127.98, 127.72, 127.66, 127.5, 127.4, 95.5, 93.5, 85.2, 76.3, 75.7, 74.2, 73.2, 72.2, 70.3, 69.2, 27.0, 19.4. HRMS [M + O2]− calcd for C46H54O11Si 810.34354, found 810.34393.
1D-1-O-(tert-butyldiphenylsilyl-2,3,6-O-tris-(benzyloxymethyl)-myo-inositol 4,5-bis-(dibenzyl phosphate) (5)
To a solution of diol 4 (0.73 g, 0.958 mmol) and 1-H tetrazole (17 mL, 7.67 mmol, 0.45 M in acetonitrile) in methylene chloride (30 mL) was added dibenzyl N,N-diisopropylphosphoramidite (1.93 mL, 5.75 mmol) under nitrogen gas, and the mixture was stirred at room temperature for 18 h. The reaction solution was cooled to − 60 °C, and m-CPBA (2.2 g, 57~86%) was added. The mixture was stirred at this temperature for 60 min and then slowly warmed to room temperature after removal of the cooling bath. The solution was diluted to 180 mL with methylene chloride, washed with saturated aqueous sodium bicarbonate (2 × 100 mL), and the aqueous phase was extracted with methylene chloride (100 mL). The combined organic phase was dried with magnesium sulfate, filtered, concentrated, and the residue was purified by chromatography on silica gel (hexanes/acetone, 3:1) giving bis-phosphodiester 5 (1.17 g, 81%) as a syrup.
[α]D296K −2.5 (c = 0.805, CHCl3); 1H NMR (400 MHz, CDCl3) δ 7.68–7.78 (m, 4H), 7.03–7.35 (m, 41H), 4.93–5.22 (m, 12H), 4.77–4.80 (m, 3H), 4.59 (d, J = 12.0 Hz, 1H), 4.39–4.47 (m, 3H), 4.28–4.32 (m, 2H), 4.16 (d, J = 6.8 Hz, 1H), 4.05 (d, J = 12.4 Hz, 1H), 3.95 (d, J = 7.2 Hz, 1H), 3.49 (s, 1H), 3.39 (dd, J = 10.0, 1.6 Hz, 1H), 1.11 (s, 9H); 13C NMR (100.5 MHz, CDCl3) δ 133.7, 132.6, 130.1, 130.0, 128.5, 128.4, 128.3, 128.2, 128.2, 128.0, 128.0, 127.9, 127.8, 127.6, 127.5, 127.3, 96.6, 95.5, 93.2, 78.4, 77.8, 75.2, 75.1, 74.0, 73.7, 70.7, 69.5, 69.5, 69.4, 69.3, 69.1, 69.1, 27.3, 19.3; 31P NMR (161.8 MHz, CDCl3) δ −0.83 (s, 1P), −1.21 (m, 1P). MALDI-HRMS [M + Na]+ calcd for C74H80O15P2SiNa 1321.4634, found 1321.4602.
1D-2,3,6-O-tris-(benzyloxymethyl)-myo-inositol 4,5-bis-(dibenzyl phosphate) (6)
A solution of fully protected inositol 5 (1.17 g, 0.90 mmol) in N,N-dimethylformamide (30 mL) was treated with tetrabutylammonium fluoride trihydrate (0.90 g, 2.85 mmol) at room temperature for 12 h. After the reaction was finished, the solution was partitioned between ethyl acetate (200 mL) and H2O (150 mL). The aqueous layer was further extracted with ethyl acetate (100 mL) and the combined organic layer was washed with brine (150 mL), dried with magnesium sulfate and filtered. Following concentration, the residue was purified by chromatography on silica gel (hexanes/acetone, 2:1) to give alcohol 6 (0.854 g, 88%) as a syrup.
[α]D296K −36.3 (c = 1.0, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.14–7.35 (m, 35H), 4.92–5.19 (m, 11H), 4.61–4.79 (m, 6H), 4.39–4.54 (m, 4H), 4.29 (s, 1H), 4.24 (d, J = 2.1 Hz, 1H), 3.88 (t, J = 9.3 Hz, 1H), 3.72 (d, J = 10.2 Hz, 1H), 3.55 (d, J = 9.3 Hz, 1H); 13C NMR (100.5 MHz, CDCl3) δ 138.0, 137.7, 136.9, 136.3, 136.2, 136.2, 136.1, 136.0, 128.7, 128.6, 128.5, 128.5, 128.4, 128.4, 128.3, 128.2, 128.1, 128.1, 128.0, 128.0, 127.9, 127.8, 127.7, 127.7, 97.3, 95.8, 95.0, 83.5, 79.5, 79.5, 78.1, 78.0, 75.8, 75.3, 70.7, 70.6, 69.9, 69.7, 69.6, 69.6, 69.6, 69.4, 69.4, 69.3; 31P NMR (161.8 MHz, CDCl3) δ −1.13 (s, 1P), −1.50 (s, 1P). MALDI-HRMS [M + Na]+ calcd for C58H62O15P2Na 1083.3456, found 1083.3410.
Benzyl (6-benzyloxycarbonylaminohexyl) 1D-2,3,6-O-tris-(benzyloxymethyl)-myo-inosit-1-yl phosphate 4,5-bis-(dibenzyl phosphate) (7)
To a solution of alcohol 6 (0.854 g, 0.805 mmol) and 1-H tetrazole (5.3 mL, 2.4 mmol, 0.45 M in acetonitrile) in methylene chloride (36 mL) was added dibenzyl diisopropylphosphoramidite (0.983 g, 2.0 mmol) under nitrogen, and the mixture was stirred at room temperature for 18 h. The reaction solution was cooled to − 60 °C, and m-CPBA (0.7 g, 57~86%) was added. The mixture was stirred at this temperature for 60 min and was then slowly warmed to room temperature after removal of the cooling bath. The solution was diluted to 100 mL with methylene chloride, washed with saturated aqueous sodium bicarbonate (2 × 60 mL), and the aqueous phase was extracted with methylene chloride (60 mL). The combined organic phase was dried with magnesium sulfate, filtered, concentrated and the residue was purified by chromatography on silica gel (hexanes/acetone, 3:1) to give tris-phosphotriester 7 (0.53 g, 45%) as a syrup.
[α]D296K −22.9 (c = 1.0, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.16–7.34 (m, 45H), 4.41–5.20 (m, 28H), 4.20–4.34 (m, 2H), 3.88–3.95 (m, 2H), 3.71 (d, J = 9.0 Hz, 1H), 3.07–3.14 (m, 2H), 1.20–1.48 (m, 8H); 13C NMR (100.5 MHz, CDCl3) δ 156.4, 138.2, 137.9, 137.5, 136.7, 136.2, 136.1, 136.0, 135.9, 135.8, 135.8, 128.6, 128.4, 128.4, 128.4, 128.3, 128.2, 128.2, 128.0, 128.0, 127.9, 127.8, 127.7, 127.7, 127.6, 127.5, 127.4, 96.6, 95.6, 94.6, 78.8, 76.4, 74.9, 74.7, 74.6, 74.5, 70.4, 70.4, 69.7, 69.7, 69.6, 69.5, 69.4, 69.3, 69.3, 68.1, 67.9, 66.5, 40.8, 30.0, 29.9, 29.7, 26.0, 24.9; 31P NMR (161.8 MHz, CDCl3) δ −1.19 (m, 3P). MALDI-HRMS [M + Na]+ calcd for C79H88NO20P3Na 1486.5005, found 1486.5027.
1-(6-aminohexyl sodium phosphate)-1D-myo-inositol-4,5-bis-(disodium phosphate) (8)
To a solution of compound 7 (0.57 g, 0.389 mmol) in methanol (50 mL) was added 20% palladium hydroxide on charcoal (0.67 g). The mixture was then stirred for 3 days at room temperature with 1 atm of hydrogen (balloon). Next, the catalyst was removed by filtration and the residue was with washed with methanol. The solvent was removed in vacuum, and the crude product was dissolved in water and stirred for 3 h with Chelex 100 resin (Sigma, Na+ form). The resin was removed by filtration, and the filtrate was lyophilized to give amino-conjugate 8 (0.245 g, 100%) as white solid.
[α]D296K −14.9 (c = 1.0, H2O); 1H NMR (300 MHz, D2O) δ 4.19–4.24 (m, 2H), 3.88–4.00 (m, 5H), 3.69 (dd, J = 9.6, 2.7 Hz, 1H), 2.99 (d, J = 7.8 Hz, 2H), 1.63–1.70 (m, 4H), 1.40–1.43 (m, 4H); 13C NMR (100.5 MHz, D2O) δ 80.0, 78.6, 78.0, 74.2, 74.1, 73.3, 68.9, 42.1, 32.0, 29.2, 27.7, 27.0; 31P NMR (121.5 MHz, D2O) δ 5.20 (s, 1P), 4.78 (s, 1P), 0.92 (s, 1P). MALDI-HRMS (free acid, [M + H]+) calcd for C12H28NO15P3H 520.0745, found 520.0729.
1-(6-(14-biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 4,5-bis-(disodium phosphate) 1
Succinimidyl ester 9 was dissolved in dimethylformamide (1 mL) and compound 8 (7 mg, 0.011 mmol), dissolved in TEAB (0.5 M, 1 mL), was added. The mixture was stirred at room temperature for 24 h. The solvent was removed under reduced pressure, and the residue was washed with acetone (4 × 2 mL) and then dried in vacuo. The resulting white solid was dissolved in water and the solution was stirred for 3 h with Chelex 100 resin (Sigma, Na+ form). The resin was removed by filtration, and the filtrate was lyophilized to give the product (13 mg, 100%) as white solid.
1H NMR (300 MHz, D2O) δ 4.56–4.58 (m, 1H), 4.38–4.41 (m, 1H), 4.04–4.25 (m, 3H), 3.82–3.95 (m, 4H), 3.58–3.74 (m, 14H), 2.93–3.40 (m, 14H), 2.72–2.85 (m, 2H), 2.24 (d, J = 6.9 Hz, 2H), 1.33–1.79 (m, 16H); 31P NMR (121.5 MHz, D2O) δ 5.79 (s, 1P), 5.70 (s, 1P), 0.87 (s, 1P). MALDI-HRMS (free acid, [M + H]+) calcd for C32H61N4O22P3SH 979.2784, found 979.2775.
Microplate analysis of PI-(4,5)-P2 headgroup–β-spectrin PH domain binding
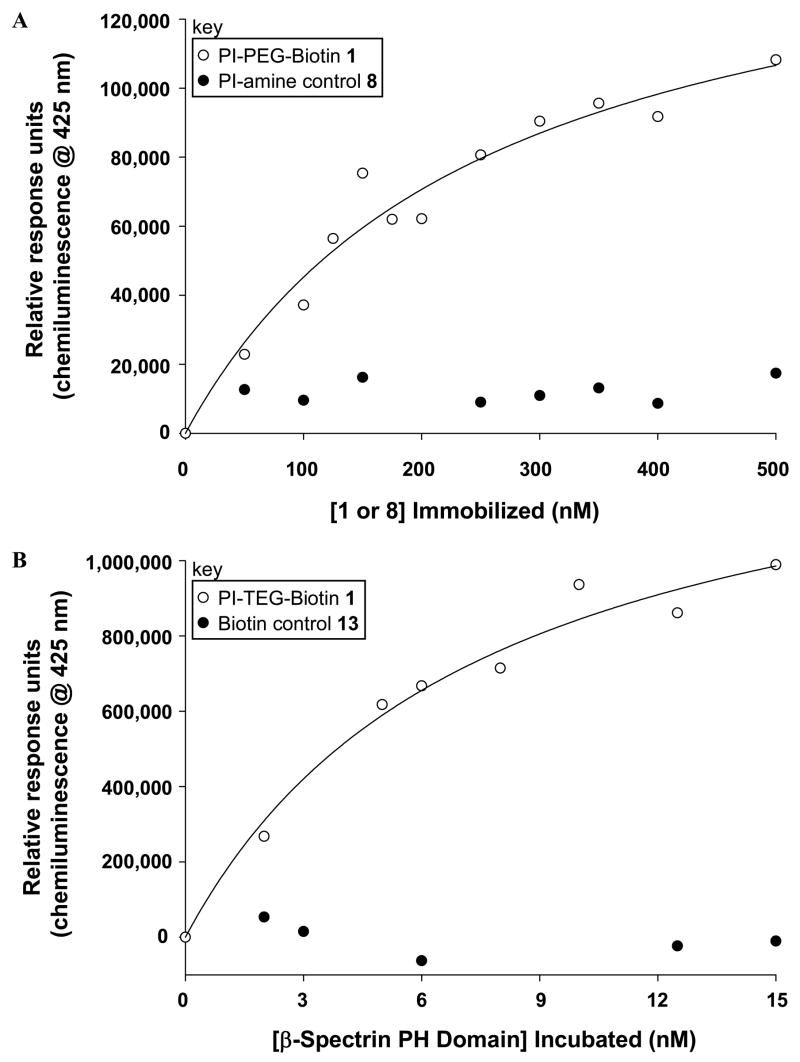
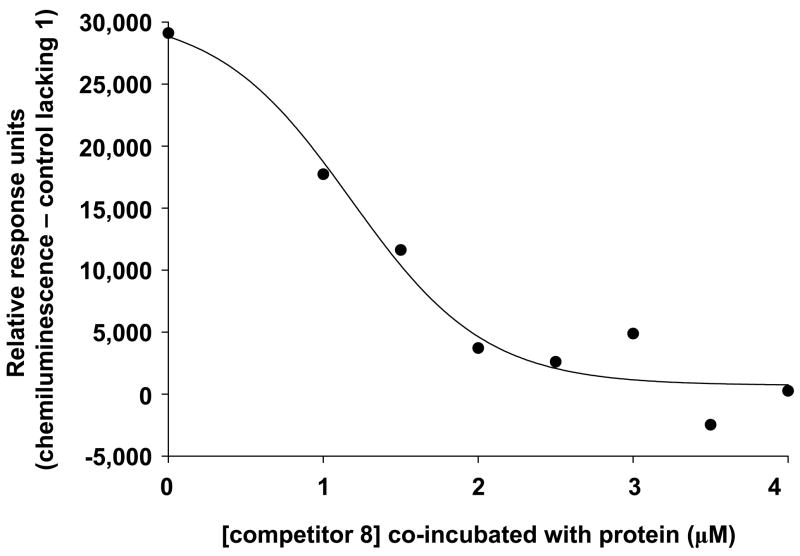
Binding assays were performed using streptavidin-coated 96-well microplates. The wash buffer that was used was 20 mM Tris buffer pH 8.0 with 0.05% tween as a blocking additive to avoid non-specific surface adsorption. In general, the wells that were to be used for analysis were washed with 200 μL of wash buffer for 30 minutes. This was removed, followed by addition of the appropriate solutions of biotin-TEG-PI-(4,5)-P2 ligand 1 or control 8 or 13 in water, and incubation for one hour. Next, these solutions were removed and the wells were washed with 3 × 250 μL wash buffer. Following this, 200 μL of GST-tagged β-spectrin PH domain solutions in wash buffer were added to each well. After a one hour incubation, the protein solution was removed and the wells were washed with 3 × 250 μL wash buffer. Next, 200 μL of a solution of goat polyclonal anti-GST tag HRP-conjugate was added to each well and allowed to incubate for one hour. This solution was then removed and the wells were washed with 3 × 250 μL wash buffer. Chemiluminescence detection of bound antibody was next performed using supersignal ELISA femto maximum sensitivity substrate. Here, a 1:1 mixture of the substrate and peroxide solutions were mixed, 100 μL of which was added to each well. The microplate was then immediately placed in the microplate reader and the chemiluminescence at 425 nm was repeatedly measured for ~ 5 minutes. Several readings that provided optimal signal were then averaged and the change in chemiluminescence was plotted verses the concentration of the varying component. The resulting curve was fit via the program SigmaPlot using the Langmuir isotherm: C = Cmax*[P]/(Kd, surf + [P]) (C = Chemiluminescence, P = Protein) to calculate dissociation constants. For Figure 1A, solutions of 1 varied from 0–500 nM, β-Spectrin PH domain added was fixed at 100 ng/mL and anti-GST Ab was added at 10 ng/mL. For Figure 1B, [1] immobilized was fixed at 500 nM, β-Spectrin PH domain solutions ranged from 0–15 nM and the anti-GST Ab solution was 100 ng/mL. For Figure 2, [1] immobilized was fixed at 500 nM, solutions of fixed β-Spectrin PH domain (200 ng/mL) and varying competitor 8 (0–4 μM) were used, and anti-GST Ab solution was 10 ng/mL.
Figure 1.
Microplate assay results obtained with A. varying concentration of ligand 1 with fixed [receptor] and B. fixed [1] with changing [receptor]. Both results indicate sensitive detection of headgroup binding.
Figure 2.
Inhibition of protein binding to the ligand-functionalized surface using solution-phase competitor 8.
RESULTS AND DISCUSSION
Due to the importance of the PIPns in cell regulation and the diversity of proteins that target these lipids in terms of structure and the details of recognition, it is imperative to develop efficient approaches for characterizing PIPn recognition at the molecular level. Current assays for detecting interactions utilize lipid-functionalized nitrocellulose membranes (such as PIP strips™, Echelon Biosciences, Salt Lake City, UT), vesicle pelleting, fluorescence, isothermal titration calorimetry (ITC), or SPR-based methods (16). However, these methods do not allow for high-throughput quantification, which is required to elucidate the grand scope of complexities associated with receptor–PIPn binding.
Microarray analysis has proven highly effective for high-throughput quantification of protein-ligand binding, including cell surface receptor–ligand binding interactions. Generally, optical detection is utilized, which benefits from high sensitivity and easy access to studies. Another advantage is that the surface-display of target ligands allows for the formation of multivalent binding interactions, which are common in membrane-based biological recognition events (17, 18), and are known to occur with certain PIPn-targeting proteins (4). Detection in a multivalent context is important as ligand preferences of receptors can differ between solution-phase studies and those using a multivalent surface (19, 20). In microarray analysis, ligand density can be directly controlled via the amount of ligand that is deposited on the surface. In fact, binding affinities have been found to vary depending upon the concentration of ligand immobilized in microarray analysis, indicating the occurrence of multivalent interactions (21, 22). The described advantages have been exhibited in the development of DNA (23, 24), protein and peptide (25–27), and carbohydrate (20, 22, 28–34) microarray systems, while lipid microarrays have only recently emerged (21, 35).
Towards our goal of a high-throughput microarray platform for efficient characterization of PIPn-binding, we have initially devised a microplate-based detection system. The first task in this pursuit was the development of a ligand-immobilization strategy, for which synthetic biotinylated PI-(4,5)-P2 headgroup analogue 1 (Scheme 1) was designed. PI-(4,5)-P2 was chosen for initial studies as it is the most abundant member of the PIPn family. The isolated headgroup motif was employed to directly investigate the extent to which effector proteins interact with this moiety outside of the membrane environment. Ligand 1 also contains a biotin group for immobilization onto streptavidin surfaces, as this high affinity non-covalent interaction enforces quantitative immobilization while avoiding potential non-specific surface attachment. Finally, a tetraethylene glycol (TEG) spacer was introduced between the headgroup and biotin to distance the ligand from the microplate surface.
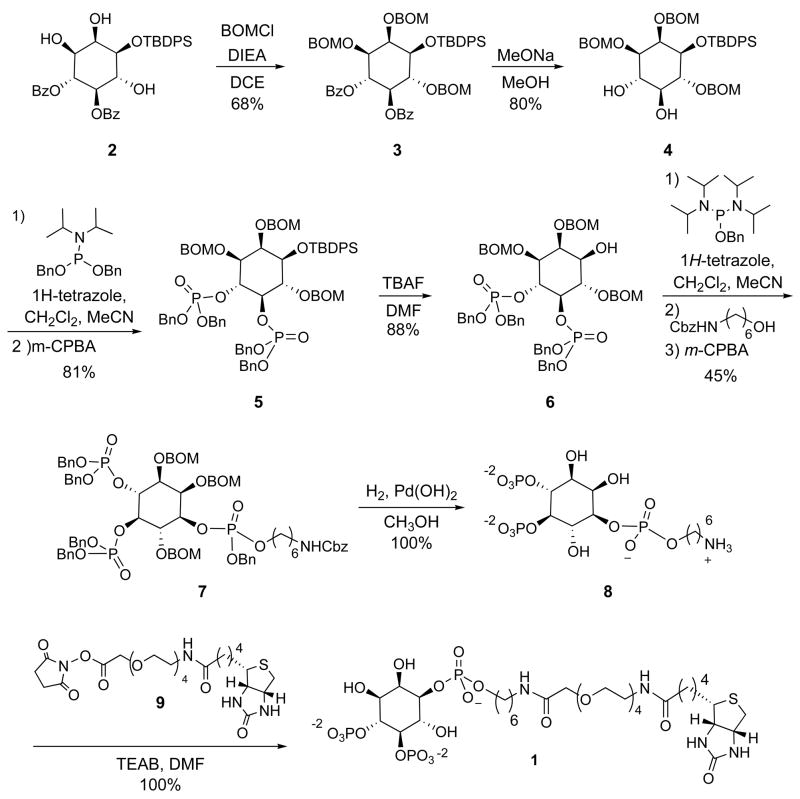
Scheme 1.
The synthesis of PI-(4,5)-P2-TEG-Biotin conjugate 1 benefited from protocols previously developed for accessing inositol headgroups (36–38), and amino-functionalized tetherable analogues thereof (39–42). Specifically, the synthetic route to protected PI-(4,5)-P2 headgroup 6 (Scheme 1) employed a procedure modified from that reported by Bruzik and co-workers (38). First, compound 2 was obtained in four steps from myo-inositol as previously described. This contains benzoyl groups at positions 4 and 5, the phosphorylation sites in the final compound, and a silyl group at the 1 position, where the phosphodiester linkage occurs in the natural lipid. At this point, we accessed compound 3 by installing benzyloxymethyl (BOM) groups at the 2, 3 and 6 positions to allow for convenient global deprotection via hydrogenolysis at the end of the synthesis. After benzoyl group removal to 4, phosphoramidite chemistry was used to access fully protected headgroup analogue 5. Following silyl deprotection to 6, a protected amine group attached via a phosphodiester linkage was introduced to generate 7. Global deprotection by hydrogenolysis then yielded conveniently functionalizable PI-(4,5)-P2-amine 8. Finally, coupling of 8 with biotin-TEG-NHS ester reagent 9 (synthesized in seven steps from TEG, see supporting information for details) provided PI-(4,5)-P2-TEG-Biotin conjugate 1.
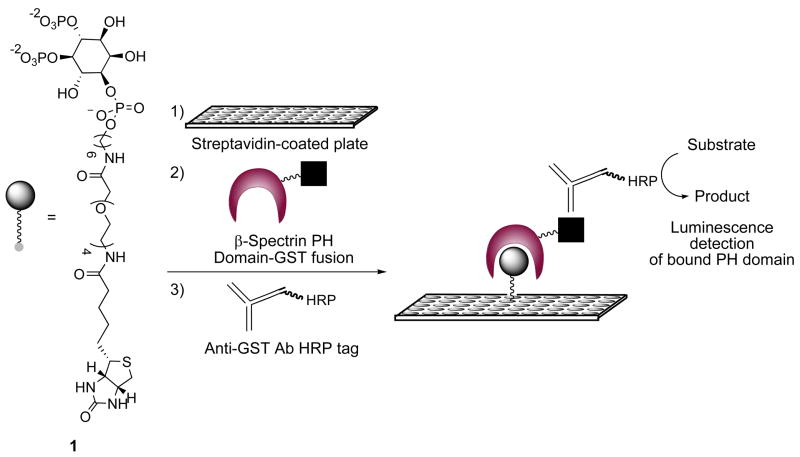
The experimental approach to PIPn microplate analysis using ligand 1 employed a surface-based antibody assay as depicted in Scheme 2. First, PI-(4,5)-P2 headgroup immobilization was achieved by incubating conjugate 1 in different wells of a streptavidin-coated 96 well microplate, followed by thorough buffer washes. Next, the ligand was incubated with the effector protein. For our initial study, we sought to characterize the PI-(4,5)-P2 headgroup-binding of the PH domain of the protein β-spectrin. To do so, we expressed and purified this domain with an appended glutathione-S-transferase (GST) tag, used for detection and purification purposes, as previously described (15). Following buffer washes to remove unbound protein, a horseradish peroxidase (HRP)-conjugated anti-GST antibody was used, followed by further washing. Finally, chemiluminescence-based detection of the secondary antibody using a microplate reader was performed to transduce the presence of bound protein.
Scheme 2.
Microplate-based assay for detection of receptor-PIPn head group binding interactions
The described microplate assay was successfully employed to probe PI-(4,5)-P2 headgroup–β-spectrin PH domain association via multiple approaches. First, analysis was performed by introducing varying concentrations of ligand 1 into the plate and incubating with fixed protein and antibody concentrations. A representative binding curve indicating enhanced protein recruitment upon increasing immobilized ligand is shown in Figure 1A. The results show highly sensitive detection of binding, as the rise in the curve corresponds to less than 50 pmoles of ligand immobilized in the plate. Also shown in Figure 1A is a control study in which PI-amine compound 8, which lacks the biotin moiety, was incubated in place of 1. This resulted in background signal, which indicates that the biotin group of 1 is required for surface deposition and rules out potential non-specific ligand and protein adsorption onto the plate.
The microplate binding assay was also carried out by depositing a fixed concentration of ligand 1 that correlates to strong protein recruitment from the previous study (500 nM) and varying the receptor concentration, for which results are shown in Figure 1B. Once again, sensitive detection of binding was obtained. Here, results were used to calculate a dissociation constant (Kd, surf) value by fitting the data to the Langmuir isotherm (22), with multiple readings yielding an average value of 6 nM ±3. Figure 1B also includes a second control in which biotin-TEG compound 13 (see supporting information for structure), which lacks the PI-(4,5)-P2 headgroup, was immobilized in place of ligand 1, again resulting in no signal. The combined results for the controls in Figures 1A and 1B show that both the PI-(4,5)-P2 headgroup and biotin moieties of compound 1 are necessary for protein binding.
The binding affinity of the GST-tagged PH domain of β-spectrin interacting with liposomes containing full PI-(4,5)-P2 lipids was previously determined (Kd = 125 nM ± 18) using a surface plasmon resonance assay (15). While the value reported herein differs from this previous study, this is to be expected since the current assay was designed to detect binding in a different environment, that in which the interaction with the lipid headgroup is studied outside of the membrane. As a result, the two assays utilize different formats, and thus each Kd value is specific to the model system employed for analysis. For example, the presentation and clustering of the PI-(4,5)-P2 headgroup will vary between microplate surfaces that are directly derivatized with the biotinylated ligand (current assay) compared with studies employing the full lipid incorporated into immobilized liposomes (previous assay). For this reason, we have qualified the dissociation constant measured herein as Kd, surf, as has been indicated in similar approaches employing surface-based binding measurements (22).
The results from this assay indicate that receptor binding domains can form high affinity interactions with PIPn headgroups outside of the membrane environment. The strong binding affinity that was observed using the isolated headgroup provides evidence that PI-(4,5)-P2-binding is primarily driven by interactions with the headgroup in the case of this particular receptor. These results are in line with previous reports in which strong interactions (Kd = 27.3 nM) between other PH domains and solubilized PIPn headgroup analogues were observed, and the fact that these soluble headgroups effectively displaced receptor–full PIPn lipid complexes (43). However, different PIPn-binding receptors are known to require the membrane context for binding (4, 6), indicating that the headgroup is not sufficient to enforce association. This diversity in receptor binding mode dictates that each protein must be analyzed independently to determine the molecular-level details of each particular receptor. Therefore, deconvolution of the complexities of receptor–PIPn affinities, specificities, and binding modes requires a high-throughput assay such as the approach described herein.
In order to extend the described microplate-based strategy, we sought to perform competitive assays to detect perturbation of protein-binding to the ligand-functionalized microplate surface. Here, microplate wells functionalized with PI-(4,5)-P2-TEG-Biotin conjugate 1 at a fixed concentration were simultaneously incubated with β-spectrin PH domain and a varying amount of soluble PI-(4,5)-P2-amine headgroup analogue 8. Figure 2 shows a resulting dose-dependant response curve, in which the extent of protein bound to the surface is diminished through incubation with increasing concentrations of competitive binding partner 8. These results further confirm the strong association of receptors with the PI-(4,5)-P2 headgroup outside of the membrane context and showcase the efficacy of our microplate assay for detecting the inhibition of receptor-PIPn interactions.
The described microplate-based approach provides highly sensitive detection of PIPn-binding as well as competitive inhibition of recognition. As such, this assay is applicable to a range of studies aimed at characterizing binding at the molecular level, including the determination of PIPn-binding specificities of receptors, the extent to which headgroup binding drives association with different proteins, and the evaluation of compounds that inhibit binding. Currently, we are extending these studies towards our goal of a high-throughput microarray assay for systematic analysis of the headgroup-binding affinities and specificities of the diverse effector domains and proteins that interact with the PIPns. The eventual goal in this regard is the implementation of biotinylated headgroup analogues of all naturally occurring PIPn isomers for simultaneous microarray analysis of receptors of interest.
Supplementary Material
Further synthetic procedures and characterizations are included. This information is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
MDB acknowledges the University of Tennessee for support and WC acknowledges funding from the National Institutes of Health (GM68849).
LITERATURE CITED
- 1.Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Ann Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH. Membrane binding domains. Biochim Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 4.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin S, Wang JY, Gambhir A, Murray D. PIP2 and proteins: Interactions, organization, and information flow. Annu Rev Biophys Biomolec Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 6.Prestwich GD. Phosphoinositide signaling: From affinity probes to pharmaceutical targets. Chem Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 8.Pendaries C, Tronchere H, Plantavid M, Payrastre B. Phosphoinositide signaling disorders in human diseases. FEBS Lett. 2003;546:25–31. doi: 10.1016/s0014-5793(03)00437-x. [DOI] [PubMed] [Google Scholar]
- 9.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: Implications for development, immunity, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 10.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 11.Samuels Y, Wang ZH, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell DM, Riggins GJ, Willson JKV, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554–554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 12.Chow LML, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 13.Flesch FM, Yu JW, Lemmon MA, Burger KN. Membrane activity of the phospholipase C-delta(1) pleckstrin homology (PH) domain. Biochem J. 2005;389:435–441. doi: 10.1042/BJ20041721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 15.Das A, Base C, Manna D, Cho W, Dubreuil RR. Unexpected complexity in the mechanisms that target assembly of the spectrin cytoskeleton. J Biol Chem. 2008;283:12643–12653. doi: 10.1074/jbc.M800094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho WW, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- 17.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem, Int Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angew Chem, Int Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horan N, Yan L, Isobe H, Whitesides GM, Kahne D. Nonstatistical binding of a protein to clustered carbohydrates. Proc Natl Acad Sci U S A. 1999;96:11782–11786. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryan MC, Fazio F, Lee HK, Huang CY, Chang A, Best MD, Calarese DA, Blixt C, Paulson JC, Burton D, Wilson IA, Wong CH. Covalent display of oligosaccharide arrays in microtiter plates. J Am Chem Soc. 2004;126:8640–8641. doi: 10.1021/ja048433f. [DOI] [PubMed] [Google Scholar]
- 21.Liang PH, Imamura M, Li X, Wu D, Fujio M, Guy RT, Wu BC, Tsuji M, Wong CH. Quantitative microarray analysis of intact glycolipid-CD1d interaction and correlation with cell-based cytokine production. J Am Chem Soc. 2008;130:12348–12354. doi: 10.1021/ja8012787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang PH, Wang SK, Wong CH. Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: Determination of surface and solution dissociation constants. J Am Chem Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 23.Stoughton RB. Applications of DNA microarrays in biology. Ann Rev Biochem. 2005;74:53–82. doi: 10.1146/annurev.biochem.74.082803.133212. [DOI] [PubMed] [Google Scholar]
- 24.Bulyk ML. DNA microarray technologies for measuring protein-DNA interactions. Curr Opin Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min DH, Mrksich M. Peptide arrays: towards routine implementation. Curr Opin Chem Biol. 2004;8:554–558. doi: 10.1016/j.cbpa.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DS, Nock S. Recent developments in protein microarray technology. Angew Chem, Int Ed. 2003;42:494–500. doi: 10.1002/anie.200390150. [DOI] [PubMed] [Google Scholar]
- 27.Kung LA, Snyder M. Proteome chips for whole-organism assays. Nat Rev Mol Cell Biol. 2006;7:617–622. doi: 10.1038/nrm1941. [DOI] [PubMed] [Google Scholar]
- 28.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays - a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Ratner DM, Adams EW, Disney MD, Seeberger PH. Tools for glycomics: Mapping interactions of carbohydrates in biological systems. Chembiochem. 2004;5:1375–1383. doi: 10.1002/cbic.200400106. [DOI] [PubMed] [Google Scholar]
- 30.Werz DB, Seeberger PH. Carbohydrates as the next frontier in pharmaceutical research. Chem Eur J. 2005;11:3194–3206. doi: 10.1002/chem.200500025. [DOI] [PubMed] [Google Scholar]
- 31.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA. Dissection of the carbohydrate specificity of the broadly neutralizing-anti-HIV-1 antibody 2G12. Proc Natl Acad Sci USA. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang CY, Thayer DA, Chang AY, Best MD, Hoffmann J, Head S, Wong CH. Carbohydrate microarray for profiling the antibodies interacting with Globo H tumor antigen. Proc Natl Acad Sci USA. 2006;103:15–20. doi: 10.1073/pnas.0509693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilobello KT, Mahal LK. Deciphering the glycocode: the complexity and analytical challenge of glycomics. Curr Opin Chem Biol. 2007;11:300–305. doi: 10.1016/j.cbpa.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Pohl NL. Fluorous tags catching on microarrays. Angew Chem, Int Ed. 2008;47:3868–3870. doi: 10.1002/anie.200704801. [DOI] [PubMed] [Google Scholar]
- 35.Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, Steinman L, Robinson WH. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nature Med. 2006;12:138–143. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- 36.Prestwich GD. Touching all the bases: Synthesis of inositol polyphosphate and phosphoinositide affinity probes from glucose. Acc Chem Res. 1996;29:503–513. [Google Scholar]
- 37.Painter GF, Grove SJA, Gilbert IH, Holmes AB, Raithby PR, Hill ML, Hawkins PT, Stephens L. General synthesis of 3-phosphorylated myo-inositol phospholipids and derivatives. J Chem Soc, Perkin Trans. 1999;1:923–935. [Google Scholar]
- 38.Kubiak RJ, Bruzik KS. Comprehensive and uniform synthesis of all naturally occurring phosphorylated phosphatidylinositols. J Org Chem. 2003;68:960–968. doi: 10.1021/jo0206418. [DOI] [PubMed] [Google Scholar]
- 39.Schafer R, Nehlssahabandu M, Grabowsky B, Dehlingerkremer M, Schulz I, Mayr GW. Synthesis and application of photoaffinity analogs of inositol 1,4,5-trisphosphate selectively substituted at the 1-phosphate group. Biochem J. 1990;272:817–825. doi: 10.1042/bj2720817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prestwich GD, Marecek JF, Mourey RJ, Theibert AB, Ferris CD, Danoff SK, Snyder SH. Tethered IP3 - synthesis and biochemical applications of the 1-O-(3-aminopropyl) ester of inositol 1,4,5-trisphosphate. J Am Chem Soc. 1991;113:1822–1825. [Google Scholar]
- 41.Marecek JF, Estevez VA, Prestwich GD. New tetherable derivatives of myo-inositol 2,4,5-trisphosphates and 1,3,4-trisphosphates. Carbohyd Res. 1992;234:65–73. doi: 10.1016/0008-6215(92)85039-3. [DOI] [PubMed] [Google Scholar]
- 42.Tegge W, Ballou CE. Syntheses of D-myo-Inositol 1,4,5-trisphosphate affinity ligands. Carbohyd Res. 1992;230:63–77. doi: 10.1016/s0008-6215(00)90513-5. [DOI] [PubMed] [Google Scholar]
- 43.Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by Pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further synthetic procedures and characterizations are included. This information is available free of charge via the Internet at http://pubs.acs.org.