Abstract
Background
Cleavage of aggrecan by ADAMTS proteinases at specific sites within highly conserved regions may be important to normal physiological enzyme functions, as well as pathological degradation.
Methods
To examine ADAMTS selectivity, we assayed ADAMTS-4 and -5 cleavage of recombinant bovine aggrecan mutated at amino acids N-terminal or C-terminal to the interglobular domain cleavage site.
Results
Mutations of conserved amino acids from P18 to P12 to increase hydrophilicity resulted in ADAMTS-4 cleavage inhibition. Mutation of Thr, but not Asn within the conserved N-glycosylation motif Asn-Ile-Thr from P6 to P4 enhanced cleavage. Mutation of conserved Thr residues from P22 to P17 to increase hydrophobicity enhanced ADAMTS-4 cleavage. A P4′ Ser377Gln mutant inhibited cleavage by ADAMTS-4 and -5, while a neutral Ser377Ala mutant and species mimicking mutants Ser377Thr, Ser377Asn, and Arg375Leu were cleaved normally by ADAMTS-4. The Ser377Thr mutant, however, was resistant to cleavage by ADAMTS-5.
Conclusion
We have identified multiple conserved amino acids within regions N- and C-terminal to the site of scission that may influence enzyme-substrate recognition, and may interact with exosites on ADAMTS-4 and ADAMTS-5.
General Significance
Inhibition of the binding of ADAMTS-4 and ADAMTS-5 exosites to aggrecan should be explored as a therapeutic intervention for osteoarthritis.
Keywords: aggrecan, interglobular domain, ADAMTS-4, ADAMTS-5, aggrecanase, proteinase, exosite
1. Introduction
Aggrecan is a large aggregating proteoglycan abundantly secreted into cartilage extracellular matrix (ECM1) that contributes to cartilage hydration and lends resistance to compressive deformation. The aggrecan core protein is composed of the G1 domain at the N-terminus followed by an extended interglobular domain (IGD), G2, KS, CS-1, CS-2 domains and the G3 domain at the C terminus. The aggrecan core protein is highly substituted with sulfated glycosaminoglycans (GAGs), mainly chondroitin sulfate (CS) attached to the core protein in the CS-1 and CS-2 domains, and to a lesser extent with keratan sulfate (KS) within the IGD and KS domains. Aggrecan is retained in the ECM by the interaction of the G1 domain with hyaluronan (HA), which is stabilized by cartilage link protein, forming a ternary complex of aggrecan, link protein, and HA referred to as the proteoglycan aggregate.
The loss of aggrecan from the cartilage ECM occurs by proteolytic cleavage at specific sites within the CS-2 domain, which in bovine aggrecan include Glu1480-Gly1481, Glu1666-Gly1667, Glu1771-Ala1772, and Glu1871-Leu1872 and within the IGD at Glu373-Ala374. This is one of the primary events observed in osteoarthritic cartilage and it is thought to be due to the action of proteinases of the ADAMTS family. ADAMTS-1, -4, -5, -9, and -15 have all been shown to be aggrecan-degrading enzymes, among which ADAMTS-4 and -5, also known as aggrecanase-1 and -2, appear to be the most active [1]. Aggrecanolysis is also a normal physiological process in the development of primary and secondary centers of ossification, and in growth plates of long bones [2]. Aggrecan fragments arising from the activity of both MMPs and aggrecanases have been detected in these growth cartilages [2–4]. Bayliss et al [5] demonstrated that aggrecanases are active in normal cartilage of different ages, with a different distribution in young than in mature cartilage. There is the potential for redundancy in the ADAMTS family of proteinases in cartilage [6]. ADAMTS-4 [7] and ADAMTS-5 [8] are potent aggrecanases. it is possible, however, that other ADAMTS family members could provide compensatory aggrecanolysis resulting in the normal growth observed in ADAMTS-4 or -5 knockout mice [9, 10].
ADAMTS-4 and ADAMTS-5 are glutamyl-endopeptidases, which cleave C-terminal to glutamate residues (P1 subsite). They are both multi-domain proteinases having an N-terminal catalytic domain, a disintegrin-like domain, a thrombospondin (TS) type I domain, a cysteine-rich region and a spacer domain at the C-terminus of ADAMTS-4, but which is followed by additional TS type I domain in ADAMTS-5. ADAMTS-4 is synthesized as a 90.2 kDa proenzyme, and is N-terminally processed by an intracellular furin-like activity to the secreted 68kDa form (p68) [1]. ADAMTS-4 is also subject to C-terminal truncation [11, 12] to generate 53kDa and 40 kDa forms of the enzyme lacking portions of the C-terminal spacer domain. In contrast to previous work [12, 13], Fushimi et al [14] reported that removal of the spacer domain is not required for full catalytic activity against the Glu373-Ala374 bond. We have also found that the 68 kDa isoform of recombinant human ADAMTS-4 used in this study, was active in cleaving the IGD site [15, 16] following initial cleavage at sites within the CS-2 domain. We have previously reported that that the 40kDa ADAMTS-4 form could readily cleave within the IGD, but had negligible cleavage activity within the CS-2 domain [15, 16].
The primary and secondary structure or post-translational modifications such as O- and N-glycosylation flanking a given cleavage site may also impact the likelihood of cleavage. The IGD region of aggrecan is predicted to lack tertiary structure. Similar “natively unfolded domains” containing low affinity protein binding motifs have been documented [17]. Westling and co-authors [18] showed that the versicanase activity of 68 kDa ADAMTS-4 was reduced by replacing valine and isoleucine with lysines at P12 and P18 sites of a versican splice variant, respectively. Similarly, clustered hydrophobic residues are also present N-terminal to aggrecan ADAMTS-4/5 cleavage sites. Additionally, some of the glycosylation sites near the ADAMTS-4/5 cleavage site may affect the interaction of ADAMTS-4 or -5 with the aggrecan substrate. Age-related variable substitution with O- or N-linked keratan sulfate at Thr352, Thr355 or Thr357, Asn368 and Thr370 has been documented [19], and it has been shown that N-linked keratan sulfate at Asn368 may potentiate [20] aggrecan core protein cleavage at Glu373-Ala374. The crystal structures of ADAMTS-4 and ADAMTS-5 [21, 22] have been recently determined. The disintegrin-like domain of ADAMT-4 and -5 is in close proximity to the active site in the three-dimensional structure of both enzymes, and could present an auxiliary substrate-binding surface sensitive to substrate topology C-terminal to the site of scission.
In the present study, we hypothesized that some of the conserved residues flanking the IGD cleavage site may affect cleavage efficiency at Glu373-Ala374 by ADAMTS-4 and ADAMTS-5, by maintaining a topology favorable for substrate recognition. We have used recombinant bovine aggrecan as an in vitro ADAMTS-4 and ADAMTS-5 substrate. We have introduced single and multiple conservative and non-conservative amino acid substitutions near the IGD cleavage site, and have analyzed ADAMTS-4 and ADAMTS-5 cleavage efficiency in response to these substitutions. Our results indicate that aggrecan core protein sites, both N- and C-terminal to the cleavage site in the IGD, affect cleavage efficiency. Furthermore, some of these residues have the potential to be differentially post-translationally modified which may in effect “fine-tune’ the susceptibility of these sites to aggrecanase-mediated cleavage. Modification of the interplay between aggrecan and enzymes involved in its degradation could be an important aspect of cartilage morphogenesis during development and regulation of growth plate dynamics. Dysregulation of this interplay may contribute to cartilage degeneration in osteoarthritic cartilage.
2. Experimental Procedures
2.1 Materials
Cell culture media, PCR primers, and lipofectAMINE PLUS reagents were purchased from Invitrogen (Carlsbad, CA). A QuikChange® site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA). Polyclonal antiserum against the G3 domain of aggrecan (LEC-7) [23] was a generous gift from Dr. Kurt Doege (Louisiana State University Health Sciences Center, Shreveport, LA). Anti-NITEGE neo-epitope antiserum [24] was a generous gift from Dr. John S. Mort (Shriners Hospital for Children, Montreal, Canada). Chondroitinase ABC, keratanase, and keratanase II were purchased from Seikagaku America, Inc. (Falmouth, MA). The recombinant human ADAMTS-4 (p68) used in this study was a generous gift from Dr. Elisabeth A. Morris (Wyeth Research, Cambridge, MA), and 40 kDa recombinant human ADAMTS-4 (p40) was purchased from Calbiochem (San Diego, CA). Recombinant human ADAMTS-5 was a generous gift from Dr. David Buttle (Division of Genomic Medicine, University of Sheffield Medical School, Sheffield, UK). We obtained recombinant human pro-MMP-13 from MP Biomedicals (Irvine, CA). Restriction enzymes were purchased from New England Biolabs (Beverly, MA). COMPLETE protease inhibitor mix (+EDTA) was purchased from Roche Applied Science (Indianapolis, IN). COS-7 cells were from ATCC (Manassas, VA). Cellgro culture media were purchased from Mediatech, Inc. (Herndon, VA). Immobilon-PSQ PVDF membranes were purchased from Millipore (Bedford, MA). M2 anti-FLAG monoclonal antibody was purchased from Sigma-Aldrich (St. Louis, MO). SDS-PAGE pre-cast gels were purchased from Bio-Rad (Hercules, CA). All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO). The construction of pBAGG64-5 and pBAGG71-28 was described previously [16]. HyperChem software was obtained from Hypercube, Inc, Gainsville FL.
2.2 Site-directed mutagenesis
Various mutations (excepting V356A) were made in the full-length aggrecan constructs, pBAGG64-5 and pBAGG71-28, by site-directed mutagenesis using the QuikChange XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) as described in the manufacturer’s protocol. A series of PAGE-purified primer sets used for mutagenesis are described in Tables 1–3. Some mutants were confirmed by direct PCR of ampicillin-resistant bacterial colonies using forward and reverse primers flanking the site of the aggrecan interglobular domain mutation (5′-CCTGACCCTTCATCCCGCTAT-3′ and 5′-GGAATGCAGTGGCCCTTACTT-3′) respectively. PCR reaction conditions were as follows: 1 min at 94 C followed by 30 cycles of (1 min at 94 °C, 1 min at 59 °C, 1 min at 72 °C), then 10 min at 72 °C. The resulting PCR products were further analyzed by restriction endonuclease digestion to identify mutant plasmid DNA using various restriction enzymes that would permit discrimination between WT and mutants. All site-directed mutations were ultimately confirmed by DNA sequencing. One mutant (p163-21; V356A) was produced using the ExSite PCR-based Site-Directed Mutagenesis Kit (Stratagene, La Jolla, Ca). A ~2.5 kb fragment of the aggrecan cDNA in vector p23-13 [16] was mutagenized using oligonucleotides described in Table 1. A portion of this mutagenized fragment, flanked by DraIII and SrfI sites, was used to replace a homologous segment in the full sized aggrecan insert of the WT (wild-type) pBAGG64-5 vector.
Table 1.
Group A: Primer sequences and template plasmids used for site-directed mutagenesis.
| Mutation(s) | Primer Sets | Primer Location | Clone Number |
|---|---|---|---|
| V356A | 5′-ACGGCGACCTGGCCTGACGTGGAGCTGCCCCTGCCCCGA-3′
5′-CTGGATGGTGATGTCCTCCTC-3′ |
1480-1518 | 163-21 |
| V361A | 5′-GACCTGGCCTGACGCCGAGCTGCCCCTGCCCCG-3′
5′-CGGGGCAGGGGCAGCTCGGCGTCAGGCCAGGTC-3′ |
1485-1517 | 190-13 |
| E362D | 5′-GCCTGACGTGGACCTGCCCCTGCCC-3
5′-GGGCAGGGGCAGGTCCACGTCAGGC-3′ |
1491-1515 | 190-3 |
| V356A-V361A-E362D | 5-′CCATCCAGACGGCCACCTGGCCTGACGCCGACCTGCCCCTGCCC-3′
5-′GGGCAGGGGCAGGTCGGCGTCAGGCCAGGTGGCCGTCTGGATGG-3′ |
1472-1515 | 304-21 |
| D360L | 5′-GACCTGGCCTCTGGTGGAGCTGC-3′
5′-GCAGCTCCACCAGAGGCCAGGTC-3′ |
1485-1507 | 196-8 |
| V361Q-E362K | 5′-CTGGCCTGACCAGAAGCTGCCCCTGC-3′
5′-GCAGGGGCAGCTTCTGGTCAGGCCAG-3′ |
1488-1513 | 306-16 |
| D360H-V361Q-E362K | 5′-GACCTGGCCTCACCAGAAGCTGCCCCTGC-3′
5′-GCAGGGGCAGCTTCTGGTGAGGCCAGGTC-3′ |
1485-1513 | 196-29,19 |
Table 3.
Group D: Primer sequences and template plasmids used for site-direct mutagenesis
| Mutation(s) | Primer sets | Primer Location | Clone Number |
|---|---|---|---|
| R375L | 5′-GAGGGTGAAGCCCTGGGCAGCGTGATCC-3′ | 1524-50 | 463-15 |
| 5′-GGATCACGCTGCCCAGGGCTTCACCCTC-3′ | |||
| S377Q | 5′-GAAGCCCGAGGCCAGGTGATCCTCACGGC-3′ | 1534-62 | 460-3 |
| 5′-GCCGTGAGGATCACCTGGCCTCGGGCTTC-3′ | |||
| S377A | 5′-GAAGCCCGAGGCGCTGTGATCCTCACGGC-3 | 1534-62 | 463-4 |
| 5′-GCCGTGAGGATCACAGCGCCTCGGGCTTC-3′ | |||
| S377T | 5′-GAAGCCCGAGGCACCGTGATCCTCACGGC-3 | 1534-62 | 463-5 |
| 5′-GCCGTGAGGATCACGGTGCCTCGGGCTTC-3′ | |||
| S377N | 5′-GAAGCCCGAGGCAATGTGATCCTCACGGC-3 | 1534-62 | 463-12 |
| 5′-GCCGTGAGGATCACATTGCCTCGGGCTTC-3′ |
2.3 Expression and purification of recombinant aggrecan
WT and mutant constructs were transfected into COS-7 cells and secreted aggrecan was purified from conditioned medium as described previously [16, 25]. Cells were transiently transfected using lipofectAMINE PLUS reagent. Seventy-two hours post-transfection, aggrecan secreted into conditioned medium was purified by Sephadex G-50 gel filtration and DEAE Sephacel ion exchange chromatography. Recombinant aggrecan was quantified relative to a standard curve based on dry-weight of cartilage-derived (A1A1D1) steer aggrecan by a dot blot western-based immunoassay with an antibody (G1–2) directed toward the G1 domain (Fig. 1). For clarity, individual amino acid residues will generally be designated by their three letter abbreviation in the text, however the single letter abbreviation will be used in the context of a mutagenized site to be consistent with abbreviations in the figures.
Fig. 1.
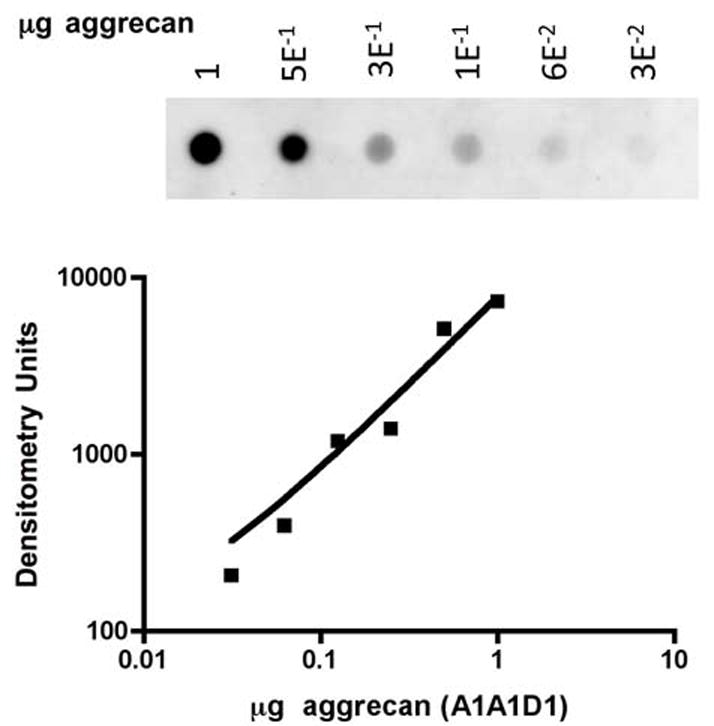
Quantification of aggrecan by dot blot western analysis. Dilutions of a standard solution of steer A1A1D1 aggrecan (shown) and similar dilutions of recombinant aggrecan preparations were dot-blotted on a PVDF membrane, reacted with anti-G1 and visualized by ECL. The concentration of recombinant aggrecan preparations was calculated from the A1A1D1 standard curve.
2.4 Protease digestion of recombinant aggrecan and Western blot analysis
Recombinant aggrecan samples (2 to 8 μg) were digested with ADAMTS-4 (p68 unless otherwise indicated) at a 1:100 enzyme to substrate ratio (w/w) in 20 mM Tris-HCl, pH 7.2; 150 mM NaCl; and 5 mM CaCl2 at 37°C for different lengths of time. Recombinant aggrecan (2 μg) was digested with 0, 50, 100, or 200 ng of ADAMTS-5 for 90 min. Reactions were terminated by the addition of EDTA to 21 mM. Aggrecan fragments were digested with chondroitinase ABC (0.01 U/0.4 pmol of aggrecan) at 37 °C for 1 hr in buffer containing 8 mM sodium acetate, 10 mM Tris/HCl (pH 8.0) with COMPLETE™ protease inhibitor mix. Experiments using pBAGG64-5 derived aggrecans were also digested with keratanase and keratanase II, which was later discontinued upon finding no detectible keratan sulfate in recombinant aggrecan. Samples were then analyzed by 4–15% SDS-PAGE/Western blot as described previously using anti-G1 domain, anti-G3 domain, anti-FLAG (M2), and anti-NITEGE as described previously [16]. All wild type and mutant pairs were electrophoresed and transferred to PVDF membranes at the same time, reacted with antibodies, washed, and processed for ECL at the same time, and exposed to film for the same length of time in the same film cassette.
3. Results
3.1 Generation of mutagenized aggrecan expression constructs
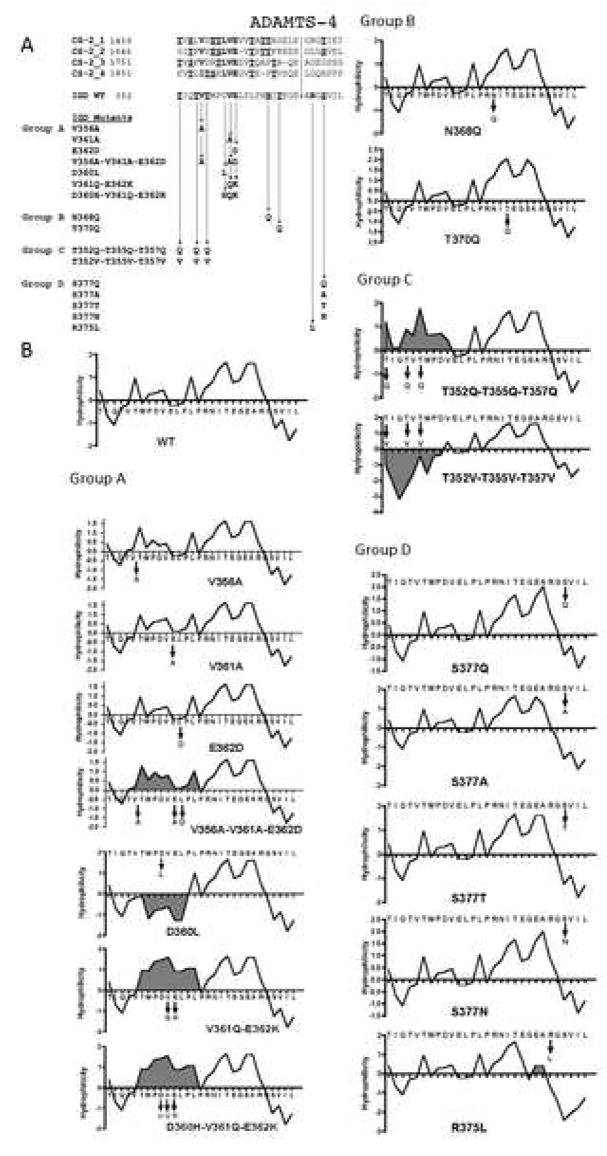
Previously, we reported the construction of two full-length recombinant aggrecan expression vectors and the characterization of recombinant aggrecan expressed in COS-7 cells [16]. We showed that the secreted recombinant aggrecan could be digested by ADAMTS-4 at known cleavage sites [26]. In the present work, we used these vectors (pBAGG64-5 and pBAGG71-28) as templates for site-directed mutagenesis to characterize the “topography” of the ADAMTS-4 cleavage site within the interglobular domain (IGD) of aggrecan. Specific amino acid residues showing high interspecies conservation, or occurring in an identical or similar position at CS-2 domain ADAMTS-4/5 sites, were targeted for site-directed mutagenesis The first set of mutants (Fig. 2A, Group A) was generated using pBAGG64-5 (no FLAG epitope tag). These are mutations at particular sites where non-glycosylated residues are conserved between the ADAMTS-4/5 cleavage sites of the IGD and CS-2 domain. The second set (Group B) was generated using both pBAGG64-5 and pBAGG71-28 (with FLAG epitope tag) and includes mutations of Asp and Thr within an N-glycosylation motif (NIT) nea the site of scission by ADAMTS-4 in the IGD. The third set (Group C) included mutations of Thr residues within a “cluster” N-terminal to the cleavage site using pBAGG71-28. Lastly, we constructed mutants within the region that is immediately C-terminal to the cleavage site (Group D). These mutants were generated using pBAGG71-28 and include mutations of residues at P2′ and P4′ subsites (Fig. 1A, Group D). Some mutants are species-mimicking mutants described later in Fig. 8A.
Fig. 2.
Summary of recombinant aggrecan mutants. (A) Mutant aggrecans were categorized into four groups having mutations within the IGD of bovine aggrecan. The first group of mutants (Group A) contains mutations of conserved amino acids in the 355Thr-Glu362 sequence found N-terminal to the IGD cleavage site. Second group (Group B) contains mutations at Thr and Asn residues, within the N-glycosylation motif (368AsnIleThr370) near the site of scission. The third group (Group C) contains mutations in a Thr-rich region (352Thr-Thr357) found N-terminal to the IGD cleavage site. The fourth group (Group D) consists of mutations of Arg375 and Ser377, C-terminal to the Glu373-Ala374 cleavage site. (B) Kyte-Doolittle hydropathy plots of WT and mutant aggrecan. Shaded areas indicate significant deviations from the WT hydropathy plot.
Fig. 8.
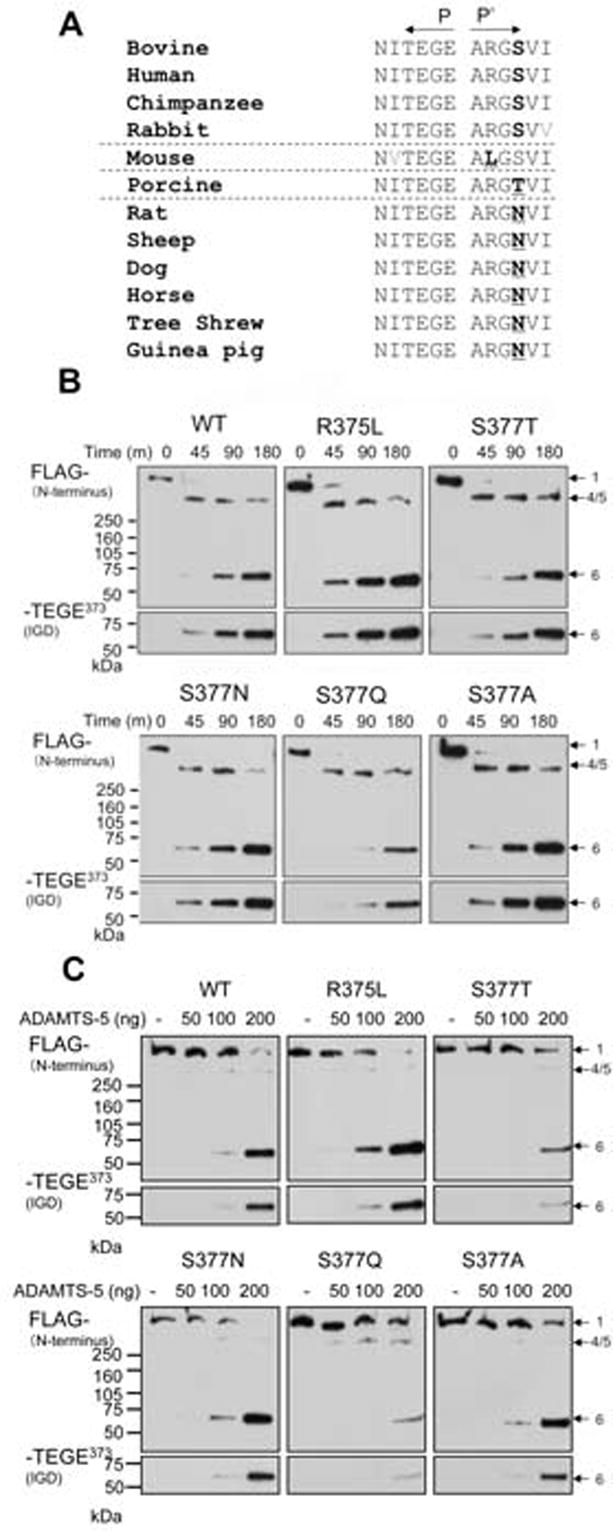
The susceptibility of recombinant aggrecan carrying species-mimicking sequence to ADAMTS-4 (p68). (A) Species specific variation found in the P2′ and P4′ position (boldface) relative to the Glu373-Ala374 cleavage site. WT and P′-mutants were digested with (B) ADAMTS-4 (p68) or with (C) ADAMTS-5, and cleavage at Glu373-Ala374 was analyzed by Western blot analysis using anti-FLAG (M2), and anti-NITEGE antibodies. (bands numbered as in Miwa et al. [16].)
3.2 Effects of conserved residues within the IGD on its susceptibility to ADAMTS-4
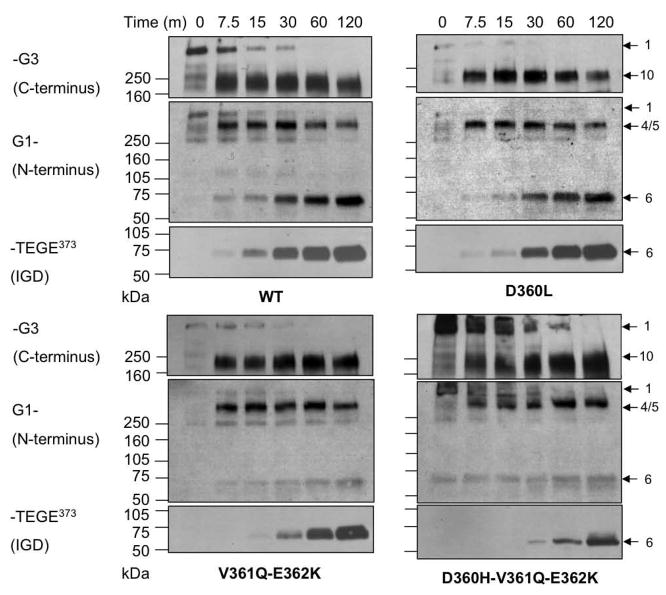
Most of the amino acid residues found at the P18 (P17 in some CS-2 cleavage sites) subsite of ADAMTS-4 cleavage sites are either Val or Ile (Fig. 2A). Furthermore, the Leu-Val-Glu motif at P14-P13-P12 (P13-P12-P11 in some CS-2 cleavage sites) is conserved at all ADAMTS-4 cleavage sites, with the exception of P14 of the IGD, which is Asp. Therefore, we decided to determine whether these residues are required for efficient cleavage by ADAMTS-4. First, we mutated Val356 at P18 to Ala (V356A), Val361 at P13 to Ala (V361A), and Glu362 at P12 to Asp (E362D) individually (single mutation) or simultaneously (triple mutation) to conserve the polarity of each amino acid and examine the changes in their susceptibility to ADAMTS-4. Susceptibility was measured by Western blot analysis of the cleavage at Glu373-Ala374 using anti-G1 domain and anti-NITEGE neoepitope antisera. Cleavage within the CS-2 domain was also monitored by analyzing fragments reactive to anti-G3 domain antiserum. The individually mutated V356A, V361A, and E362D mutants were cleaved at a similar rate compared to the WT aggrecan (Fig. 3A). However, when all of these residues were simultaneously mutated (V356A-V361A-E362D), this mutant was extremely resistant to cleavage at Glu373-Ala374, relative to the WT control digested at the same time. Whereas IGD cleavage to generate the NITEGE neoepitope was evident after 2 hrs in the WT control, an anti-NITEGE reactive fragment appeared only after 24 hr of digestion in the triple mutant (Fig. 3B). Although the individual mutations did not change the overall hydrophilicity of this region relative to the WT control, the triple mutant showed a cumulative increase in hydrophilicity (Fig. 2B). This result suggests that the hydropathy of the amino acid side chain at these conserved locations can affect susceptibility to ADAMTS-4. We also noted that appearance of fragments 4 and 5, which have the G1 domain at the N-terminus and Glu1666 or Glu1480, respectively at the C-terminus, were also delayed in the triple mutant relative to wild-type (Fig. 3B). The appearance of fragment 10, which has Gly1481 at the N-terminus and the G3 domain at the C-terminus was also delayed.
Fig. 3.
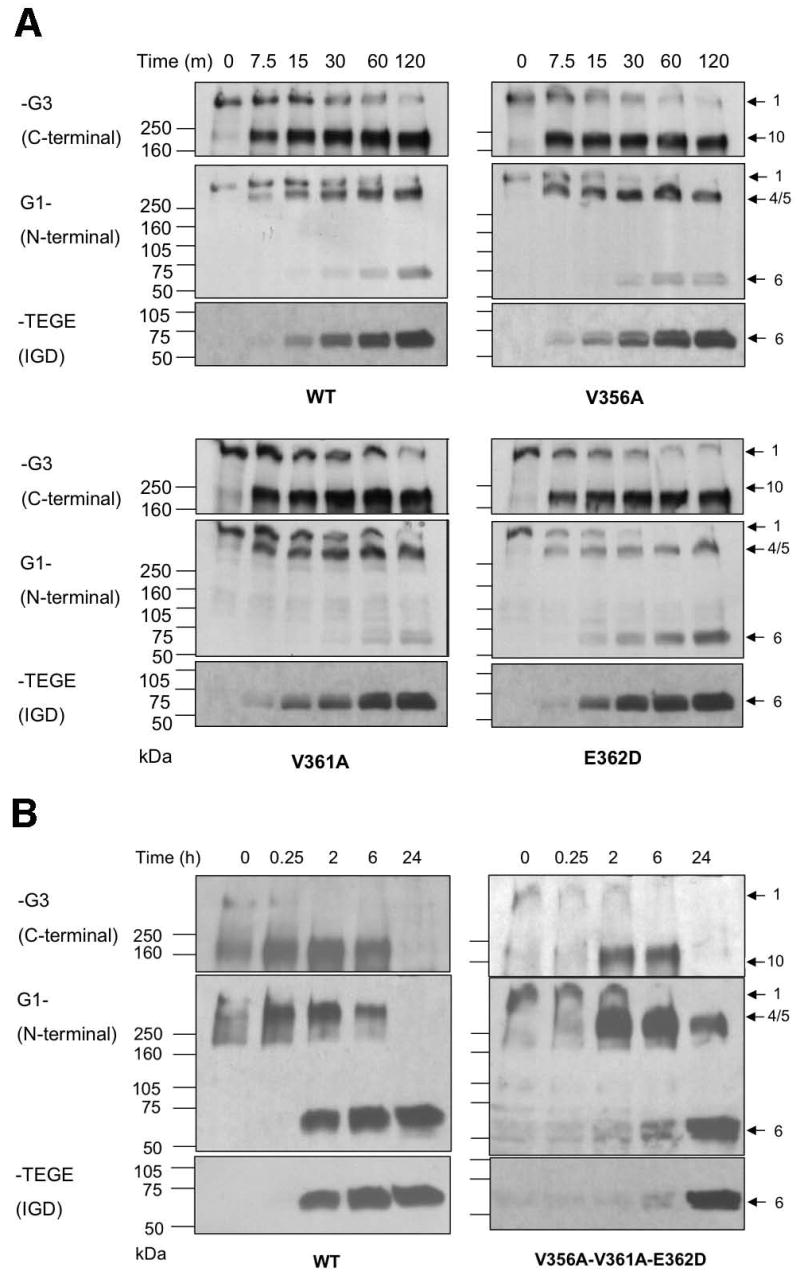
Western blot analysis of fragments derived from WT and mutant recombinant aggrecan: Effects of mutation of conserved Val356, Val361, and Glu362 residues on cleavage by ADAMTS-4 (p68). (A) WT, and V356A, V361A, and E362D mutant aggrecan and ADAMTS-4-generated fragments produced at intervals up to 120 minutes. (B) WT and V356A-V361A-E362D triple mutant aggrecan were digested with ADAMTS-4 for up to 24 h. Antibody used for each Western blot is indicated at the left. Intact aggrecan and fragments generated by ADAMTS4 digestion are numbered at right as described in Miwa et al. [16]. The band labeled 4/5 are unresolved fragments having TFKEEE1666 and TAGELE1480 C-termini.
To further examine the role of the 360AspValGlu362 sequence in the IGD and its interaction with ADAMTS-4, we mutated Asp360 at P14 to Leu (D360L). All of the cleavage sites in the bovine aggrecan CS-2 domain, as well as in other species, have a conserved LeuValGlu motif N-terminal to the ADAMTS-4 cleavage site, whereas in the IGD, the conserved triplet is AspValGlu (Fig. 2A). Since cleavage sites in the CS-2 domain were originally reported to be more favorable than those in the IGD, we reasoned that the D360L mutation might enhance cleavage within the IGD. This mutation, however, did not enhance susceptibility at Glu373-Ala374 (Fig. 4). Similar to the WT, cleavage at Glu373-Ala374, producing fragment 6, followed cleavage at Glu1480-Gly1481 and Glu1666-Gly1667 (fragments marked 4/5). Replacing Asp with Leu at this position does not appear to account for the cleavage preference given to sites within the CS-2 domain. We then mutated Val361 and Glu362 non-conservatively to Gln and Lys (V361Q-E362K), respectively, (Fig. 2A). A triple mutant with Asp360 mutated to His, in addition to V361Q and E362K (D360H-V361Q-E362K), was also generated. The V361Q-E362K mutant significantly increased the overall hydrophilicity of this region (Fig. 2B), and showed a slight reduction in its susceptibility at Glu373-Ala374 (Fig. 3). The additional non-conservative D360H mutation (D360H-V361Q-E362K), also showed increased hydrophilicity relative to WT (Fig. 2B), and brought about the most significant cleavage reduction (Fig. 4). Overall, these results suggest that the 360Leu/AspValGlu362 sequence is important for enzyme-substrate recognition and cleavage at Glu373-Ala374. Whereas a shift to greater hydrophobicity (D360L) mutant, Fig. 2B) was not sufficient to increase cleavage efficiency, mutations producing greater hydrophilicity (V356A-V361A-E362D, V361Q-E362K and D360H-V361Q-E362K, Fig. 2B) resulted in diminished cleavage (Figs. 3 and 4). As we observed for the V356A-V361A-E362D mutant (Fig. 3), the appearance of fragments 4/5, and fragment 10, were also delayed in the time-course digestion of the D360H-V361Q-E362K mutant (Fig. 4).
Fig. 4.
Effects of non-conservative mutation of the WT 360AspValGlu362 (DVE) motif on cleavage by ADAMTS-4 (p68). The significance of the DVE sequence in the IGD was examined by producing the mutant D360L to generate LVE, the sequence preceding the more susceptible sites in the CS-2 domain. The WT (DVE) was also mutated to 360AspGlnLys362 (V361Q-E362K) and 360HisGlnLys362 (D360H-V361Q-E362K).
3.3 Role of N-glycosylation upon IGD cleavage
Glycosylation at 368AsnIleThr370 near the IGD cleavage site may affect cleavage efficiency. N-linked KS is found at this site in steer aggrecan, but not newborn calf aggrecan. [19] and N-linked KS at Asn368 may potentiate aggrecanase-mediated cleavage at Glu373-Ala374 [20]. We did not expect an effect of KS at the Asn368 site, since COS-7 cell-derived recombinant bovine aggrecan is not substituted with KS [16]. We made mutants (Fig. 2A, Group B) to prevent N-glycosylation at Asn368 (N368Q and T370Q) the first of which retains a potentially O-glycosylated Thr370. Digestion of both mutants with rhADAMTS-4 generated a 62 kDa band reactive to both FLAG and NITEGE antibodies, instead of the 67 kDa band seen in the WT aggrecan, consistent with absence of the N-linked oligosaccharide normally present (Fig. 5). A commercially-obtained p40 preparation, which cleaves recombinant aggrecan efficiently in the IGD [15], was used for this analysis to conserve p68 aggrecanase. Since this fragment can be detected with both the anti-FLAG and anti-NITEGE antisera, the mutation within the NITEGE motif does not appear to affect anti-NITEGE binding. When the rhADAMTS-4 (p68), susceptibility at Glu373-Ala374 of the N368Q and T370Q mutants were compared with WT aggrecan, N368Q (Fig. 6A) showed little difference from WT, but the T370Q mutant showed significantly enhanced cleavage (Fig. 6B). The more rapid accumulation of fragment 6 in the T370Q mutant digestion is accompanied by the lack of accumulation of the fragments 4 and 5, from which fragment 6 is generated. Interestingly, fragment 10, generated by cleavage in the CS-2 domain, disappeared more rapidly than in the WT control.
Fig. 5.
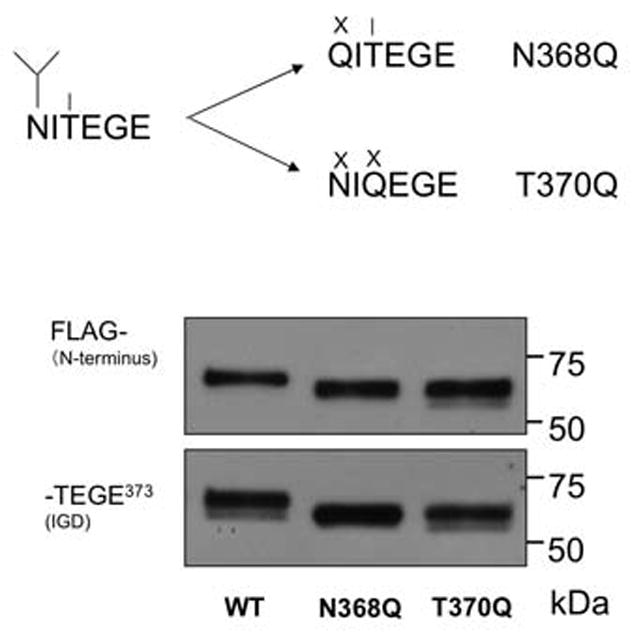
The effect of N-linked glycosylation at Asn368 on aggrecan cleavage by ADAMTS-4 (p40) at Glu373-Ala374. (A) Mutations at Asn368 and Thr370 remove a potential N-linked site (fork symbol). In N368Q the potentially O-glycosylated Thr (line symbol) remains available. (B) Mutation at Asn368 and Thr370 both result in reduction of the molecular mass of anti-FLAG and anti-NITEGE reactive fragment following digestion with ADAMTS-4 (p40) consistent with elimination of N-linked glycosylation at Asn368.
Fig. 6.
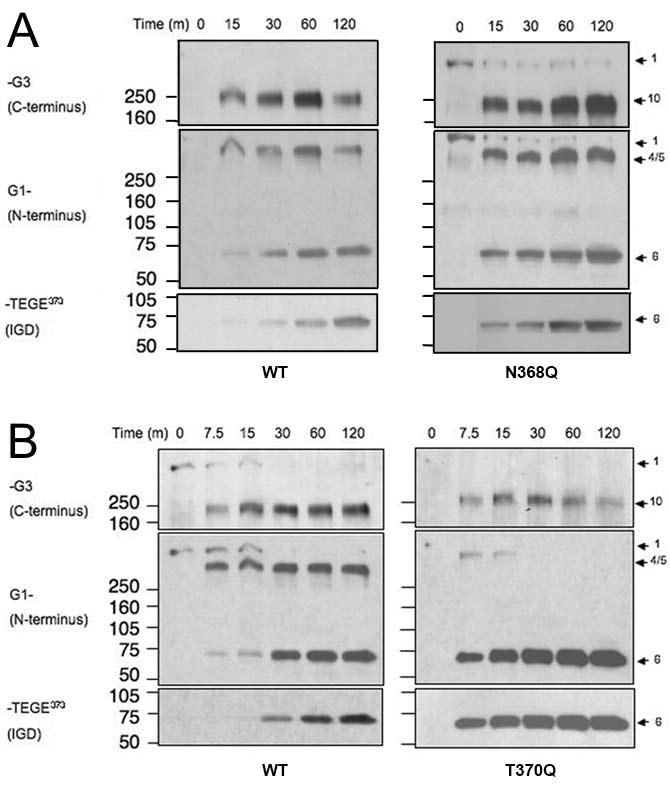
The effect of the N-linked oligosaccharide motif upon ADAMTS-4 cleavage. WT and mutant aggrecans were digested with ADAMTS-4 (p68) to compare the cleavage efficiency at the Glu373-Ala374 bond. (A) N368Q mutation, (B) T370Q mutation.
3.4 Effect of residues within conserved threonine cluster upon ADAMTS-4 cleavage
Comparison of sequence near the ADAMTS cleavage sites within the IGD and CS-2 domain (Fig. 2A) shows that each site has an abundance of Thr and Ser residues. In the bovine aggrecan IGD, Thr residues are clustered in the P22-P17 region N-terminal to the Glu373-Ala374 scission site (Thr352, Thr355 and Thr357), two out of three of which have been shown to be substituted with KS [19] in native bovine aggrecan.
We have conservatively mutated these Thr residues simultaneously to Gln. We also simultaneously mutated them non-conservatively to Val (Fig. 2A, Group C). We then compared the relative rate of cleavage by rhADAMTS4 (p68) within the IGD. It was apparent that the T352Q-T355Q-T357Q mutant was cleaved somewhat less rapidly than WT (Fig. 7A). In contrast, the T352V-T355V-T357V mutant was cleaved more rapidly at Glu373-Ala374 as determined by increased accumulation of anti-NITEGE product relative to WT aggrecan (Fig. 7B). This result suggests that ADAMTS-4 (p68) interacts with this region of recombinant aggrecan via hydrophobic interactions, which could be further enhanced by simultaneous mutations to Val.
Fig. 7.
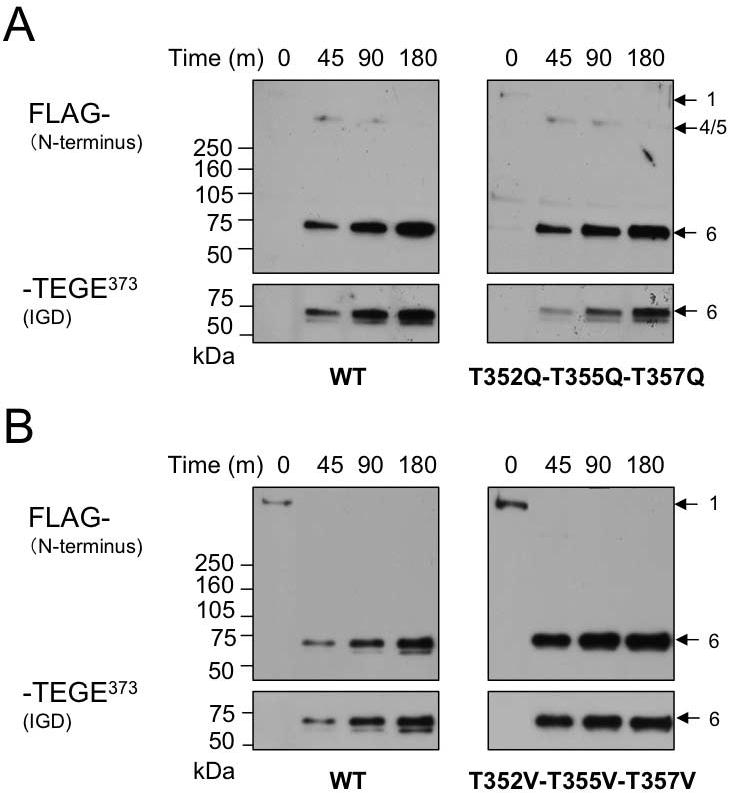
The effect of a cluster of threonine residues on aggrecan cleavage by ADAMTS-4 (p68) at Glu373-Ala374. A cluster of threonines (Thr352, Thr355, and Thr357) were simultaneously mutated to glutamine or valine (A, B). WT and mutant aggrecans were digested with ADAMTS-4 (p68) to compare the cleavage efficiency at the Glu373-Ala374 bond by the appearance of anti-NITEGE reactive fragments within the time-course assay. The molecular mass of the major band in each digestion is approximately 66.5 kDa.
3.5 Sequence C-terminal to the ADAMTS-4 cleavage site influences cleavage efficiency
To determine other potential exosite interactions with ADAMTS-4, we have also mutagenized amino acid residues C-terminal to the Glu373-Ala374 cleavage site. Most of the residues in this region, at the P1′, 2′, 3′, 5′ and 6′ position are highly conserved. Interestingly, the amino acid at the P4′ subsite is variable between species (Ser, Thr or Asn), a relatively conservative difference (Fig. 8A). For the 12 different species compared in Fig. 8A, the only other variable residue is in the P2′ position, where the mouse has the non-conservative substitution of Leu for Arg.
The P2′ position of the bovine aggrecan construct was mutated (R375L), as in mouse aggrecan. Surprisingly, this non-conservative substitution at a site one residue distant from the site of scission did not result in an observable difference in ADAMTS-4 susceptibility (Fig. 8B). We observed that mutations in recombinant bovine aggrecan at Ser377 to Thr or Asn, as found in other species (Fig. 8A), did not result in any significant difference in ADAMTS-4 cleavage. The S377Q mutation, however, resulted in a significant reduction of cleavage by ADAMTS-4 (p68) (Fig. 8B). Interestingly, the S377Q mutant was susceptible to cleavage by the C-terminally truncated ADAMTS-4 (p40) (result not shown). Substitution of Ala for Ser is generally considered to be a neutral difference for extracellular proteins [27]. We produced an S377A mutant, and found that it was cleaved similarly to WT (Fig. 8B).
ADAMTS-5 has been identified as a proteinase responsible for onset of osteoarthritis in murine models [9, 10]. Since both ADAMTS-4 and ADAMTS-5 exhibit similar substrate specificity, we also examined the susceptibility of the same set of mutants to ADAMTS-5 (Fig. 8C). We found that the ADAMTS-5 preparation used in this study was heat-labile, and could not be used for prolonged time-course digestion. Therefore, aggrecan was digested for 90 min. with different amounts of ADAMTS-5. As was observed with ADAMTS-4, the S377Q mutant was extremely resistant to cleavage at Glu373-Ala374 by ADAMTS-5. However, cleavage of S377T with ADAMTS-5 was also significantly inhibited compared to WT. The other P′-mutant digests were similar to WT. We conclude that Gln or Thr at the P4′ position are inhibitory to ADAMTS-5 cleavage.
4. Discussion
4.1 Substrate specificity of ADAMTS-4 against recombinant aggrecan
Although aggrecanases have high catalytic activity towards proteoglycans of the hyalectan family, including aggrecan, brevican, and versican, the only residue common to all of the known cleavage sites is Glu at the P1 site. Nevertheless, aggrecanases are quite specific, and previous studies have indicated that this specificity may be due to substrate interactions with exosites on the ADAMTS proteinase [28–31]. The present study was designed to use mutagenized recombinant aggrecan to identify elements of the aggrecanase recognition motif near the cleavage site. When comparing the primary sequence of aggrecan between species, it is apparent that regions of the protein proximal to aggrecanase cleavage sites are conserved [32]. In the present work, we focused on the aggrecanase cleavage site within this region, which may be important for normal physiological cleavage as well as pathological degradation of aggrecan.
4.2 The interaction of ADAMTS-4 with a conserved motif within the aggrecan core protein
We first investigated the role of three highly conserved residues, Val356, Val361, and Glu362, within the sequence Thr355 through Glu362, located 12–14 amino acids N-terminal to the ADAMTS-4 cleavage site within the IGD of the aggrecan core protein. Seven different variations within this region (Group A in Fig. 2) were produced by site-directed mutagenesis, ranging from a single conservative substitution to completely non-conservative simultaneous substitutions for as many as three residues. We found that single conservative substitutions V356A, V361A, and E362D had little apparent effect upon cleavage by ADAMTS-4. On the other hand, simultaneous conservative substitutions at all three sites drastically decreased ADAMTS-4 cleavage at the IGD site.
We expected that non-conservative mutation Asp360Leu within the 360AspValGlu362 sequence in the IGD might actually facilitate cleavage, since Leu is present within each of the similar triplet (LeuValGlu) motifs at the P14 or P13 position of each apparently more “favored” sites within the CS-2 domain. Interestingly, the Asp360Leu mutant did not appear to be cleaved more rapidly than the WT (Fig. 4). Apparently, the favored cleavage previously observed [16, 26] in the CS-2 domain relative to the IGD site is not likely to involve a simple interaction with the first residue of the triplet motif of 360AspValGlu362 in the IGD or the four LeuValGlu motifs in the CS-2 region at positions 1469–1471, 1655–1657, 1760–1762, 1860–1862. The apparent preference for CS-2 cleavage sites may be due to the binding of ADAMTS ancillary domain exosites to chondroitin sulfate [15].
Further analysis of this motif, however, does suggest that it interacts with ADAMTS-4. Simultaneous double (V361Q-E362K) or triple (D360H-V361Q-E362K) non-conservative mutations within the triplet 360AspValGlu362 diminished ADAMTS-4 cleavage, most obviously in the triple mutant (Fig. 4). This is interesting, since the Val-Glu pair found at the P13 and P12 position occur at the same position C-terminal to the highly favored CS-2 domain cleavage site at Glu1481-Gly1482 and in the P12 and P11 positions of the other three CS-2 domain sites. It is likely that these residues are important to aggrecanase-aggrecan recognition at these sites. We observed inhibition of cleavage within the CS-2 domain, when cleavage within the IGD was inhibited. It is possible that ADAMTS-4 exosite(s) bind certain “inhibiting” IGD mutants with a higher affinity and lower rate of dissociation than WT. Conversely, the proteinase exosite may bind “enhancing” IGD mutants with a lower affinity and and increased rate of dissociation than WT. Additional studies should be done to validate or disprove this model.
4.3 Role of N-glycosylation upon IGD cleavage
Asparagine-linked (N-linked) glycosylation is a co-translational protein modification that occurs in the endoplasmic reticulum at the consensus sequence Asn-X-Thr/Ser [33]. In glycoproteins, this modification has been shown to have diverse functions, and plays a role in intermolecular recognition, protein folding, and resistance to proteolytic cleavage [34]. To explore the role of glycosylation within this motif upon the aggrecan-ADAMTS-4 interaction, mutations (Fig. 2, group B) were generated to change the first and the third residue within the N-linked oligosaccharide motif 368AsnIleThr370, which is found in the P6 to P4 position relative to the site of ADAMTS-4 scission in the IGD. We expect that the Asn368Gln mutant would lack N-glycosylation, but could potentially be O-glycosylated at Thr370. The T370Q mutant, however, would be neither N- nor O-glycosylated. The mutant which conservatively changed the first residue of the N-glycosylation motif (N368Q) showed no apparent enhancement of ADAMTS-4 susceptibility, relative to WT. The other mutant (T370Q), however, showed enhanced susceptibility to cleavage at Glu373-Ala374. Although this mutation is near the C-terminal neoepitope (368Asn to Glu373) of the N-terminal aggrecan fragment, it does not appear to affect antibody reactivity, confirmed by reactivity of the same band with the anti-G1 polyclonal antiserum. The T370Q mutation, likely to be near the catalytic cleft in the enzyme/substrate complex, may influence ADAMTS-4 binding to the substrate. We observed in the time-course digestion of the T370Q mutant, that fragment 10, generated by cleavage within the CS-2 domain, was also cleaved more rapidly than WT. Substrate binding to ADAMTS-4 or -5 exosites may affect the association and dissociation rates, binding “enhancing” IGD mutants with a lower association rate and an increased rate of dissociation than WT. This would result in greater availability, in this closed system, of ADAMTS-4 to the CS-2 cleavage sites, which may subsequently enhance CS-2 cleavage.
The G1-E373 fragments following ADAMTS-4 cleavage of the N368Q and T370Q mutants migrated on an SDS-PAGE gel with apparently reduced molecular mass, consistent with the absence of an N-linked oligosaccharide at Asn368. We infer from our data that in WT recombinant bovine aggrecan, glycosylation of Asn368 does occur. The 368AsnIleThr370 motif is very highly conserved between species, suggesting functional importance. N- or O-linked oligosaccharides at this site may influence proteolytic cleavage within the IGD of aggrecan. Our data suggests a greater role for Thr370 in mutagenized aggrecan expressed in COS7 cells. In native aggrecan, this cis-acting effect could be further modulated by varying N- and O-linked KS on the oligosaccharide on Asn368 or O-linked to Thr370. It has been previously shown that N- or O-linked KS are found at Asn368 and/or Thr370 in the steer aggrecan IGD [19]. We conclude that whereas the Thr370Gln mutant would have no glycosylation at Asn368 or Thr370, the Asn368Gln mutant retains a Thr residue that is potentially O-glycosylated. Since prevention of N-glycosylation at Asn368 did not significantly enhance cleavage, we conclude that O-glycosylation at Thr370 may be the inhibitory component of the 368AsnIleThr370 motif. This putative O-glycosylation in recombinant bovine aggrecan is not likely to be KS, as we have determined previously [16]. Although we have shown evidence that inhibition of cleavage may result from O-glycosylation at T370, Poon et al. [20] reported that N-linked KS substitution, likely at Asn368, enhances cleavage of a recombinant peptide substrate by ADAMTS-4. The potential for variable substitution of the 368AsnIleThr370 motif suggests that oligosaccharide(s) at this motif may variably affect cleavage within the IGD.
4.4 The potential role of the Thr352-Thr357 sequence on ADAMTS-4 cleavage of aggrecan
In bovine aggrecan, there are 4–7 Thr or Ser residues N-terminal to the site of scission near each of the ADAMTS-4 cleavage sites. We generated a two triple mutants (Group C) substituting Gln and Val for the three Thr residues in this cluster within the IGD. The T352Q-T355Q-T357Q. mutant showed a small reduction in its susceptibility to ADAMTS-4 compared with WT aggrecan. Substitution of the Thr by Gln residues increased the overall hydrophilicity of this region. Although the V356A-V361A-E362D mutant in Group A showed a somewhat smaller increase in regional hydrophilicity, it showed a much greater decrease in cleavage. This suggests that the hydropathy of specific residues rather than the additive effect of multiple residues may be important for enzyme-substrate recognition.
The T352V-T355V-T357V mutant was significantly more susceptible than WT to cleavage by ADAMTS-4 at Glu373-Ala374. The introduction of Val residues greatly increased the hydrophobicity in this region, suggesting that the introduction of additional hydrophobic residues in this region of aggrecan may strengthen interaction of this region with an ADAMTS-4 exosite. In fact, sequences N-terminal to the ADAMTS-4 cleavage sites within the IGD and CS-2 domain are Val-rich, suggesting that hydrophobic residues may be important for exosite interactions at other aggrecanase sites as well. Westling et al. [18] showed with mutated recombinant versican that replacing the hydrophobic Val residue with hydrophilic Lys at P18 resulted in a reduction in ADAMTS-4 cleavage, consistent with our results. The hydropathy of specific amino acid residues in the region N-terminal to the site of scission may be a key element of the substrate topography determining the specificity of ADAMTS-4 recognition or binding. It should be noted, however, that in native cartilage aggrecan, 2/3 of the Thr residues from P22–P17 are normally substituted with negatively charged KS, which would effectively increase the hydrophilicity of this region. The mutants we have constructed in recombinant aggrecan should therefore be considered proof of principle that increased hydrophobicity at particular sites enhances the interaction with ADAMTS-4. This type of interaction is not unique to aggrecan and aggrecanases. Other examples of hydrophobic contacts between substrate and protease exosites include protease-activated receptor 3 (PAR3) [35], and the interaction between thrombin and fibrin [36].
Most of the mutations studied in our analysis involve residues in the well-conserved amino acid sequence N-terminal to the site of scission in the IGD. Horber et al. [30] have observed that truncation of a COS-7 cell-derived recombinant aggrecan IGD substrate N-terminal to the site of cleavage profoundly inhibited cleavage. These truncated regions of the IGD contained multiple conserved and/or potentially glycoslyated residues. Our site-specific mutants in this region support a role for these conserved amino acid motifs in the aggrecan-ADAMTS-4 interaction.
4.5 ADAMTS-4 and ADAMTS-5 interact with a substrate residue C-terminal to the site of scission
In addition to the conserved motifs on the N-terminal side of the aggrecanase site, there also appears to be a region of the aggrecan IGD, C-terminal (P′) to the site of scission, that is involved in the aggrecan-ADAMTS-4 interaction. The motif surrounding the IGD aggrecanase cleavage site is rigidly conserved in the twelve species shown in Fig. 8A. However, there is limited variability of the amino acid at the P4′ site, which is occupied by Ser, Thr or Asn. The hydropathy plots for the Ser377(Gln, Ala, Thr, Asn) mutants and WT are very similar. Since Asn is common at this site in multiple species, we anticipated that a conservative S377Q substitution at the bovine aggrecan P4′ site would not result in a global change in conformation. It is likely, therefore, that the inhibition of cleavage that we subsequently observed with the S377Q mutant arose from a specific incompatible interaction of ADAMTS-4 with the P4′ residue.
It is interesting that a similar inhibition of cleavage was observed when the conservative S377Q mutant was digested with ADAMTS-5. In addition, ADAMTS-5 cleavage of the conservative S377T mutant was inhibited. Taken together, these results suggest that a limited range of amino acid residues are permissible at the P4′ position. The difference in cleavage susceptibility with the S377GQ substitution in both the ADAMTS-4 and -5 digests, and the S377T substitution in the ADAMTS-5 digest, may be the result of steric hindrance of substrate binding to the catalytic cleft or an exosite. Furthermore, we conclude that ADAMTS-4 and ADAMTS-5 are not identical in their substrate specificity. We are currently investigating other sites of ADAMTS-4 cleavage in the CS-2 domain (Glu1481 and Glu1666) which also have conserved Ser or Thr in the P4′ position. As a component of the P′ ADAMTS-4 motif in phage-displayed peptides, Arg or Lys were frequently found in the P2′ or P3′ position [29]. Occupation of the P2′ site by Arg is nearly invariable between species in the IGD, but an exception to this conservation is seen in murine aggrecan, where the P2′ residue is Leu. A murine species-mimicking mutation of bovine aggrecan at the P2′ position (R375L) showed no apparent difference in cleavage from WT bovine aggrecan. This result suggests that there may be a narrow range for variability between species permitted in this position, perhaps restricted by structural features of the ADAMTS-4 recognition site.
4.6 A proposed mechanism for ADAMTS-4 and ADAMTS-5 recognition and cleavage within the IGD
Based on our findings, we propose a model (Fig. 9) for the interaction between ADAMTS-4 or ADAMTS-5 and the aggrecan IGD. This model is consistent with a two step interaction recently proposed by Wittwer et al [28], wherein a remote region of the substrate binds at an enzyme exosite prior to binding the region surrounding the site of scission to the enzyme active site. It is also consistent with the earlier work of Horber et al [30], who used a recombinant substrate consisting of the IGD of human aggrecan to produce a series of deletion mutants N-and C-terminal to the site of scission at Glu373-Ala374. When the N-terminal region was reduced to 16 amino acids, eliminating the Thr352-Thr357 region examined in the Group C mutants, cleavage by ADAMTS-4 was completely inhibited. A C-terminal deletion leaving only 13 amino acids had no effect upon cleavage. Their findings suggested that the IGD has exosite-binding residues N-terminal to the site of scission. Our study extends these findings, to determine which amino acid residues N-terminal to the scission site may be involved in the exosite-substrate interaction.
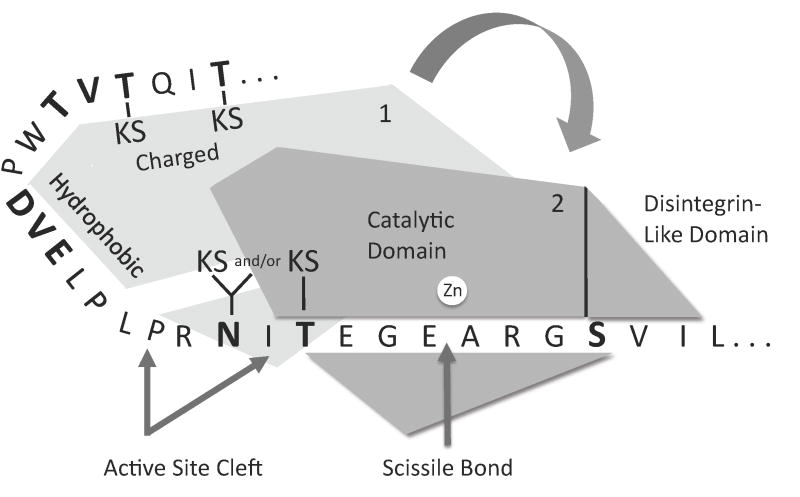
Fig. 9.
Model for interactions between aggrecan IGD and ADAMTS-4 or ADAMTS-5. Initial docking of the proteinase may occur via hydrophobic interactions between exosite residues and a region of the IGD remote from the cleavage site. In the second step, the active site cleft of the exosite-bound proteinase may dock with the cleavage site of the IGD substrate. Additional interactions may occur between Ser373 and the catalytic or disintegrin-like domain. Binding to the catalytic cleft is followed by chain scission at Glu373-Ala374 and dissociation of the exosite-bound and catalytic cleft-bound polypeptides. The indicated KS-substituted Thr and Asn residues are found in cartilage-derived aggrecan, and absent from recombinant aggrecan. Residues that were mutated in this study are shown in boldface.
In Fig. 9, the enzyme and the length of the IGD polypeptide are drawn approximately to scale. The crystal structures of ADAMTS-4 and ADAMTS-5 are known [21], and we have modeled (not shown) the secondary (PredictProtein server [37]) and tertiary (Hyperchem, HyperCube Inc.) structure of the IGD. Polypeptide chain directionality is based on the modeling of enzyme-substrate complexes for MMP-3, ADAM-9, and ADAM-10 whose catalytic domains are homologous to those of ADAMTS-4 and ADAMTS-5 [38]. In the region N-terminal to the site of scission, three closely spaced Pro residues would result in chain rotation and would impart three distinct “kinks” in the polypeptide. There are also a number of other residues (Glu, Thr, Asn, Leu, Asp) that would permit rotation around the amino acid N-Cα and Cα-C bonds [39], conferring conformational flexibility upon this region. Our finding that the hydrophobicity of residues N-terminal to the site of scission potentiates cleavage suggests that these residues interact with an ADAMTS-4 or ADAMTS-5 exosite in the first of the two binding interactions. The exosite-substrate interaction in the model is speculative, since the actual location of the enzyme exosite is unknown. In the model, exosite binding precedes the second binding event between the active site cleft and the region surrounding the scissile bond at Glu373-Ala374. This is followed by dissociation between the N-terminal region of the substrate and the enzyme exosite. Horber et al [30] observed no cleavage in the deletion mutant which excluded Thr352 to Thr357. This Thr-rich region is predicted to be in a β-strand conformation [40] consistent with a functional role. This region may be particularly important to the first binding interaction. Although our mutant recombinant aggrecan contained no KS in this region, native aggrecan is KS substituted at two of the three Thr residues [19]. KS appears to be invariably substituted at Thr357, and is also present at either Thr352 or Thr354. On the other hand, there might also be a hydrophobic interaction involving Val353 and Ile356 within the Thr-rich region. In our study, the Thr352Val, Thr354Val and Thr357Val mutant showed significantly enhanced cleavage, suggesting that this region may have access to a hydrophobic patch on the enzyme. If not involved in binding to charged residues of the exosite, KS chains at this site may negatively influence hydrophobic interactions. Our data suggested that N-glycosylation at Asn368 neither enhanced nor inhibited cleavage, but that O-glycosylation at Thr370 negatively influenced cleavage. In native bovine aggrecan, both Asn368 and Thr370 may be KS substituted, possibly enhancing cleavage [20]. The S377Q mutant inhibits cleavage by ADAMTS-4 (p68) but not by ADAMTS-4 (p40), suggesting that the C-terminal ancillary domains in the p68 isoform may affect substrate binding. Both the S377Q or S377T mutants inhibit cleavage by ADAMTS-5. From its proximity to the cleavage site, we conclude that Ser377 may be an important in the second binding event between the substrate and the catalytic cleft, and may interact with the catalytic domain. Alternatively, it may bind to an exosite within the distintegrin-like domain. Future work will be directed toward proof or disproof of this hypothetical mechanism.
We conclude that ADAMTS-4 may have multiple specific hydrophobic or non-hydrophobic interactions with the substrate that may occur N- (this work) or C-terminal (this work and Hills et al [29]) to the site of cleavage. This information may be useful in prediction of cleavage sites in other protein or proteoglycan substrates degraded by the ADAMTS family of proteinases. Additionally, this work provides an experimental basis for interfering with the interactions between aggrecan and exosites of ADAMTS-4 or ADAMTS-5 as a mechanism for therapeutic intervention in osteoarthritis.
Table 2.
Groups B and C: Primer sequences and template plasmids used for site-direct mutagenesis.
| Mutation(s) | Primer sets | Primer location | Template Plasmid | Clone Number |
|---|---|---|---|---|
| N368Q | 5′-CCTGCCCCGACAGATCACTGAGG-3′ | 1509-31 | 64-5 | 192-9 |
| 5′-CCTCAGTGATCTGTCGGGGCAGG-3′ | 71-28 | 459-1 | ||
| T370Q | 5′-CCCCGAAATATCCAGGAGGGTGAAGCC-3′ | 1513-39 | 64-5 | 196-34,36 |
| 5′-GGCTTCACCCTCCTGGATATTTCGGGG-3′ | 71-28 | 459-5 | ||
| T352Q | 5′-GAGGAGGACATCCAGATCCAGACGGTGACCTG-3′ | 1459-1490 | 71-28 | 457-24 |
| 5′-CAGGTCACCGTCTGGATCTGGATGTCCTCCTC-3′ | ||||
| T352Q-T357Q | 5′-CATCCAGACGGTGCAGTGGCCTGACGTG-3′ | 1473-1500 | 457-24 | 460-6 |
| 5′-CACGTCAGGCCACTGCACCGTCTGGATG-3′ | ||||
| T352Q-T355Q-T357Q | 5′-CATCCAGATCCAGCAGGTGCAGTGGCCTG-3′
5′-CAGGCCACTGCACCTGCTGGATCTGGATG-3′ |
1467-95 | 460-6 | 461-4 |
| T352V | 5′-GAGGAGGACATCGTCATCCAGACGGTG-3′ | 1459-1490 | 71-28 | 458-3 |
| 5′-CACCGTCTGGATGACGATGTCCTCCTC-3′ | ||||
| T352V-T357V | 5′-CATCCAGACGGTGGTCTGGCCTGACGTG-3′ | 1473-1500 | 458-3 | 459-24 |
| 5′-CACGTCAGGCCAGACCACCGTCTGGATG-3′ | ||||
| T352V-T355V-T357V | 5′-CATCACCATCCAGGTGGTGACCTGGCC-3′
5′-GGCCAGGTCACCACCTGGATGGTGATG-3′ |
1467-95 | 459-24 | 460-11 |
Underlined letters indicate changed bases. Amino acid residue numbers (minus 19 residues of the signal peptide) and primer location refer to numbering in Hering et al. [32]. Triple mutants were generated by mutating a previous single and double mutant.
Acknowledgments
We are grateful for the technical assistance of Diane Kocka and Patrick Klepcyk for DNA sequencing.
Funding was provided by NIH grants AG17303, AR47892, and AR20618.
Footnotes
The Abbreviations used are: ADAMTS, A Disintegrin And Metalloproteinase with Thombospondin Motifs, ADAMTS-4 (p68) and ADAMTS-4 (p40), ADAMTS-4 isoforms having molecular masses of 68 kDa and 40 kDa, respectively. “ADAMTS-4” indicates the p68 isoform unless otherwise indicated. APMA, 4-aminophenylmercuric acetate; CS, chondroitin sulfate; ECM, extracellular matrix; EDTA, ethylene diamine tetraacetic acid; G1 domain, N-terminal globular domain of aggrecan; G2 domain, second globular domain of aggrecan; G3 domain, C-terminal globular domain of aggrecan, GAG, glycosaminoglycan; GnHCl, guanidine hydrochloride; HA, hyaluronan; IGD, interglobular domain; KS, keratan sulfate; MMP, matrix metalloproteinases; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; PVDF, polyvinylidine fluoride; ECL, enhanced chemiluminescence.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hughes CE, Caterson B, Little DB, Wainwright SW. Aggrecanases 1 and 2. In: RN, Barrett AJ, Woesneer JF, editors. Handbook of proteolytic enzymes. Elsevier Academic Press; London: 2004. pp. 740–746. [Google Scholar]
- 2.Lee ER, Lamplugh L, Davoli MA, Beauchemin A, Chan K, Mort JS, Leblond CP. Enzymes active in the areas undergoing cartilage resorption during the development of the secondary ossification center in the tibiae of rats ages 0–21 days: I. Two groups of proteinases cleave the core protein of aggrecan. Dev Dyn. 2001;222:52–70. doi: 10.1002/dvdy.1168. [DOI] [PubMed] [Google Scholar]
- 3.Lee ER, Lamplugh L, Leblond CP, Mordier S, Magny MC, Mort JS. Immunolocalization of the cleavage of the aggrecan core protein at the Asn341-Phe342 bond, as an indicator of the location of the metalloproteinases active in the lysis of the rat growth plate. Anat Rec. 1998;252:117–132. doi: 10.1002/(SICI)1097-0185(199809)252:1<117::AID-AR10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Mort JS, Flannery CR, Makkerh J, Krupa JC, Lee ER. Use of anti-neoepitope antibodies for the analysis of degradative events in cartilage and the molecular basis for neoepitope specificity. Biochem Soc Symp. 2003:107–114. doi: 10.1042/bss0700107. [DOI] [PubMed] [Google Scholar]
- 5.Bayliss MT, Hutton S, Hayward J, Maciewicz RA. Distribution of aggrecanase (ADAMts 4/5) cleavage products in normal and osteoarthritic human articular cartilage: the influence of age, topography and zone of tissue. Osteoarthritis Cartilage. 2001;9:553–560. doi: 10.1053/joca.2001.0425. [DOI] [PubMed] [Google Scholar]
- 6.East CJ, Stanton H, Golub SB, Rogerson FM, Fosang AJ. ADAMTS-5 deficiency does not block aggrecanolysis at preferred cleavage sites in the chondroitin sulfate-rich region of aggrecan. J Biol Chem. 2007;282:8632–8640. doi: 10.1074/jbc.M605750200. [DOI] [PubMed] [Google Scholar]
- 7.Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, Rosenfeld SA, Copeland RA, Decicco CP, Wynn R, Rockwellk A, Yang F, Duke JL, Solomon K, George H, Bruckner R, Nagase H, Itoh Y, Ellis DM, Ross H, Wiswall BH, Murphy D, Hillman MCJ, Hollis GF, Newton RC, Magolda RL, Trzaskos JM, Arner EC. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 8.Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Duke JL, George HJ, Hillman MC, Jr, Murphy K, Wiswall BH, Copeland RA, Decicco CP, Bruckner R, Nagase H, Itoh Y, Newton RC, Magolda RL, Trzaskos JM, Burn TC, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 9.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, Majumdar MK, Morris EA. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 10.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, Fourie AM, Fosang AJ. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 11.Flannery CR, Zeng W, Corcoran C, Collins-Racie LA, Chockalingam PS, Hebert T, Mackie SA, McDonagh T, Crawford TK, Tomkinson KN, LaVallie ER, Morris EA. Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J Biol Chem. 2002;277:42775–42780. doi: 10.1074/jbc.M205309200. [DOI] [PubMed] [Google Scholar]
- 12.Gao G, Westling J, Thompson VP, Howell TD, Gottschall PE, Sandy JD. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J Biol Chem. 2002;277:11034–11041. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- 13.Kashiwagi M, Enghild JJ, Gendron C, Hughes C, Caterson B, Itoh Y, Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 14.Fushimi K, Troeberg L, Nakamura H, Lim NH, Nagase H. Functional differences of the catalytic and non-catalytic domains in human ADAMTS-4 and ADAMTS-5 in aggrecanolytic activity. J Biol Chem. 2008;283:6706–6716. doi: 10.1074/jbc.M708647200. [DOI] [PubMed] [Google Scholar]
- 15.Miwa HE, Germen TA, Hering TM. Effects of covalently attached chondroitin sulfate on aggrecan cleavage by ADAMTS-4 and MMP-13. Matrix Biol. 2006;25:534–545. doi: 10.1016/j.matbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Miwa HE, Germen TA, Huynh TD, Flory DM, Hering TM. Mammalian expression of full-length bovine aggrecan and link protein: formation of recombinant proteoglycan aggregates and analysis of proteolytic cleavage by ADAMTS-4 and MMP-13. Biochim Biophys Acta. 2006;1760:472–486. doi: 10.1016/j.bbagen.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Dafforn TR, Smith CJ. Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep. 2004;5:1046–1052. doi: 10.1038/sj.embor.7400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westling J, Gottschall PE, Thompson VP, Cockburn A, Perides G, Zimmermann DR, Sandy JD. ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem J. 2004;377:787–795. doi: 10.1042/BJ20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry FP, Rosenberg LC, Gaw JU, Gaw JU, Koob TJ, Neame PJ. N- and O-linked keratan sulfate on the hyaluronan binding region of aggrecan from mature and immature bovine cartilage. J Biol Chem. 1995;270:20516–20524. doi: 10.1074/jbc.270.35.20516. [DOI] [PubMed] [Google Scholar]
- 20.Poon CJ, Plaas AH, Keene DR, McQuillan DJ, Last K, Fosang AJ. N-linked keratan sulfate in the aggrecan interglobular domain potentiates aggrecanase activity. J Biol Chem. 2005;280:23615–23621. doi: 10.1074/jbc.M412145200. [DOI] [PubMed] [Google Scholar]
- 21.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, Kriz R, Reifenberg EL, Collins-Racie LA, Corcoran C, Freeman B, Zollner R, Marvell T, Vera M, Sum PE, Lavallie ER, Stahl M, Somers W. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shieh HS, Mathis KJ, Williams JM, Hills RL, Wiese JF, Benson TE, Kiefer JR, Marino MH, Carroll JN, Leone JW, Malfait AM, Arner EC, Tortorella MD, Tomasselli A. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- 23.Sandy JD, Thompson V, Doege K, Verscharen C. The intermediates of aggrecanase-dependent cleavage of aggrecan in rat chondrosarcoma cells treated with interleukin-1. Biochem J. 2000;351:161–166. doi: 10.1042/0264-6021:3510161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sztrolovics R, Alini M, Roughley PJ, Mort JS. Aggrecan degradation in human intervertebral disc and articular cartilage. Biochem J. 1997;326:235–241. doi: 10.1042/bj3260235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagishita M, Midura RJ, Hascall VC. Proteoglycans: isolation and purification from tissue cultures. Methods Enzymol. 1987;138:279–289. doi: 10.1016/0076-6879(87)38023-1. [DOI] [PubMed] [Google Scholar]
- 26.Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, Burn T, Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4) J Biol Chem. 2000;275:18566–18573. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- 27.Barnes M, editor. Bioinformatics for geneticists: a bioinformatics primer for the analysis of genetic data. 2. John Wiley & Sons Ltd; West Sussex: 2007. [Google Scholar]
- 28.Wittwer AJ, Hills RL, Keith RH, Munie GE, Arner EC, Anglin CP, Malfait AM, Tortorella MD. Substrate-dependent inhibition kinetics of an active site-directed inhibitor of ADAMTS-4 (Aggrecanase 1) Biochemistry. 2007;46:6393–6401. doi: 10.1021/bi7000642. [DOI] [PubMed] [Google Scholar]
- 29.Hills R, Mazzarella R, Fok K, Liu M, Nemirovskiy O, Leone J, Zack MD, Arner EC, Viswanathan M, Abujoub A, Muruganandam A, Sexton DJ, Bassill GJ, Sato AK, Malfait AM, Tortorella MD. Identification of an ADAMTS-4 cleavage motif using phage display leads to the development of fluorogenic peptide substrates and reveals matrilin-3 as a novel substrate. J Biol Chem. 2007;282:11101–11109. doi: 10.1074/jbc.M611588200. [DOI] [PubMed] [Google Scholar]
- 30.Horber C, Buttner FH, Kern C, Schmiedeknecht G, Bartnik E. Truncation of the amino-terminus of the recombinant aggrecan rAgg1mut leads to reduced cleavage at the aggrecanase site. Efficient aggrecanase catabolism may depend on multiple substrate interactions. Matrix Biol. 2000;19:533–543. doi: 10.1016/s0945-053x(00)00113-x. [DOI] [PubMed] [Google Scholar]
- 31.Mercuri FA, Maciewicz RA, Tart J, Last K, Fosang AJ. Mutations in the interglobular domain of aggrecan alters matrix metalloproteinase and aggrecanase cleavage patterns: Evidence that matrix metalloproteinase cleavage interfers with aggrecanase activity. J Biol Chem In Press. 2000 doi: 10.1074/jbc.275.42.33038. [DOI] [PubMed] [Google Scholar]
- 32.Hering TM, Kollar J, Huynh TD. The complete coding sequence of bovine aggrecan: comparative structural analysis. Archives Biochem Biophys. 1997;345:259–270. doi: 10.1006/abbi.1997.0261. [DOI] [PubMed] [Google Scholar]
- 33.Yan A, Lennarz WJ. Unraveling the mechanism of protein N-glycosylation. J Biol Chem. 2005;280:3121–3124. doi: 10.1074/jbc.R400036200. [DOI] [PubMed] [Google Scholar]
- 34.Lee RT, Lauc G, Lee YC. Glycoproteomics: protein modifications for versatile functions. Meeting on glycoproteomics. EMBO Rep. 2005;6:1018–1022. doi: 10.1038/sj.embor.7400556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bah A, Chen Z, Bush-Pelc LA, Mathews FS, Di Cera E. Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4. Proc Natl Acad Sci U S A. 2007;104:11603–11608. doi: 10.1073/pnas.0704409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL, Medved L. Crystal structure of the complex between thrombin and the central “E” region of fibrin. Proc Natl Acad Sci U S A. 2004;101:2718–2723. doi: 10.1073/pnas.0303440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manzetti S, McCulloch DR, Herington AC, van der Spoel D. Modeling of enzyme-substrate complexes for the metalloproteases MMP-3, ADAM-9 and ADAM-10. J Comput Aided Mol Des. 2003;17:551–565. doi: 10.1023/b:jcam.0000005765.13637.38. [DOI] [PubMed] [Google Scholar]
- 39.Huang F, Nau WM. A conformational flexibility scale for amino acids in peptides. Angew Chem Int Ed Engl. 2003;42:2269–2272. doi: 10.1002/anie.200250684. [DOI] [PubMed] [Google Scholar]
- 40.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]