Abstract
Xeroderma pigmentosum variant (XPV) patients carry germ-line mutations in DNA polymerase η (polη), a major translesion DNA synthesis (TLS) polymerase, and exhibit severe sunlight sensitivity and high predisposition to skin cancer. Using a quantitative TLS assay system based on gapped plasmids we analyzed TLS across a site-specific TT CPD (thymine-thymine cyclobutane pyrimidine dimer) or TT 6-4 PP (thymine-thymine 6-4 photoproduct) in three pairs of polη-proficient and deficient human cells. TLS across the TT CPD lesion was reduced by 2.6–4.4 fold in cells lacking polη, and exhibited a strong 6–17 fold increase in mutation frequency at the TT CPD. All targeted mutations (74%) in polη-deficient cells were opposite the 3’T of the CPD, however, a significant fraction (23%) were semi-targeted to the nearest nucleotides flanking the CPD. Deletions and insertions were observed at a low frequency, which increased in the absence of polη, consistent with the formation of double strand breaks due to defective TLS. TLS across TT 6-4 PP was about twofold lower than across CPD, and was marginally reduced in polη-deficient cells. TLS across TT 6-4 PP was highly mutagenic (27–63%), with multiple mutations types, and no significant difference between cells with or without polη. Approximately 50% of the mutations formed were semi-targeted, of which 84–93% were due to the insertion of an A opposite the template C 5’ to the 6-4 PP. These results, which are consistent with the UV hypermutability of XPV cells, highlight the critical role of polη in error-free TLS across CPD in human cells, and suggest a potential involvement, although minor, of polη in TLS across 6-4 PP under some conditions.
1. Introduction
UV light is an established mutagen and carcinogen in humans, and deficiencies in the ability to repair or tolerate UV lesions cause the severe hereditary disease xeroderma pigmentosum (XP), characterized by extreme sunlight sensitivity and high predisposition to skin cancer [1]. The two major biologically significant UV lesions are the cyclobutane pyrimidine dimers (CPD), and the 6-4 photoproducts (6-4 PP). While 6-4 PP are primarily repaired by error-free nucleotide excision repair (NER), CPD are weak substrates for NER, and therefore frequently escape error free repair [2–4]. During replication CPD are tolerated by translesion DNA synthesis (TLS), which is carried out by DNA polymerase η (polη) with relatively high fidelity. This conclusion is based on the biochemical properties of purified polη [5,6], and on the UV hyper-mutability of cells from patients with the variant form of XP (XPV) [7]. In these patients the lack of polη activity causes clinical symptoms similar to the NER-deficient forms of XP, including high predisposition to skin cancer [1,8]. The slight UV sensitivity and the UV hyper-mutability of cells from XPV patients are generally explained by the activity of an alternative TLS DNA polymerase, most likely polτ, which bypasses the CPD with lower efficiency and higher error frequency compared to polη [9–13], leading to increased sensitivity and hyper-mutability, respectively. Although cells from XPV patients were extensively investigated, to date the direct effects of the lack of polη on TLS across defined CPD and 6-4 PP in cells was not determined. Here we present a quantitative analysis of TLS across site specific TT CPD and TT 6-4 PP lesions using a TLS assay system based on tranfection of cultured cells with gapped plasmids. This was done in three different pairs of polη-proficient and polη-deficient human cells, and included DNA sequence analysis of over a 1000 unselected TLS events. Our results show, for the first time, that TLS across a TT CPD in XPV polη-deficient cells is less efficient and more mutagenic than in polη-proficient cells, and it exhibits a typical mutational signature. In contrast, TLS across a TT 6-4 PP is hardly affected by the absence of polη.
2. Materials and Methods
2.1. Materials
Dulbecco’s phosphate buffered saline without calcium chloride and magnesium chloride (PBS), and Minimum Essential Medium Eagle (MEM) were from Sigma. Roswell Park Memorial Institute medium (RPMI 1640) with 2 mM L-glutamine was from GIBCO/BRL. Trypsin-EDTA, 100 mM glutamine, and a mixture of penicillin and streptomycin for cell culture were from Biological Industries (Beit Haemek, Israel). Fetal Bovine Serum (FBS) was from HyClone. JetPEI was from Polyplus-transfection (Illkirch, France).
2.2. Cell cultures
GM00495 (normal) and GM03055 (XPV) human fibroblasts were purchased from Corriell cell repository. The SV40-transformed human fibroblasts, MRC5 (normal) and XP30RO (sv) (XPV; also designated GM3617 were gifts from A. R. Lehmann (University of Sussex, Brighton, UK). The human fibroblast cells were cultured in MEM supplemented with 2 mM L-glutamine, 100 units/ml of penicillin, 100 µg/ml of streptomycin, and 15% FBS. The polη-null derivative of the type I Burkitt's lymphoma BL2 cell lines was obtained by gene-targeted inactivation as described [14]. The lymphoma cells were cultured in RMPI 1640 supplemented with 2 mM L-glutamine, 100 units/ml of penicillin, 100 µg/ml of streptomycin, and 10% FBS. The cells were incubated at 37 °C in a 5% CO2 atmosphere.
2.3. Construction of DNA substrates and plasmids
DNA oligonucleotides without a lesion were supplied by Sigma-Genosys. The construction of the control gapped plasmid GP20 was previously described [15]. A gap-lesion plasmid with a site-specific TT CPD was prepared using the site specifically modified 11-mer oligonucleotide 5'-GCAAGTTGGAG-3' containing a single site-specific TT CPD (underlined) that was generated as previously described [16]. It was extended to a 53-mer oligonucleotides by ligating to its 5’ end the 21-mer 5’-ACCGCAACGAAGTGATTCCTC-3’, and to its 3’ the 21-mer 5’-CTGGCTACTTGAACCAGACCG-3’, using as a scaffold the 33-mer 5’-AAGTAGCCAGCTCCAACTTGCGAGGAATCACTT-3’. The resulting 53-mer was separated from the scaffold and excess 21-mers on a 12% denaturing polyacrylamide gel containing 8 M urea, and used to construct the gap-lesion plasmid GP-TT-CPD. A gap-lesion plasmid with a site-specific TT 6-4 PP was prepared using the site specifically modified 11-mer oligonucleotide 5'-GCAAGTTGGAG-3' containing a single site-specific TT 6-4 PP (underlined) that was generated as previously described [17]. It was extended to a 53-mer oligonucleotides by ligating to its 5’ end the 21-mer 5’-ACCGCAACGAAGTGATTCCTC-3’, and to its 3’ the 21-mer 5’-GAGGCTACTTGAACCAGACCG-3’, using as a scaffold the 33-mer 5’- AAGTAGCCTCCTCCAACTTGCGAGGAATCAC-3’. The resulting 53-mer was separated from the scaffold and excess 21-mers on a 12% denaturing polyacrylamide gel containing 8 M urea, and used to construct the gap-lesion plasmid GP-TT-6-4 PP.
2.4. In vivo TLS assay
The TLS assay has been previously described [18–20]. Briefly, cells were co-transfected with a DNA mixture containing 100ng of a gapped-lesion plasmid (GP-TT-CPD, or GP-TT-6-4 PP; 6 kanR), 100ng of a control gapped plasmid without a lesion (GP20; cmR), and 5µg of the carrier plasmid pUC18, using jetPEI/DNA complexes for the human fibroblast cells or the NucleofectorTM system (Amaxa, GmbH, Köln, Germany), for the BL2 cell lines. The cells were incubated 24 hours in order to allow repair, and then the DNA was extracted using alkaline lysis conditions and used to transform E. coli reporter strains. The percentage of plasmid repair, of which most occurs by TLS, was calculated by dividing the number of transformants obtained from the gap-lesion plasmid (number of kanR colonies) by the number of transformants obtained from the control gapped-plasmid (number of cmR colonies). A small fraction of gap-lesion plasmids can be repaired by non-TLS events, which involve most likely formation as a double stranded break as an intermediate. These are observed as plasmid isolates with large deletions or insertions. To obtain precise TLS extents, the plasmid repair extents were multiplied by the fraction of TLS events out of all plasmid repair events, as obtained by DNA sequence analysis. To determine the DNA sequence changes that have occurred during plasmid repair, sequence analysis was carried using the TempliPhi DNA Sequencing Template Amplification Kit from GE Healthcare and the BigDye Terminator v1.1 Cycle Sequencing Kit from Applied Biosystems. Reactions were analyzed by capillary electrophoresis on an ABI PRISM™ 3130xl Genetic Analyzer from Applied Biosystems.
3. Results
3.1. The experimental system
To analyze TLS across defined and site-specific UV lesions in a quantitative manner we used an assay system previously developed in our lab, based on tranfection of cultured mammalian cells with gapped plasmids carrying site-specific lesions opposite the gap [18–21]. Briefly. The cells were transiently transfected with a mixture of a gap-lesion plasmid (kanR) carrying the site-specific UV lesion, a control gapped plasmid without the lesion (cmR), and a carrier plasmid (ampR). After allowing time for gap repair in the mammalian cells, the plasmids were extracted under alkaline conditions, such that only plasmids that have been completely filled in and ligated remained intact. These were used to transform E. coli recA cells, which were plated in parallel on LB-kanamycin plates to select for filled in gap-lesion plasmids, and LB-chloramphenicol plates to select for filled in control plasmids. The ratio of kanR/cmR colonies is an indication of the extent of TLS in the mammalian cells. Plasmids were extracted from kanR cells, and the DNA sequence was determined across the original damage site to reveal any sequence changes caused during gap filling. The plasmids do not replicate in the mammalian cells, enabling quantification of the extent of gap filling, but meaning that TLS was assayed uncoupled from replication and in the context of a plasmid. Studies previously performed in our lab have shown that this assay system is useful for studying TLS: It is affected by the cellular composition of DNA polymerases, and is subjected to at least some of the regulatory mechanisms controlling chromosomal TLS [18–21].
3.2. TLS across a TT CPD is reduced in cells lacking polη
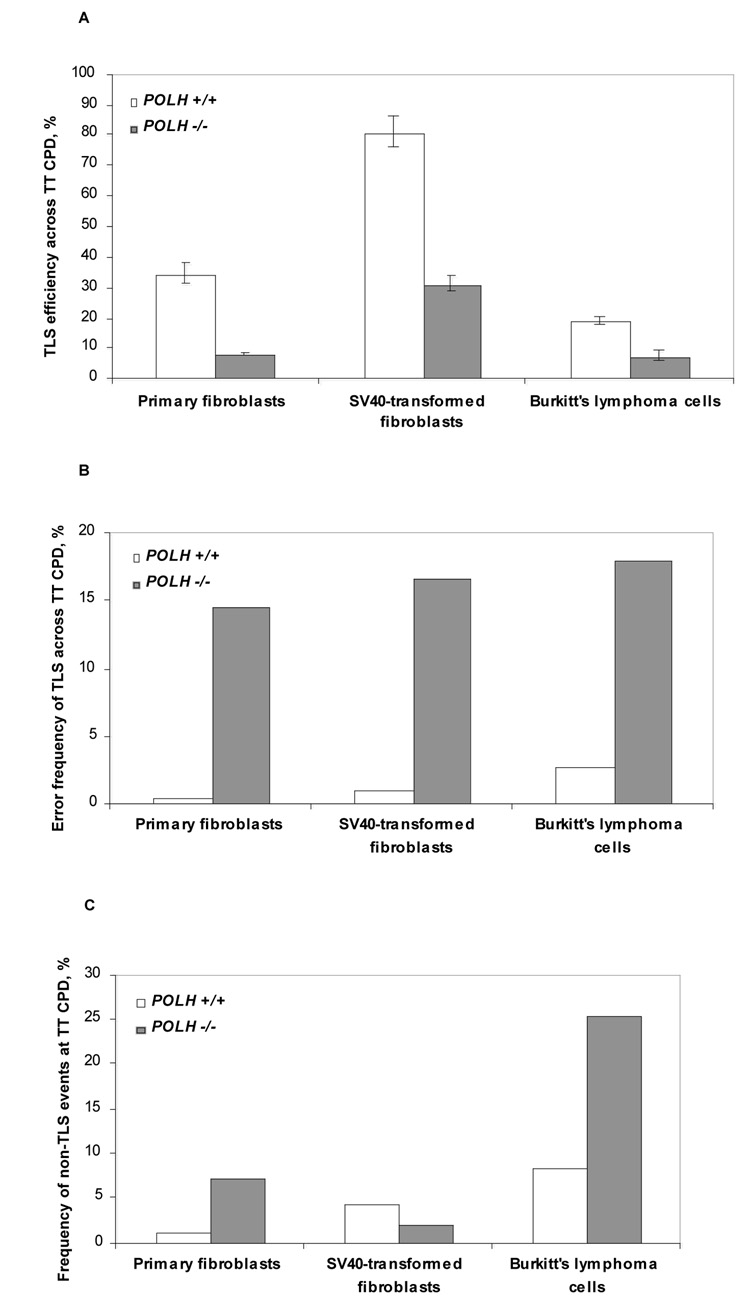
TLS was assayed using a gapped plasmid with a site-specific TT CPD in three pairs of polη-proficient and polη-deficient human cell types: Primary fibroblasts from a normal individual and from an XPV patient, SV40-transformed fibroblasts from a normal individual and from and an XPV patient, and Burkitt lymphoma cell line BL2 and its polη-deficient derivative. As can be seen in Fig. 1A and Table 1, the extent of TLS across the TT CPD in polη-proficient primary fibroblasts was 35%, compared to 8% in primary fibroblasts obtained from an XPV patient, representing a 4.4-fold decrease in the extent of TLS in the absence of polη. In SV40-transformed polη-proficient fibroblasts TLS across the TT CPD was high, reaching 81%, compared to 31% in SV40-transformed fibroblasts obtained from an XPV patient (Fig. 1A and Table 1), representing a 2.6-fold decrease in TLS in the absence of polη. TLS extents in the lymphoma cell lines were 19% and 7% for the polη-proficient and polη-deficient cells, respectively (Fig. 1A and Table 1), representing a 2.7-fold lower TLS in the absence of polη. Thus, despite the presence of multiple additional DNA polymerases in the human cells, the absence of polη caused a significant deficiency in the ability to perform TLS across a TT CPD. The smaller effects in the SV40-transformed fibroblasts and the BL2 lymphoma cells may be due to deregulation of TLS, allowing 'unauthorized' DNA polymerases to perform TLS across the TT CPD, which is usually bypassed by polη.
Fig. 1. TLS across a TT CPD in polη-proficient and polη-deficient cells.
A. The efficiency of TLS. B. The error frequency of TLS. C. The frequency of non-TLS events consisting of large deletions and insertions of sequences from the control plasmid. The data for the graph in panels A was taken from Table 1, and for the graphs presented in panels B and C from Table 2 & Table 3.
Table 1.
TLS across a site-specific TT CPD in polη-proficient and polη-deficient human cells
| Cell type | Transformants |
Plasmid repair, % | TLS, % | ||
|---|---|---|---|---|---|
| KanR | CmR | ||||
| Primary fibroblasts | POLH+/+ | 121 | 357 | 35±3 | 35±3 |
| POLH−/− (XPV) | 100 | 1066 | 9±1 | 8±1 | |
| SV40-transformed fibroblasts | POLH+/+ | 307 | 345 | 85±5 | 81±5 |
| POLH−/− (XPV) | 523 | 1707 | 32±3 | 31±2 | |
| Burkitt's lymphoma cells | POLH+/+ | 223 | 1045 | 21±2 | 19±1 |
| POLH−/− | 105 | 972 | 10±2 | 7±2 | |
Plasmid mixtures containing GP-TT-CPD (kanR) along with the control GP20 (cmR) and the carrier plasmids were introduced into the indicated human cells, after which the DNA was extracted and introduced into an E. coli indicator strain. The extent of plasmid repair was calculated by the kanR/cmR ratio, and the extent of TLS was obtained by calculating this value by the percentage of TLS events out of all repair events, as determined by DNA sequence analysis, and shown in Table 2 and Table3. The colony counts are of a representative experiment. The efficiencies of plasmid repair and TLS were obtained by averaging the results of at least three experiments.
3.3. TLS across a TT CPD is much more mutagenic in cells lacking pol η
To examine the fidelity and mutation types produced during TLS across the TT CPD we extracted kanR colonies from the TLS experiments described above, and subjected them to DNA sequence analysis. As can be seen in Table 2 and Table 3 and Fig. 1B, the vast majority of gap filling events in primary fibroblasts (78/79 events; 99%) are consistent with gap filling by TLS, and all (78/78) had the correct sequence AA inserted opposite the TT CPD. This represents an error frequency of <1.3%, which is consistent with the in vitro error frequency determined for purified human polη ([22,23]). The picture was generally similar in the polη-proficient SV40-transformed fibroblasts and the BL2 lymphoma cell line, where accurate TLS accounted for 94% and 89%, respectively, however the fraction of mutagenic TLS was slightly higher (Table 2 & Table 3, Fig. 1B). The accuracy of TLS across the TT CPD significantly changed in primary fibroblasts from XPV patients, where the fraction of errors made by TLS increased by >11-fold (15% of all isolates; P=0.002; Table 3 & Fig. 1B). Large increases in mutagenic TLS at the CPD were also observed in SV40-transformed fibroblasts from XPV patients, and in the BL2 polη-deficient lymphoma cell line, reaching error frequencies 16-fold (P=0.0003) and 6.4-fold (P=0.003) higher, respectively, than in their cognate polη-proficient cells (Table 3 and Fig. 1B). Thus, in the absence of polη, TLS across the TT CPD becomes more mutagenic by approximately an order of magnitude.
Table 2.
DNA sequence analysis of descendants of a gap-lesion plasmid carrying a site-specific TT CPD after repair in polη-proficient and polη-deficient human cells
| DNA sequence opposite TT CPD |
Primary fibroblasts | SV40-transformed fibroblasts |
Burkitt's lymphoma | |||
|---|---|---|---|---|---|---|
| POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | |
| Number of isolates (%) | ||||||
| C-AA-C | 78 (99) | 54 (78) | 84 (94) | 73 (81) | 64 (89) | 44 (56) |
| A-AA-C | - | 2 (3) | - | 1 (1) | 1 (1.5) | - |
| G-AA-C | - | 1 (1) | - | 1 (1) | - | - |
| C-CA-C | - | - | - | 1 (1) | - | - |
| C-GA-C | - | 3 (4) | - | 5 (6) | - | 6 (8) |
| C-TA-C | - | 2 (3) | - | 5 (6) | - | 7 (9) |
| C-AA-A | - | 2 (3) | - | 1 (1) | - | 1 (1) |
| Δ-AA-C | - | - | 1 (1) | - | - | - |
| C-AT-A | - | - | - | 1 (1) | - | - |
| C-TA-A | - | - | - | - | 1 (1.5) | - |
| Non-TLS events | 1 (1) | 5 (7) | 4 (4) | 2 (2) | 6 (8) | 20 (26) |
| Total | 79 (100) | 69 (100) | 89 (100) | 90 (100) | 72 (100) | 78 (100) |
Plasmids were extracted from kanR colonies obtained in the experiments described in Table 1, and subjected to DNA sequence analysis. The sequence opposite the site of the TT CPD is shown in the 5' to 3' direction. Accurate TLS is represented by the sequence 5'-CAAC-3'. The underlined nucleotides are those located opposite the original TT CPD. Mutations are presented by bold type. Δ represents a single-nucleotide deletion. Non-TLS events include big deletions and insertion of sequences from the control plasmid.
Table 3.
Categories of molecular events occurring during TLS across a TT CPD in polη- proficient and polη-deficient human cells
| Cell type: | Primary fibroblasts | SV40-transformed fibroblasts | Burkitt's lymphoma cells | ||||
|---|---|---|---|---|---|---|---|
| POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | ||
| Category of molecular event | Number of isolates (%) | ||||||
| Error-free TLS | 78 (99) | 54 (78) | 84 (94) | 73 (81) | 64 (89) | 44 (56) | |
| Error-prone TLS | 0 (<1) | 10 (15) P=0.002 | 1 (1) | 15 (17) P=0.0003 | 2 (3) | 14 (18) P=0.003 | |
| Targeted mutations | |||||||
| Transitions at 3' T | 3 | 5 | 6 | ||||
| Transversions at 3' T | 2 | 6 | 7 | ||||
| Small deletions | 1 | ||||||
| Semi-targeted mutations | |||||||
| 3' to the lesion | 3 | 2 | 1 | ||||
| 5' to the lesion | 2 | 1 | 1 | ||||
| Tandem double | 1 | ||||||
| Non-tandem double | 1 | ||||||
| Non-TLS events (insertions or deletions) | 1 (1) | 5 (7) P=0.06 | 4 (4) | 2 (2) P=0.4 | 6 (8) | 20 (26) P=0.005 | |
| Total | 79 (100) | 69 (100) | 89 (100) | 90 (100) | 72 (100) | 78 (100) | |
Based on the data presented in Table 2. The statistical significance of the difference in mutant frequencies between pairs of POLH+/+ and POLH−/− cells was calculated using the χ-square test, and the P values are presented.
3.4. Mutagenic signature of TLS across a TT CPD in XPV cells
Generally the mutational patterns were similar in each of the three polη-deficient cell types (Table 2 & Table 3). First, somewhat unexpectedly, a significant fraction of 23% of all the mutants (9/39) were semi-targeted, appearing not opposite the site of the CPD, but at its nearest flanking nucleotides (roughly equally distributed between the template nucleotides 3' and 5' to the CPD; Table 2 & Table 3). Second, nearly all targeted mutations occurred opposite the 3' T of the CPD (the first to be encountered by the replicating polymerase). This included 97% (29/30) of the targeted mutations, which comprise 74% (29/39) of all mutagenic TLS events. One occurrence was a tandem double base substitution, which included the 5' T of the CPD, and the template G 5' to it.
3.5. Non-TLS events are increased at a TT CPD in cells lacking pol η
Some of the DNA sequences obtained contained deletions and insertions at the original gap-lesion site in the plasmid, which were clearly the result of non-TLS events. The deletions were typically 9–70 bp long, and the insertions were typically 5–50 bp long, originating from the control plasmid GP20. They represent, most likely, events initiated by breakage of the plasmid at the ssDNA region, creating effectively a double strand break (DSB). Such DSB can be repaired by non-homologous end joining (NHEJ) or homologous recombination (HR), as previously shown ([1,24] and Covo and Livneh, in preparation). The non-TLS events are infrequent in normal fibroblasts (1/79; 1.3%), however they increased by approximately 5-fold (5/69; 7%; P=0.06) in primary fibroblasts from XPV patients (Table 3 & Fig. 1C). Similarly, a significant fraction of DSB-related events were observed in BL2 polη-deficient cells, amounting to 26% (20/78) of all isolates, which was 3-fold higher than in the parental BL2 polη-proficient cell line (Table 3 & Fig. 1C). In the SV40-transformed fibroblast cell lines the fraction of DSB-related events was similar with and without polη, but the numbers of events were too small to draw firm conclusions. The increase in the putative DSB-related events observed in cells lacking polη is consistent with strand breakage caused by a difficulty in synthesizing across the CPD due to the deficiency in TLS.
3.6. TLS across a site-specific TT 6-4 PP is marginally affected by the lack of pol η
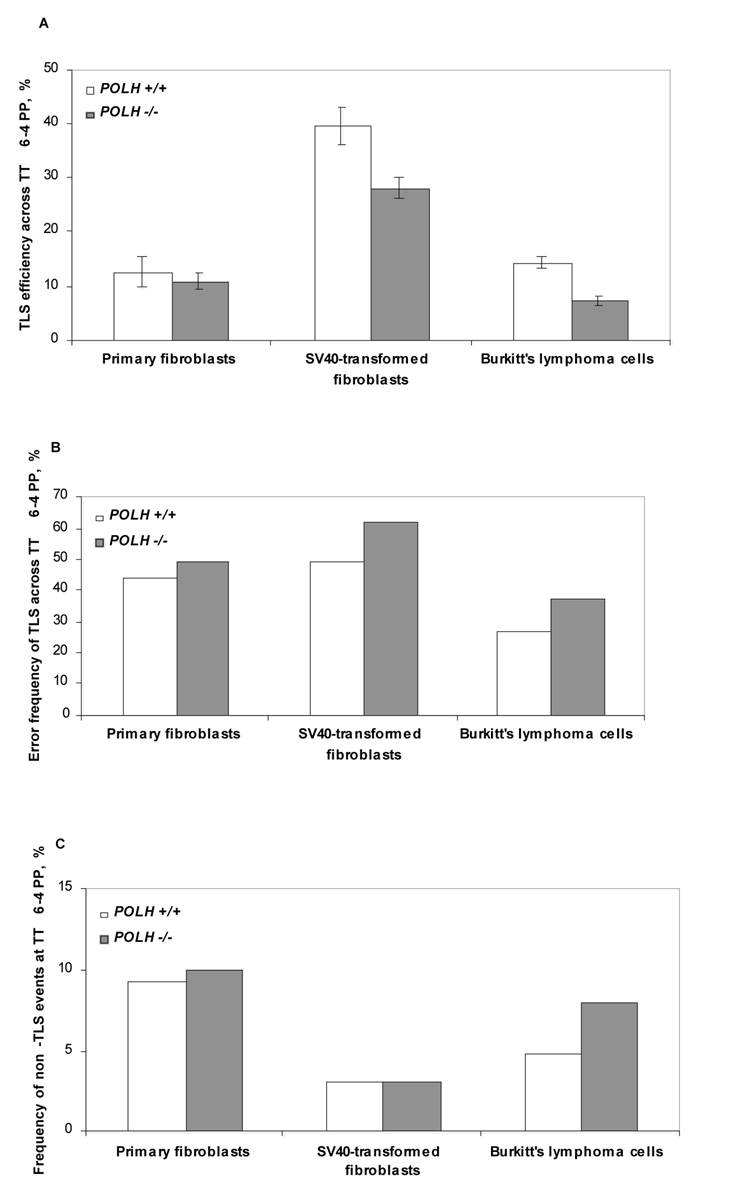
In order to examine TLS across a TT 6-4 PP we performed a similar set of TLS experiments with a gapped plasmid carrying a site-specific TT 6-4 PP in the three pairs of polη-proficient and polη-deficients cell types. As can be seen in Fig. 2A and Table 4, the extent of TLS across the TT 6-4 PP in primary fibroblasts was lower than across TT CPD, suggesting that the TLS machinery copes less well with this type of lesion, perhaps due to the larger deformation that it causes in DNA, and the lack of a specific DNA polymerase which is able to bypass it. When assayed in primary fibroblasts from XPV patients, TLS across TT 6-4 PP was the same as in primary fibroblasts from normal individuals (Fig. 2A & Table 4), suggesting that polη does not play a significant role in TLS across this lesion. In the pair of SV40-transformed fibroblasts the extent of TLS across this lesion was higher than in primary fibroblasts, and there appears to be a slight decrease in TLS in the XPV cell line compared to the polη-proficient line (Fig. 2A and Table 4). Interestingly, in the BL2 polη-deficient cell line, TLS across the TT 6-4 PP was nearly twofold lower than in its polη-proficient parental cell line (Fig. 2A & Table 4). This might be significant, since unlike the previous cell type pairs, this pair of cells is isogenic. Thus, it is possible that at least under some conditions, perhaps when TLS is deregulated, polη might be involved in bypass across a TT 6-4 PP.
Fig. 2. TLS across a TT 6-4 PP in polη-proficient and polη-deficient cells.
A. The efficiency of TLS. B. The error frequency of TLS. C. The frequency of non-TLS events consisting of large deletions and insertions of sequences from the control plasmid. The data for the graph in panels A was taken from Table 4, and for the graphs presented in panels B and C from Table 5 & Table 6.
Table 4.
TLS across a site-specific TT 6-4 PP in polη-proficient and polη-deficient human cells
| Cell type | Transformants |
Plasmid repair, % |
TLS, % | ||
|---|---|---|---|---|---|
| KanR | CmR | ||||
| Primary fibroblasts | POLH+/+ | 323 | 2064 | 14±3 | 13±3 |
| POLH−/− (XPV) | 274 | 2207 | 12±2 | 11±2 | |
| SV-40 transformed fibroblasts | POLH+/+ | 154 | 332 | 41±4 | 40±4 |
| POLH−/− (XPV) | 342 | 1022 | 29±2 | 28±2 | |
| Burkitt's lymphoma cells | POLH+/+ | 88 | 566 | 15±2 | 14±1 |
| POLH−/− | 68 | 790 | 8±1 | 7±1 | |
The experiments were performed as described in the legend to Table 1, except that the gap-lesion plasmid was GP-TT-6-4-PP (kanR). The extent of plasmid repair was calculated by the kanR/cmR ratio, and the extent of TLS was obtained by calculating this value by the percentage of TLS events out of all repair events, as determined by DNA sequence analysis, and shown in Table 5 and Table 6. The colony counts are of a representative experiment. The efficiencies of plasmid repair and TLS were obtained by averaging the results of at least three experiments.
3.7. TLS across TT 6-4 PP is highly mutagenic, with a complex spectrum that is unaffected by the absence of pol η
To establish the type of mutations formed during TLS across a TT 6-4 PP, we performed DNA sequence analysis of plasmids isolated from kanR colonies obtained in TLS experiments with gapped plasmid carrying the site-specific TT 6-4 PP. As can be seen in Fig. 2B and Table 5 & Table 6, TLS across the TT 6-4 PP was highly mutagenic in primary fibroblasts, with 45% (57/128) of the bypass products carrying point mutations. Very similar results were obtained with primary fibroblasts from XPV patients. Also in the two other cell pairs TLS across the TT 6-4 PP was highly mutagenic, with no significant difference between cells with or without polη (Fig. 2B and Table 5 & Table 6).
Table 5.
DNA sequence analysis of descendants of a gap-lesion plasmid carrying a sitespecific TT 6-4 PP after repair in polη-proficient and polη-deficient human cells
| DNA sequence opposite TT CPD | Primary fibroblasts | SV40-transformed fibroblasts | Burkitt's lymphoma cells | ||||
|---|---|---|---|---|---|---|---|
| POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | ||
| Number of isolates (%) | |||||||
| C-AA-C | 59 (46) | 40 (40) | 43 (47) | 22 (34) | 86 (68) | 54 (55) | |
| A-AA-C | - | - | 1 (1) | - | 2 (2) | - | |
| T-AA-C | 1 (1) | 1 (1) | 7 (8) | 1 (2) | - | - | |
| C-CA-C | 1 (1) | - | 1 (1) | 2 (3) | - | 1 (1) | |
| C-GA-C | 5 (4) | 5 (5) | 2 (2) | 2 (3) | 2 (2 ) | 1 (1) | |
| C-TA-C | 2 (2) | 3 (3) | 1 (1) | 2 (3) | - | 2 (2) | |
| C-AC-C | 2 (2) | 1 (1) | 3 (3) | 2 (3) | 1 (1) | - | |
| C-AG-C | 3 (2) | - | - | 2 (3) | 3 (2) | 1 (1) | |
| C-AT-C | 3 (2) | - | 3 (3) | 1 (2) | 1 (1) | 1 (1) | |
| C-AA-A | 25 (20) | 32 (32) | 22 (24) | 17 (27) | 11 (9) | 15 (15) | |
| C-AA-G | - | 1 (1) | - | - | - | - | |
| C-AA-T | - | - | - | 1 (2) | - | 1 (1) | |
| C-ΔA-C | - | - | 4 (4) | 1 (2) | 1 (1) | 3 (3) | |
| C-AΔ-C | 1 (1) | - | - | - | 5 (4) | 8 (8) | |
| C-AA-Δ | - | - | - | 1 (2) | - | - | |
| C-AΔ-Δ | - | - | - | - | - | 1 (1) | |
| A-CA-C | - | 1 (1) | - | - | - | - | |
| A-GA-C | - | - | - | 1 (2) | - | - | |
| A-TA-C | 1 (1) | - | - | - | - | - | |
| T-CA-C | 1 (1) | - | - | - | - | - | |
| C-CC-C | - | 1 (1) | - | - | - | - | |
| C-GC-C | 1 (1) | - | - | - | - | - | |
| C-TG-C | 1 (1) | - | - | - | 1 (1) | - | |
| C-TT-C | - | 2 (2) | - | 1 (2 ) | 1 (1) | 1 (1) | |
| A-AG-C | - | - | 1 (1) | - | - | - | |
| C-GA-A | 3 (2) | 2 (2) | - | 4 (6) | 4 (3) | 2 (2) | |
| C-TA-A | 7 (5) | - | - | 2 (3) | 2 (2) | - | |
| Non-TLS events | 12 (9) | 10 (10) | 3 (3) | 2 (3) | 6 (5) | 8 (8) | |
| Total | 128 (100) | 99 (100) | 91 (100) | 64 (100) | 126 (100) | 99 (100) | |
Plasmids were extracted from kanR colonies obtained in the experiments described in Table 4, and subjected to DNA sequence analysis. The sequence opposite the site of the TT 6-4 PP is shown in the 5' to 3' direction. Accurate TLS is represented by the sequence 5'-CAAC-3'. The underlined nucleotides are those located opposite the original TT 6-4 PP. Mutations are presented by bold type. Δ represents a one-nucleotide deletion. Non-TLS events include big deletions and insertion of sequences from the control plasmid.
Table 6.
Categories of molecular events occurring during TLS across a TT CPD in polη-proficient and polη-deficient human cells
| Cell type: | Primary fibroblasts | SV40-transformed fibroblasts | Burkitt's lymphoma cells | |||
|---|---|---|---|---|---|---|
| POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | POLH+/+ | POLH−/− | |
| Category of molecular event | Number of isolates (%) | |||||
| Error-free TLS | 59 (46) | 40 (40) | 43 (47) | 22 (34) | 86 (68) | 54 (55) |
| Error prone TLS | 57 (45) | 49 (50) | 45 (50) | 40 (63) | 34 (27) | 37 (37) |
| Targeted mutations | ||||||
| Transitions | ||||||
| At 3' T | 5 | 5 | 2 | 2 | 2 | 1 |
| At 5' T | 3 | 2 | 3 | 1 | ||
| Transversions | ||||||
| At 3' T | 3 | 3 | 2 | 4 | 3 | |
| At 5' T | 5 | 1 | 6 | 3 | 2 | 1 |
| Small deletions | 1 | 4 | 2 | 6 | 12 | |
| Semi-targeted mutations | ||||||
| 3' to the lesion | 1 | 1 | 8 | 1 | 2 | |
| 5' to the lesion | 25 | 33 | 22 | 18 | 11 | 16 |
| Tandem double | 4 | 4 | 2 | 2 | 1 | |
| Non-tandem double | 10 | 2 | 1 | 6 | 6 | 2 |
| Non-TLS events (insertions or deletions) | 12 (9) | 10 (10) | 3 (3) | 2 (3) | 6 (5) | 8 (8) |
| Total | 128 (100) | 99 (100) | 91 (100) | 64 (100) | 126 (100) | 99 (100) |
Based on the data presented in Table 5.
The DNA sequence changes caused by TLS across the TT 6-4 PP exhibited a large variety of mutations, with no significant difference between cells with or without polη (Table 5 and Table 6). Notably, the most abundant mutation was a G→T transversion semi-targeted to the template G located 5' to the TT 6-4 PP. It accounted for 44% (25/57) of the mutations in primary normal fibroblasts, with similar abundance in the other cell types. Targeted base substitutions were formed opposite each of the two T bases of the TT 6-4 PP, and small but significant fractions of tandem and no-tandem double base substitutions, and minus 1 deletions, were observed (Table 5 & Table 6).
4. Discussion
The involvement of polη in the relatively error-free TLS across UV lesions, specifically CPD, is well established [5,6,23]. Still previous studies have not measured in a direct and quantitative fashion the effect of the absence of polη on TLS across defined UV lesions. In order to ensure that effects observed in cells lacking polη were indeed due to the absence of this polymerase, we performed our study in three pairs of polη-proficient and polη-deficient human cells. As cell lines frequently accumulate genetic changes that might affect a variety of normal pathways, of the cells used, primary fibroblasts are perhaps the closest to the definition of a normal cell. Primary fibroblasts lacking polη that were obtained from an XPV patient, obviously originated from a different person, and thus were not isogenic to the control normal fibroblasts. Likewise, the second pair of SV40-transformed fibroblasts is not isogenic. In contrast, the third pair was isogenic: the cell line BL2, derived from a Burkitt lymphoma patient, and a polη-knockout derivative of it. The fact that in all 3 cell-pairs TLS across the TT CPD in the polη-deficient cell was less efficient and more mutagenic than in its cognate polη-proficient cell indicates that the these effects are indeed mainly due to the lack of polη.
There was a variation among the three polη-proficient cell types in the extent of TLS across the TT CPD, varying from 21% to 85%. A similar variation, from 15% to 41% was observed for TLS across the TT 6-4 PP. The reasons for these variations are not clear, but they may stem, at least in part from differences in the regulation or performance of the TLS machinery. For example, the higher TLS in SV40-transformed fibroblasts, in which p53 is inactivated [25], is consistent with the finding that p53 suppresses TLS [20]. This correlation does not hold for the BL2 cell lines, which overexpress Mdm2, thereby causing a deficiency in p53 [26]. In any case, for all three cell-pairs, TLS across TT CPD was lower in the polη-deficient cell compared to its polη-proficient control by 2.6-4.4 fold. Thus, despite the presence of a multiplicity of other DNA polymerases in the cell, the lack of polη cannot be fully compensated as far as efficiency of TLS is concerned.
When the accuracy of TLS across the TT CPD is considered, in all three cell-pairs there was a dramatic increase of about an order of magnitude in the mutagenicity of TLS across a TT CPD in cells lacking polη compared to their polη-proficient counterparts. Examination of the mutations caused by the TT CPD in cells lacking polη revealed nearly equal numbers of transversions and transitions, most of which were targeted to the 3’ T of the TT CPD. Most interestingly, nearly a quarter of the mutations were semi-targeted to the two nearest nucleotides flanking the TT CPD. To our knowledge, this is the first demonstration that TLS across a defined TT CPD is much more mutagenic in mammalian cells lacking polη in general, and cells from XPV patients in particular, and the first report on the specificity of its mutagenicity.
The mutagenic spectrum of the 6-4 adducts is complex, and includes nearly every possible point mutation opposite the 6-4 PP, and the two nearest bases flanking it. This highlights the big structural deviation of this lesion from the structure of two adjacent normal T bases, and the difficulty of fitting a particular base in the active site of the polymerase performing the insertion opposite the lesion. It is therefore remarkable that in 35–59% of the TLS events the correct AA nucleotides are inserted opposite the TT 6-4 PP. However, this might not represent the ability of the TLS system to correctly pair the dAMP residues opposite the TT 6-4 PP, but rather a fortuitous insertion according to the ‘A rule’, whereby an A is preferentially inserted opposite non-coding lesions such as an abasic site [18,27,28] or a –(CH2)3- insert [21]. The hotspot of the semi-targeted G→T transversion at the template G located 5' to the TT 6-4 PP may be another manifestation of the local deformation caused by the 6-4 PP, which affects the base pairing at neighboring template base, as are the tandem double and non-tandem double point mutations in this site.
Previous studies have used site specific TT CPD and TT 6-4 PP in various forms of vectors in mammalian cells, however, as the assays in those studies were not quantitative, the efficiency of TLS could not be estimated [29,30]. As far as mutational specificity is concerned, the mutational spectrum of a TT CPD was previously not determined in XPV cells. In contract, the mutability of a TT 6-4 PP was determined in monkey cells, using a replicating plasmid [29,30]. Our results are comparable to those obtained in the COS cells, including the semi-targeted hot spot. Interestingly, while we found essentially no difference in insertion specificity opposite TT 6-4 PP in human cells with or without polη, the situation in S. cerevisiae might be different. Thus, it was reported that in this organism mutagenic TLS across TT 6-4 PP was strongly suppressed in a rad3 Δstrain lacking polη [31]. A second study, using a different sequence context around the lesion, found only a very small difference [32]. These differences may reflect differences in DNA sequence context, or a difference between humans and S. cerevisiae in the DNA polymerases involved in TLS across TT 6-4 PP, reflecting perhaps the fact that the former have more TLS polymerases [33].
Several studies suggested that CPD rather than 6-4 PP are the main premutagenic lesions, perhaps because 6-4 PP are both less abundant, and repaired much better than CPD [34–37]. However, the identity of the polymerase that causes CPD-induced mutations in wild-type cells is not clear. The main candidate is polη, since it is responsible for the majority of TLS across CPD in human cells [38]. Most UV induced mutations occur at cytosines, suggesting involvement of cytosine-containing CPD. The latter have been shown to undergo facilitated spontaneous deamination, which converts them into U-containing CPD, i.e., UT, TU and UU [39,40] which exhibit coding properties similar to TT CPD [41,42]. Thus, the combination of spontaneous deamination of C-containing CPD, followed by bypass of the resultant U-containing CPD by polη provides a potent mutagenic pathway for UV induced C→T transitions. It has been estimated that C-containing CPD in DNA deaminate in vitro with a half-life of approximately 5 h [40]. Thus, the contribution of this process to UV mutagenesis might be significant, depending on the in vivo rates of deamination on one hand, and TLS on the other hand. In any case, it should be kept in mind that other polymerases can substitute for polη in bypass across CPD, and with a much higher error frequency as shown above, a process that might also contribute to the mutagenesis of UV.
Analysis of UV mutations in the HPRT gene in human cells showed that 33% and 42% occurred at a template T, in normal and XPV fibroblasts, respectively, for which the results obtained with the TT CPD are directly relevant [43]. In that study one of the main differences in the UV mutational spectra of normal and XPV cells, was a strong increase in the occurrence of transversions at the expense of transitions in the latter. Thus, in XPV cells 62% of the mutations were transversions, whereas in normal cells they accounted to only 14%. When mutations at only T are concerned, the frequency was 58% transversions versus 14%, respectively [43]. Similar results were obtained also in assay systems based on shuttle vectors [44]. Our results presented above showed that in XPV cells 52% (15/29) of the base substitutions targeted to the lesions were transversions, and the majority of those were T→A, which may explain the occurrence of this mutation in about 50% of the transversions at AT pairs in the HPRT system.
Failure to bypass DNA lesions may lead to the formation of DSB, and subsequent deletions and chromosomal abnormalities. In particular, it was shown that in XPV cells, UV light, which does not directly form DSB in DNA, caused the appearance of DSB, presumably due to the inability to replicate across UV lesions, which may be responsible, at least in part, for the UV sensitivity of these cells [45,46]. Interestingly, although the gapped plasmid system does not involve replication, we have observed that when the TT CPD-containing gapped plasmid was analyzed in cells lacking polη, the fraction of non-TLS events consisting of large deletions and insertions significantly increased. The simplest explanation for these events is the formation of a break in the ssDNA region of the plasmid due to failure of TLS. This is practically a DSB in the plasmid, and it may be repaired by either NHEJ, with the accompanying formation of deletions, or homologous recombination repair with the control intact plasmid [47,48]. This effect appears to be specifically the result of failure to bypass the TT CPD, since it was not observed for the TT 6-4 PP lesions, in which there was no difference in the frequency of deletions/insertions TLS events between polη-proficient and deficient cells.
The skin cancer predisposition of XPV patients is explained by a germ-line mutation in the POLH gene that renders them polη-deficient, combined with the exposure to sunlight that produces DNA lesions, leading to increased mutagenesis and therefore facilitated carcinogenesis. Indeed, it is long known that cells from XPV patients exhibit UV sensitivity and hypermutability [1]. The identification of CPD as the major mutagenic UV lesion, along with the ability of purified polη, which is lacking in XPV patients, to efficiently and relatively accurately bypass CPD, led to the explanation that much of the phenotypes of XPV cells can be explained by a deficiency in TLS across CPD [1]. The results presented here provide direct supporting evidence to this explanation, by demonstrating the TLS across a TT CPD in the absence of polη is less efficient, and much more mutagenic.
Acknowledgements
We thank A. Lehmann (Falmer, Brighton, UK) for the MRC5 and XP30RO cells. This work was supported by grants to ZL from the Flight Attendant Medical Research Institute, Florida, USA, the Israel Science Foundation (no. 564/04), and the M.D. Moross Institute for Cancer Research, Weizmann Institute of Science. We thank Dr. A. Kolbanovskiy for the synthesis of some of the TT CPD & TT 6-4 PP lesions, work supported by NIH/NCI Grant CA099194 at New York University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington DC: ASM Press; 2006. [Google Scholar]
- 2.Vreeswijk MP, van Hoffen A, Westland BE, Vrieling H, van Zeeland AA, Mullenders LH. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J. Biol. Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 3.Tung BS, McGregor WG, Wang YC, Maher VM, McCormick JJ. Comparison of the rate of excision of major UV photoproducts in the human HPRT gene of normal and xeroderma pigmentosum variant cells. Mutat. Res. 1996;362:65–74. doi: 10.1016/0921-8777(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima S, Lan L, Kanno S, Takao M, Yamamoto K, Eker AP, Yasui A. UV light-induced DNA damage and tolerance for the survival of nucleotide excision repair-deficient human cells. J. Biol. Chem. 2004;279:46674–46677. doi: 10.1074/jbc.M406070200. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 7.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 9.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, Masutani C, Hanaoka F. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol. Cell. Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. USA. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 12.Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota. EMBO J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Yuan F, Wu X, Taylor J-S, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucl. Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA. Role of human DNA polymerases eta, iota, and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008 doi: 10.1016/j.dnarep.2008.05.012. In Press. [DOI] [PubMed] [Google Scholar]
- 15.Tomer G, Livneh Z. Analysis of unassisted translesion replication by the DNA polymerase III holoenzyme. Biochemistry. 1999;38:5948–5958. doi: 10.1021/bi982599+. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee SK, Christensen RB, Lawrence CW, LeClerc JE. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc. Natl. Acad. Sci. USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeClerc JE, Borden A, Lawrence CW. The thymine-thymine pyrimidine-pyrimidone(6-4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3' thymine-to-cytosine transitions in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avkin S, Adar S, Blander G, Livneh Z. Quantitative Measurement of Translesion Replication in Human Cells: Evidence for Bypass of Abasic Sites by a Replicative DNA Polymerase. Proc. Natl. Acad. Sci. USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells. The Role of DNA polymerase k. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 20.Avkin S, Sevilya Z, Toube L, Geacintov NE, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Adar S, Livneh Z. Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair (Amst) 2006;5:479–490. doi: 10.1016/j.dnarep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 23.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 24.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 25.Bryan TM, Reddel RR. SV40-induced immortalization of human cells. Crit. Rev. Oncog. 1994;5:331–357. doi: 10.1615/critrevoncog.v5.i4.10. [DOI] [PubMed] [Google Scholar]
- 26.Capoulade C, Bressac-de Paillerets B, Lefrère I, Ronsin M, Feunteun J, Tursz T, Wiels J. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt's lymphoma cells. Oncogene. 1998;16:1603–1610. doi: 10.1038/sj.onc.1201702. [DOI] [PubMed] [Google Scholar]
- 27.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 28.Strauss BS. The 'A rule' of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 29.Gentil A, Le Page F, Margot A, Lawrence CW, Borden A, Sarasin A. Mutagenicity of a unique thymine-thymine dimer or thymine-thymine pyrimidine pyrimidone (6-4) photoproduct in mammalian cells. Nucleic Acids Res. 1996;24:1837–1840. doi: 10.1093/nar/24.10.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya H, Iwai S, Kasai H. The (6-4) photoproduct of thymine-thymine induces targeted substitution mutations in mammalian cells. Nucl. Acids Res. 1998;26:2611–2617. doi: 10.1093/nar/26.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bresson A, Fuchs RPP. Lesion bypass in yeast cells: Pol eta participates in a multi- DNA polymerase process. EMBO J. 2002;21:3881–3887. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutyl dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 34.Asahina H, Han Z-B, Kawanishi M, Kato T, Jr, Ayaki H, Todo T, Yagi T, Takebe H, Ikenaga M, Kimura SH. Expression of a mammalian DNA photolyase confers light-dependent repair activity and reduces mutations of UV-irradiated shuttle vectots in xeroderma pigmentosum cells. Mutat. Res. 1999;435:255–262. doi: 10.1016/s0921-8777(99)00051-8. [DOI] [PubMed] [Google Scholar]
- 35.You Y-H, Lee D-H, Yoon J-H, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 36.Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Human Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
- 37.Pfeifer GP, You Y-H, Besaratinia A. Mutations induced by ultraviolet light. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 38.Stary A, Kannouche P, Lehmann AR, Sarasin A. Role of DNA polymerase eta in the UV mutation spectrum in human cells. J. Biol. Chem. 2003;278 doi: 10.1074/jbc.M211838200. 18767-18718-18775. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Rubio M, Bockrath R. On the possible role of cytosine deamination in delayed photoreversal mutagenesis targeted at thymine-cytosine dimers in E. coli. Mutation Res. 1989;210:93–102. doi: 10.1016/0027-5107(89)90048-1. [DOI] [PubMed] [Google Scholar]
- 40.Barak Y, Cohen-Fix O, Livneh Z. Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA: Significance for UV light mutagenesis. J. Biol. Chem. 1995;270:24174–24179. doi: 10.1074/jbc.270.41.24174. [DOI] [PubMed] [Google Scholar]
- 41.Jiang N, Taylor JS. In vivo evidence that UV-induced C-->T mutations at dipyrimidine sites could result from the replicative bypass of cis-syn cyclobutane dimers or their deamination products. Biochemistry. 1993;32:472–481. doi: 10.1021/bi00053a011. [DOI] [PubMed] [Google Scholar]
- 42.Takasawa K, Masutani C, Hanaoka F, Iwai S. Chemical synthesis and translesion replication of a cis-syn cyclobutane thymine-uracil dimer. Nucleic Acids Res. 2004;32:1738–1745. doi: 10.1093/nar/gkh342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang YC, Maher VM, Mitchell DL, McCormick JJ. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters HL, Seetharam S, Seidman MM, Kraemer KH. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J. Invest. Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]
- 45.Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Polymerase eta deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl. Acad. Sci. USA. 2000;97:7039–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Squires S, Coates JA, Goldberg M, Toji LH, Jackson SP, Clarke DJ, Johnson RT. p53 prevents the accumulation of double-strand DNA breaks at stalled-replication forks induced by UV in human cells. Cell Cycle. 2004;3:1543–1557. doi: 10.4161/cc.3.12.1272. [DOI] [PubMed] [Google Scholar]
- 47.van den Bosch M, Lohman PH, Pastink A. Double strand-break repair by homologous recombination. Biol. Chem. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- 48.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]