Abstract
Background
It has been shown that mutations in at least four myotubularin family genes (MTM1, MTMR1, 2 and 13) are causative for human neuromuscular disorders. However, the pathway and regulative mechanism remain unknown.
Methodology/Principal Findings
Here, we reported a new role for Mtmr8 in neuromuscular development of zebrafish. Firstly, we cloned and characterized zebrafish Mtmr8, and revealed the expression pattern predominantly in the eye field and somites during early somitogenesis. Using morpholino knockdown, then, we observed that loss-of-function of Mtmr8 led to defects in somitogenesis. Subsequently, the possible underlying mechanism and signal pathway were examined. We first checked the Akt phosphorylation, and observed an increase of Akt phosphorylation in the morphant embryos. Furthermore, we studied the PH/G domain function within Mtmr8. Although the PH/G domain deletion by itself did not result in embryonic defect, addition of PI3K inhibitor LY294002 did give a defective phenotype in the PH/G deletion morphants, indicating that the PH/G domain was essential for Mtmr8's function. Moreover, we investigated the cooperation of Mtmr8 with PI3K in actin filament modeling and muscle development, and found that both Mtmr8-MO1 and Mtmr8-MO2+LY294002 led to the disorganization of the actin cytoskeleton. In addition, we revealed a possible participation of Mtmr8 in the Hedgehog pathway, and cell transplantation experiments showed that Mtmr8 worked in a non-cell autonomous manner in actin modeling.
Conclusion/Significance
The above data indicate that a conserved functional cooperation of Mtmr8 with PI3K regulates actin filament modeling and muscle development in zebrafish, and reveal a possible participation of Mtmr8 in the Hedgehog pathway. Therefore, this work provides a new clue to study the physiological function of MTM family members.
Introduction
PTEN (phosphatase and tensin homolog deleted on chromosome ten) and MTM (myotubularin myopathy) family factors are members of the growing class of dual-specificity phosphatases (DSPs), which can dephosphorylate the products of phosphoinositide 3-kinase (PI3K) and antagonize downstream effectors using 3-phosphoinositides as ligands [1], [2]. They have been known to contribute to diverse processes that include cellular adhesion, signal transduction, and cell-cycle regulation[3]. Myotubularin-related (MTMR) phosphatases display a conserved active site motif CX5R, and an invariant sequence ‘CSDGWDR’ exists commonly in all the enzymatically active members[4]–[6]. Eight active members, including MTMR8, have been found in the MTM family. Several studies have further demonstrated that the PH/G domain functions to localize MTM to different subcellular compartments in the cell[7]–[9], and its deletion leads to activity loss of MTMR3 in vitro [10].
Mutations or altered expression of PTEN or MTM family members have been observed in human cancers[11]–[13] and in genetic development defects[14], [15]. Recent studies using RNA interference have revealed an unexpected role for several MTMs (including MTMR8) in promoting cell proliferation and survival [16], [17]. MTM1 is mutated in individuals with X-linked recessive myotubular myopathy [14]. MTM1 is adjacent to MTMR1 on the X chromosome, which plays a role in muscle formation and represents abnormal mRNA splicing in myotonic dystrophy [18]. Function loss of MTMR2 and mutation of MTMR13 causes Charcot–Marie–Tooth disease type 4B1 and 4B2[17], [19], a severe demyelinating neuropathy characterized by muscle weakness and sensory loss in the lower extremities beginning in early childhood. In addition, MTMR6 and MTMR9 were identified genetically to be required in Caenorhabditis elegans for endocytosis[20], [21]. Recently, the cell-therapy strategies, which use cultured myoblasts[22] or stem cells, have had notable successes in dystrophic mouse models[23] and DMD muscle[24]–[26], because transplanting whole cells have the potential to correct the symptoms of disease although each has its inherent disadvantages[27].
PI3K is conserved across eukaryotic organisms and regulates many facets of pathways involving cellular growth, survival, metabolism, vehicle trafficking, and chemotaxis[28]. The pathways controlling random movement parallel those of the amplification step of chemotaxis that is controlled through a regulatory loop containing Ras, PI3K, PTEN, and F-actin[29]. The initial activation of Ras and PI3K is independent of F-actin polymerization. However, F-actin polymerization is essential for amplifying the signal and stabilizing the leading edge in neutrophils and Dictyostelium [30], [31]. In Dictyostelium, F-actin recruits additional PI3K to the newly forming leading edge, enhancing the PIP3 response and downstream effector function [29], [30]. Cells with decreased PI3K activity exhibit a decrease in the second peak of RacB activation and F-actin polymerization, which has been linked to pseudopod extension.
To reveal the biological functions of MTM family members, we identified Mtmr8 from the model animal zebrafish. Based on its expression pattern predominantly in the eye field and somites during early somitogenesis, we analyzed its physiological roles in embryo development by morpholino-mediated knockdown. We firstly showed that loss-of-function of Mtmr8 led to defects in somitogenesis, and further examined the possible underlying mechanism and the PH/G domain function. Moreover, we investigated the cooperation of Mtmr8 with PI3K in actin filament modeling and muscle development, and revealed a possible participation of Mtmr8 in the Hedgehog pathway. The findings revealed a new role of Mtmr8 and its functional mechanism in neuromuscular development.
Results
Molecular characterization and expression pattern of Mtmr8 in zebrafish embryos
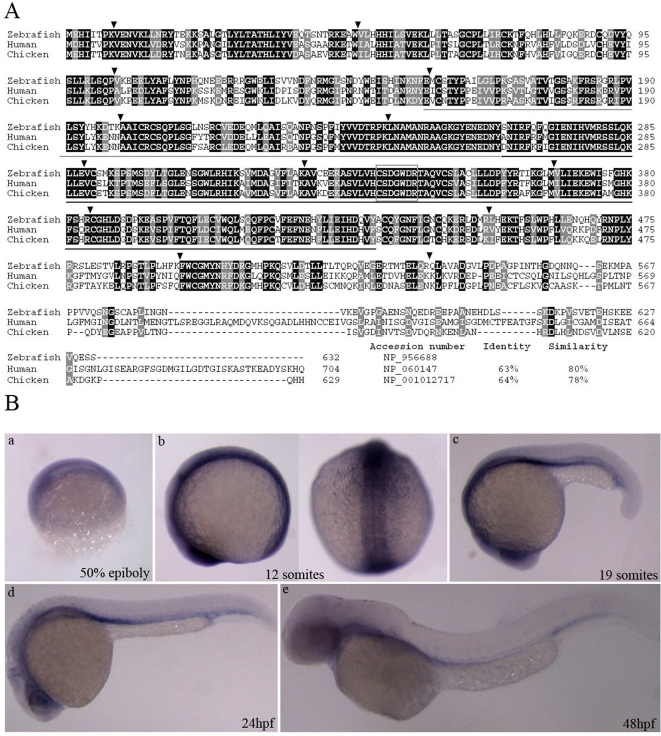
The complete ORF for the Mtmr8 encodes polypeptides of 632 amino acids, which contains 14 exons and 13 introns (identical to human MTMR8). An amino acid sequence alignment of zebrafish, chicken and human Mtmr8 polypeptides is shown in Fig. 1A. Zebrafish Mtmr8 exhibit 63% and 64% identity, and 80% and 78% similarity, with human and chicken Mtmr8 respectively, and higher identities exist in the Myotub-related and PTPc_DSPc motifs (amino acids 155–263 and 264–432 of zebrafish Mtmr8). The high homology implies that the zebrafish Mtmr8 may have the same functions as in human.
Figure 1. Sequence comparison and expression pattern of the deduced zebrafish Mtmr8.
(A) Amino acid alignment of Mtmr8 between zebrafish with human and chicken. Similar and identical amino acids are highlighted in grey and black boxes. And the percentages of identities and similarities in Mtmr8 were shown compared zebrafish with others. Arrowheads indicate the location of introns and are flanked by the corresponding exon numbers. The region encompassing the Myotub-related and PTPc_DSPc domain is underlined in grey and black lines. The rectangular box indicates the CX5R active site motif of enzymatically active members in the MTM family. (B) Expression pattern of zebrafish Mtmr8. Whole-mount RNA in situ hybridization were performed using a Mtmr8 specific antisense riboprobe on embryos at the indicated stages. The arrows indicate the signals in the anterior and head. Embryos in panels are lateral view with the animal pole toward the top, and the right picture of panel b is dorsal view. The embryos in other panels are lateral views, with dorsal toward the top and anterior to the left. All scale bars are 100 µm.
Whole-mount in situ hybridization was used to analyze the expression pattern of Mtmr8 during zebrafish embryogenesis. The expression distribution of Mtmr8 was same to the result reported by Thisse B and C [32]. As shown in Fig. 1B, Mtmr8 mRNA is expressed in prechordal plate and eye field at 50% epiboly (Fig. 1B-a). Between 1–13 somites, Mtmr8 transcript becomes restricted to eye field and somites (Fig 1B-b). At 19 somites, Mtmr8 expression is shown in the eye, telencephalon and ventral mesoderm (Fig 1B-c). At 24hpf, Mtmr8 is expressed predominantly in the eye and vasculature (Fig 1B-d). Later, Mtmr8 is expressed in the vasculature at 48hpf (Fig 1B-e).
Targeted knockdown of zebrafish Mtmr8 impaired embryo development
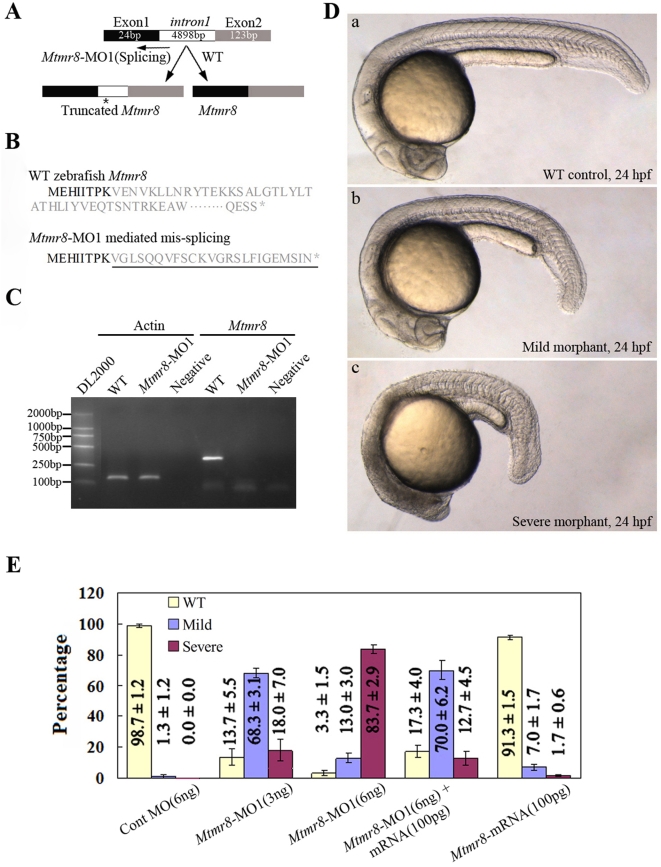
To determine the physiological effect of Mtmr8, we undertook loss-of-function experiments in zebrafish by using morpholinos. Firstly, we designed a splice junction morpholino targeted against the first coding exon-intron boundary to evaluate the knockdown effect and efficacy in vivo. As shown in Fig. 2A and Fig. 2B, the exon-intron morpholino (MO1) introduces intron 1 (about 5 kb) into the altered transcript, and includes a premature termination codon. Because the altered transcript is more than 5kb in its sequence, it could not be amplified from the morpholino-injected embryos in one minute by RT-PCR (Fig. 2C). In the control embryos, the amplified PCR product is 310 bp by the same primers designed from the first and third extrons (Fig. 2C). And, the altered transcript sequence was further amplified and verified by bi-directional DNA sequencing. Microscope observation indicated that the morpholino-mediated Mtmr8 knockdown resulted in dramatic phenotypic abnormalities in somitogenesis (Fig 2D). At 24hpf, control experiments in which embryos were injected with Cont morpholino did not alter the wild-type phenotype throughout zebrafish development (Fig 2D-a), whereas the Mtmr8-MO1 morphants exhibited dose-dependent effect on embryos and resulted in mild and severe phenotype defects. In mild defects, morpholino-injected embryos showed delayed development, shorter and curled trunk and tail compared to wild-type or to control MO-injected embryos (Fig. 2D-b). In severe defects, the morphants were marked by small heads, abnormal tail fins, and U-shaped somites (Fig. 2D-c). Fig. 2E shows the percentages of normal, mild and severe defect embryos under different injection doses of morpholino. Moreover, an increasing percentage of normal or mild defect embryos was observed when the embryos were co-injected with 100 pg capped Mtmr8 RNA and 6 ng Mtmr8 MO1 (Fig. 2E). However, the defects could not be reduced when co-injected with 100 pg of capped GFP RNA and Mtmr8 MO1 (Data not shown). In addition, overexpression of zebrafish Mtmr8 by injection of capped RNA (100 pg) did not cause a visible phenotype. We used 100 pg doses in all the gain-of-function experiments described below if not indicated otherwise.
Figure 2. Targeted knockdown of Mtmr8 using splicing morpholino in zebrafish embryos.
(A) Diagram of splicing junction morpholino targeted against Mtmr8 exon-intron boundary. (B) Amino acid sequences of wild type Mtmr8 and Mtmr8-MO1 mediated mis-splicing Mtmr8. Sequencing of the RT-PCR products revealed the mis-splicing transcript leading to a premature stop (asterisk) and causing a truncation in the protein (exon 1 in bold, the other exons in plain, and intron in plain and underlined). (C) RT-PCR detection of Mtmr8 transcript at 24hpf in WT and Mtmr8-MO1 (6 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (D) Live morphology of WT control zebrafish embryo (a) and Mtmr8-MO1 knockdown embryo (b, c) at 24hpf. Injection volume was about 2 nL at 1-cell stage embryos. All scale bars are 100 µm. (E) Statistical data of three independent experiments on Mtmr8 knockdown as well as its overexpression and Mtmr8 mRNA rescue. Results are represented as mean±SD of three separate experiments (60 embryos in each experiment).
Role of Mtmr8 in regulating PI3K/Akt signaling pathway
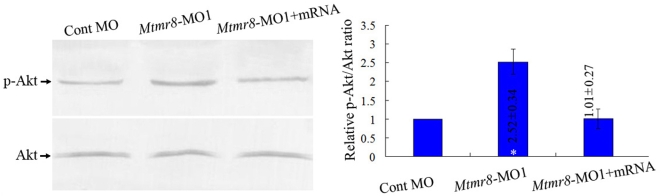
Recent studies indicated that several MTMs might control PI3K/Akt activation by virtue of its ability to dephosphorylate PIP3[33]. However, there was no any direct report about the PIP3 lipid phosphatase activity of Mtmr8. Because knockdown of Mtmr8 expression gave distinct developmental phenotypes, we asked whether the PIP3 lipid phosphatase activity of Mtmr8 was conserved in the development of zebrafish. To confirm it, we compared the level of phosphorylated Akt (pAkt) to total Akt (Akt) in whole fish lysates prepared from wild type zebrafish and Mtmr8 morphant embryos. As shown in Fig 3, knockdown of Mtmr8 expression produces a significant increase in the relative amount of pAkt compared to Cont MO injected embryos, whereas the elevated level of p-Akt could be reduced by coinjection with Mtmr8 mRNA. These results indicate that zebrafish Mtmr8 exhibits PIP3 lipid phosphatase activity and functions to negatively regulate the PI3-Kinase/AKT pathway.
Figure 3. Phosphatase activity of zebrafish Mtmr8.
Different extracts from Cont MO embryos, Mtmr8 morphants and Mtmr8 morphants co-injected with Mtmr8 mRNA were prepared at 24 hpf (30 embryos/group) and subjected to Western blot detection with anti-Akt antibody or anti-phospho-Akt (pAkt) antibody. The corresponding histograms (right panels) plot the relative Ser(P)473-Akt to total Akt ratio (Relative p-Akt/Akt ratio), determined by band intensities that were analyzed by Scion software, with the ratio in experiment extracts normalized to the ratio determined in Cont MO injected embryo extracts. Results represent mean±SD of three separate experiments. *P<0.05.
Mtmr8 PH/G domain plays a critical role with PI3K in regulating zebrafish embryogenesis
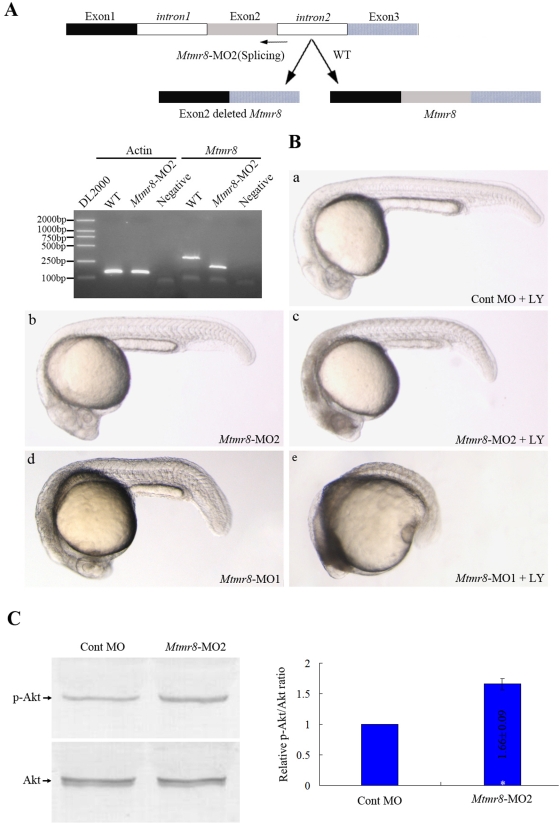
PH/G domain plays an important role in the activity of MTMR family [10]. To delete the PH/G domain of zebrafish Mtmr8, we designed a splice junction morpholino targeted against the second coding exon-intron boundary. In the Mtmr8-MO2 morphants, the targeted exon was eliminated, which encodes partial of the PH/G domain, as shown by a smaller PCR transcript which was verified by bi-directional DNA sequencing (Fig 4A). However, unlike Mtmr8-MO1 morphants (Fig 4B-a), the Mtmr8-MO2 morphants display no obvious defects, except mild abnormal in the head (Fig 4B-b). Intriguingly, previous studies showed that the defects resulted by homozygous single or double mutants of ptena−/− and ptenb−/−, were rescued by treatment with the phosphatidylinositol-3-kinase inhibitor, LY294002[34]. In order to assay the connections between Mtmr8 and PI3-Kinase/AKT pathway during embryo development, we inhibited the PI3K activity with 10 µM LY294002 between 10 hpf and 24 hpf, when transcript of Mtmr8 became restricted to eye field and somites. When treated with 10 µM LY294002, the defects of Mtmr8-MO1 morphant embryos could be further deteriorated (Fig 4B-d), and the Mtmr8-MO2 morphants had severe defects (Fig 4B-e), comparing with Cont morpholino injected morphants which did not alter the wild-type phenotype throughout development 4B-c). In addition, loss of PH/G domain could also produce a significant increase in the relative amount of pAkt compared to Cont MO injected embryos (Fig 4C). The results indicate that the PH/G domain is essential for Mtmr8's function and that protein encoded by the splice variant may lose part of its functions.
Figure 4. PH/G domain of Mtmr8 is required to cooperate with PI3K regulating embryo development and essential for the phosphatase activity of zebrafish Mtmr8 gene.
(A) Splice junction morpholino targeted against Mtmr8 exon-intron boundary resulted to loss of PH/G domain. RT-PCR of Mtmr8 transcript at 24hpf in WT and Mtmr8-MO2 (6 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (B) Embryo phenotypes observed under the microscope. LY represents the PI3K inhibitor, LY294002. Each picture represents typical results out of three separate experiments (30 embryos in each experiment). All scale bars are 100 µm. (C) Immunoblot of different extracts from Cont MO embryos and Mtmr8-MO2 morphants at 24 hpf (30 embryos/group). Relative p-Akt/Akt ratio was determined by band intensities that were analyzed by Scion software, with the ratio in experiment extracts normalized to the ratio determined in Cont MO injected embryo extracts. Results represent mean±SD of three separate experiments. *P<0.05.
Mtmr8 cooperation with PI3K regulates actin filament modeling
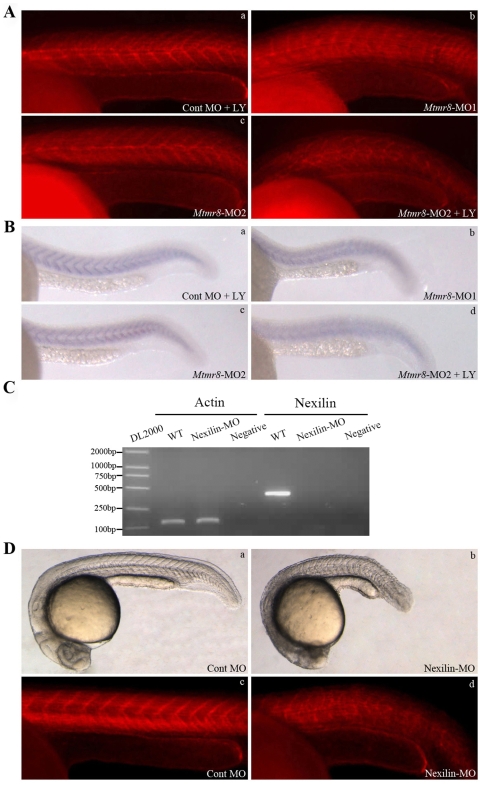
Mtmr8 is a member of the growing class of dual-specificity phosphatases (DSPs) as PTEN, which controls cell random movement through the Ras, PI3K, PTEN, and F-actin regulatory loop [29]. To examine a possible requirement for Mtmr8 in integrity of the actin cytoskeleton, we used phalloidin-TRITC to stain filamentous actin in embryos at 24 hpf. In almost all the embryos treated with Mtmr8-MO1, we observed obvious defect of the actin cytoskeleton in a proportion of skeletal muscle fibres (Fig 5A-b), compared to the Mtmr8-MO2 morphant (Fig 5A-c) and control embryos (Fig 5A-a). However, when treated with LY294002, the actin cytoskeleton of Mtmr8-MO2 morphant was severely disturbed (Fig 5A-d).
Figure 5. Mtmr8 cooperates with PI3K regulating actin modeling.
(A) Staining of embryos with phalloidin (F-actin) at 24hpf. (B) Nexilin mRNA expression at 24 hpf. (C) RT-PCR of Nexilin transcript at 24hpf in WT and Nexilin-MO (3 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (D) The effects of Nexilin knockdown on embryo development (a, b) and F-actin (c, d). The pictures are representative of at least three experiments (30 embryos in each experiment). All scale bars are 100 µm.
Nexilin, a novel actin filament (F-actin)-binding protein, which was strongly expressed at somites at 24hpf [32]. We performed whole-mount in situ hybridization with zebrafish Nexilin riboprobes, and revealed that nexilin was significantly reduced and disorganized in the 24hpf Mtmr8-MO1 morphants (Fig 5B- b), However, the expression of nexilin was not obviously changed in Mtmr8-MO2 morphants (Fig 5B-c) and Cont MO morphants treated with LY294002 (Fig 5B-a). To further understand the function of Nexilin in integrity of the actin cytoskeleton, we performed morpholino to knockdown the zebrafish Nexilin. As shown by RT-PCR, in the control embryos, the amplified PCR product is 382 bp by primers designed from the first and second extrons. However, in the Nexilin-MO morphants, because of inclusion of intron 1, the product is longer than 3 kb, which could not be amplified in one minute (Fig 5C). Embryos injected with the 3 ng Nexilin MO and stained with phalloidin, did show severe skeletal muscle detachments (Fig 5d-b,d), compared to the control (Fig 5d-a,c).
Mtmr8 is essential for the hedgehog pathway to regulate muscle development during embryogenesis
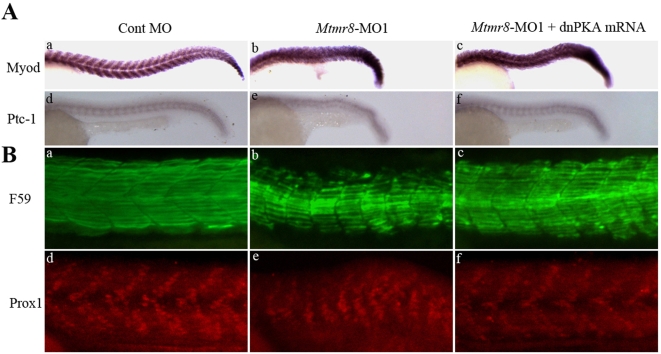
The Mtmr8 morphants have morphological traits in common with Hedgehog pathway mutants, such as U-shaped somite boundaries. To verify that the defects in Mtmr8 morphants are correlated with the changes in Hh signaling, we examined the expression of myod and patched1 (ptc1), a downstream target gene and receptor of Sonic hedgehog [35], [36]. Compared with the control (Fig 6A-a,d), Myod and ptc1 expression is severely reduced and disorganized in Mtmr8 morphants (Fig 6A-b,e). Dominant negative PKA (dnPKA) mRNA, when injected into embryos, results in a broader expression of Hh target genes and a rescue of the Myod and ptc1 expression (Fig 6A-c,f). These results support the conclusion that Mtmr8 is required for Hedgehog signaling in zebrafish development.
Figure 6. The effect of Mtmr8 knockdown on hedgehog pathway and slow muscle development.
(A) The expression of myod and ptc1 at 24hpf in the Cont MO (a, d), Mtmr8-MO1 morphant (b, e) and it injected with dnPKA mRNA (c, f). (B) Whole-mount staining of 24 hpf embryo with F59 antibody (green) and Prox1 antibody (red) and to reveal slow muscle cells. The pictures are representative of at least three experiments (30 embryos in each experiment). All scale bars are 100 µm.
To study the function of Mtmr8 in slow muscle development, the embryos were then stained with antibody F59, which detects mostly slow myofibrils, although it also reacts weakly with fast myofibrils. While slow fibrils in Cont MO embryos were well organized with striation at 24 hpf (Fig 6B-a), fibrils of Mtmr8 morpholino-injected embryos were shorter and reduced (Fig 6B-b). When injecting Mtmr8 morpholino together with dnPKA mRNA, we detected myofibrils increased and longer than those of Mtmr8 morphants (Fig 6B-c). Labeling with anti-Prox1, a slow muscle nuclear marker, shows that the numbers of muscle pioneer and slow muscle cells are reduced in Mtmr8 morphant embryos (Fig 6B-e) compared to control embryos (Fig 6B-d), which is rescued by coinjection of dnPKA mRNA (Fig 6B-f). These findings indicate that the expression of Mtmr8 is dependent on Hh signaling.
Mtmr8 acts non-cell autonomously to F-actin modeling
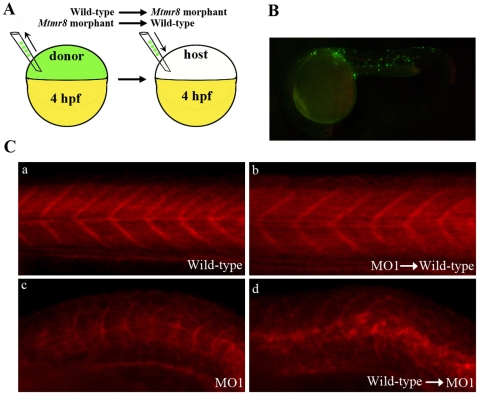
Previous studies have demonstrated that the transplanted labeled cells at the prospective mesodermal region (as shown in Fig 7A) will mainly differentiated into several regions including the neural tube, brain, and somites at 24 hpf when the muscle cells are easily distinguished from other cell types morphologically [37]. To determine whether Mtmr8 acts in a cell autonomous or non-cell autonomous manner in muscle development, reciprocal cell transplantation experiments were carried out between wild-type and Mtmr8 morphant embryos. We transplanted donor fluorescein-labeled cells into age-matched host embryos at 4 hpf. At 24 hpf, the phenotype of the embryos were observed under the microscope. The embryos with transplanted cell in the somites (Fig 7B) were chosen for phalloidin staining. In wild-type embryos, cells transplanted from Mtmr8 morphant embryos could not change wild-type phenotype and F-actin modeling (Fig 7C-a, b, Table 1). In the reciprocal experiment, when wild-type donor cells were implanted into a Mtmr8 morphant host, the phenotype was consistent with Mtmr8 morphant, the U-shaped somites were still existed and the signal of F-actin was disorganized (Fig 7C-c, d, Table 1). We conclude that Mtmr8 protein acts non-cell autonomously within the actin assembly to promote slow fibres development.
Figure 7. Mtmr8 controls actin modeling non-cell-autonomously.
(A) Schematic depiction of cell transplant experiments. (B) Representative picture of a chimeric embryo at 24 hpf. The labeling cells were mostly located in somites of trunk and tail regions. (C) The effects of cell transplantation on F-actin.
Table 1. Mtmr8 is non-cell autonomous for muscle F-actin modeling.
| Donor genotype | Host genotype | Result |
| Mtmr8 morphant | Wild-type | 15/15 Wild-type (100%) |
| 10/10 Wild-type (100%) | ||
| 13/13 Wild-type (100%) | ||
| Wild-type | Mtmr8 morphant | 12/12 Severe defect (100%) |
| 13/13 Severe defect (100%) | ||
| 12/12 Severe defect (100%) |
Donor embryos were labeled with fluorescein dextran. At 24 hpf, donor and host embryos were observed under the microscope, and then fixed for phalloidin labeling to determine genotype (as shown in Fig 7C). The phenotype of the transplanted embryos (number and percentage) is reported.
Discussion
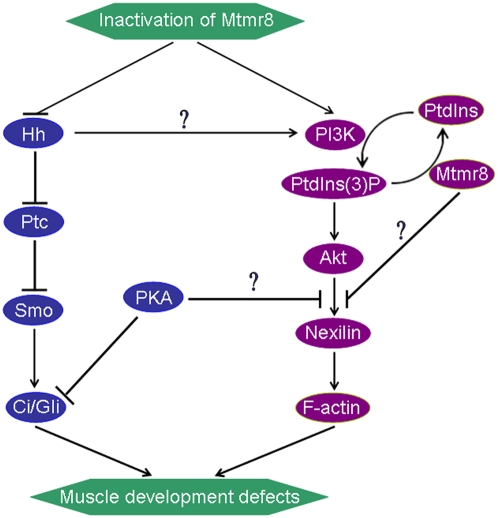
Recent studies clearly showed that myotubularin specifically dephosphorylates phosphatidylinositol 3-monophosphate (PI3P). The action of myotubularin on PI3P levels may implicate two parallel pathways by acting both as a protein phosphatase decreasing PI3P level by down-regulating PI3K activity and, a lipid phosphatase directly degrading PI3P than PI4P in vivo[38]. Such a dual activity has also been suggested for PTEN[39]. Mutations in MTMR family proteins are associated with the human neuromuscular disorders X-linked myotubular myopathy (myotubularin and MTMR1) and type 4B Charcot-Marie-Tooth neuropathy (MTMR2 and MTMR13). These diseases arise from impaired development and/or maturation of skeletal muscle cells and myelinating Schwann cells, respectively. Although a significant body of evidence has linked MTMR to endocytic membrane trafficking events, their role in neuromuscular development is currently unclear. Our findings in zebrafish demonstrated that Mtmr8 is an essential component, together with PI3K, nexilin and F-actin, of the positive-feedback cycle that maintains the normal development of muscle. According to previous studies[40]–[42] and our current studies, a diagram is depicted to explain Mtmr8 function and the relationship between Mtmr8 and the hedgehog pathway (Fig 8). Until now, because of no available means to prevent myotubular myopathy, our findings for the first time provided a new clue to study MTM family related disease in vivo.
Figure 8. A hypothesized signal pathway that Mtmr8 regulates zebrafish muscle development.
Knockdown of Mtmr8 promotes PI3K/Akt pathway and prevents Hh signaling pathway. The two pathways may interact with each other in regulating F-actin modeling and muscle development.
In this study, we presented evidence that the PH/G domain is indeed a function domain of Mtmr8 that contributes to the development of zebrafish. Deletion of the PH/G domain could result in increasing the relative amount of pAkt (Fig. 4C). Akt is activated by various growth factors and hormones such as IGF-1 and insulin. Activation occurs at the plasma membrane after PI3K-dependent generation of PIP3. Thereby, Akt is recruited to the plasma membrane via its PH (pleckstrin homology) domain and is activated by phosphorylation at Thr308 by PDK1 followed by phosphorylation at Ser473 by the mTOR complex[43]. Previous studies have demonstrated that the PH/G domain in a number of different MTMs bind phosphoinositides and also function to localize MTMs to different subcellular compartments in the cell[7]–[9]. All these indicated that Mtmr8 could inhibit PI3K/Akt activation through its functional PH/G domain, loss of which results in apoptotic signaling and cell death. Moreover, identifying the specific binding partners of the PH/G domains on the MTMs will provide important clues to the specific functions regulated by other MTMs as well as the mechanisms whereby loss of some MTMs lead to disease.
Recently studies demonstrated the existence of PI3K-dependent and -independent pathways for F-actin polymerization during chemoattractant-stimulated lamella extension in the human neutrophil. One pathway is dependent on PI3Kγ activation and downstream is dependent on PKCδ and Akt/PKB. This pathway controls the formation of 70% to 80% of the F-actin in the lamella region [44]. In vitro studies, activation of PI3K activity alone is sufficient to remodel actin filaments to increase cell migration through the activation of Akt and p70S6K1 in CEF cells [45]. However, in vivo, the F-actin polymerization and modeling is also Mtmr8-dependent. Although the PI3K/Akt was activated in Mtmr8 knocked embryos, it was not enough to model actin filaments alone. Nexilin is a F-actin binding protein and mediates cell motility, over-expression of which promoted cell migration and adhesion[46]. However, the mechanism of its function is not clear. In Mtmr8 deficient embryos, the expression of Nexilin was reduced, which induced the defect of F-actin modeling. Inhibition of PI3K with LY294002 did not alter the initial formation of these F-actin-rich cup structures at the plasma membrane but it did prevent Akt/PKB recruitment to these cups and their subsequent fusion into the large rings characteristic of normal[47]. However, the effects of inhibiting PI3K on the embryo cytoskeleton are not well characterized. Low dose of LY294002 didn't obviously affect F-actin, whereas it severely disturbed the modeling of F-actin in Mtmr8 morphant and PH/G losing embryos. These indicated that the increasing of pAkt is a very important recovery mechanism for Mtmr8 deficient, although it could not completely rescue the defects.
The model about the mechanisms underlying the Hh signalling pathway is clearly shown by Masai et al. [41]. And Sonic hedgehog (Shh) has been reported to act as a mitogen and survival factor for muscle satellite cells. PI3K/Akt pathway is essential for Shh's actions and directly involved in adult muscle cell proliferation and differentiation. Shh induces Akt phosphorylation in adult muscle cells and influences the PI3K/Akt pathways in a manner similar to IGF-I [48]. Although an increase in Akt phosphorylation could be detected in response to knockdown of Mtmr8, it was not enough to rescue the expression of Myod and Ptc1. Over-expression of dnPKA mRNA, a downstream of Shh, could reverse the defect induced by Mtmr8 deficient. Previous findings provide a basis for the synergistic role of PI3-kinase/Akt in Hh signaling in embryonic development and Hh-dependent tumors [49]. The PI3K inhibitor LY294002 inhibited migrating neo cells and was able to further inhibit residual dnPKA cell migration. PKA may play an important role in the signaling processes that lead to motility. Either inhibition or hyper-activation of PKA may inhibit cell migration, F-actin polymerization and synthesis [42], [50]–[52]. Knockdown of Mtmr8 led to hyper-activation of PKA, which caused the reduction expression of Nexilin and F-actin. All these suggested that Mtmr8 and PI3K/Akt play a synergistic role in regulation of Hh signaling in embryonic muscle development.
Currently, many potential therapies are evaluated by introducing fluorescently tagged cells into a diseased animal and then following their fate using fluorescence to determine if the introduced cells localize to muscle and participate in the repair process[27]. Mutation or knockdown of several genes in zebrafish have been shown to be the underlying basis of many muscular defects[53]–[56]. Cell transplantation experiments could confirm that the gene function is required cell-autonomously or non-cell-autonomously within the muscle development, which may be as a model to evaluate the possibility to use cell therapy for human genetic muscle disease during early fetus. Mtmr8 acts in non-cell-autonomous manner during embryos muscle development, which may suggest that cell therapy is not an efficient way to rescue the defect during early embryo development. The effectiveness of cell therapy could also be evaluated in human neuromuscular disorders caused by mutations in other myotubularin family genes.
From our studies, we have demonstrated that Mtmr8 have the same function as lipid phosphatases PTEN to dephosphorylate the PI3K products by down-regulating PI3K activity, and to regulate actin modeling and muscle development of zebrafish [57]. However, it is different from tumor suppressor gene, PTEN. Mtmr8 is not only an antagonist of PI3K, but also as a partner to balance the PI3K expression in embryo development regulation of zebrafish.
Materials and Methods
Maintenance of zebrafish
A breeding colony of zebrafish (Danio rerio) were maintained at 28.5°C on a 14 h (hour) light/10 h dark cycle [58]. All embryos were collected by natural spawning and staged according to Kimmel et al. [59]. Kinase inhibitors, LY294002 were dissolved in DMSO at stock concentration of 50 mM, and then diluted to final concentration of 10 µM in embryo media from 10 hpf to 24 hpf, which had no obvious effect on embryos survival, activity and health. Control embryos were treated with the equivalent amount of DMSO solution.
RT-PCR and Western blotting
Total RNAs were isolated by SV RNA Isolation Reagent (Promega) from different stages and tissues, and the concentration and quality were determined by agarose electrophoresis and spectrophotometer. After treating with DNase I (RNase-free, Promega), the RNAs (about 1 µg) were reverse-transcribed with MMLV (Gibco) at 37°C with oligo(dT)15 primer. β-actin was used as internal control genes. All samples were analyzed in triplicates. Phosphatase activity of zebrafish Mtmr8 and Western blot detection were performed according to previous reports[60], [61]. Images of blots were captured with a scanner, and quantitative densitometric analysis was performed using Scion Image.
Antisense morpholino and mRNA microinjection
Morpholinos were synthesized by GeneTools LLC (Philomath, OR). Following are the sequences for various morpholinos: Mtmr8-MO1 (Splicing antisense), 5′-CACCTGCTGACTCAGACCTACCTTC-3′; Mtmr8-MO2 (Splicing antisense), 5′-GGCCAACATTACCCATGTTTCTTTG-3′; Nexilin-MO (Splicing antisense), 5′-ATAGCCTCTTCATACTTTACCTCTT-3′; Standard control MO (Cont MO), 5′-CCTCTTACCTCAGTTACAATTTATA-3′. For injection, MOs were injected into fertilized zebrafish eggs at the one-cell stage at a concentration of about 6 ng each embryo. After injection, embryos were incubated at 28.5°C in Embryo Medium [58].
Plasmids pCS2-Mtmr8 and pCS2-dnPKA were linearized for in vitro transcription. Capped sense RNAs were synthesized using SP6 RNA polymerase and the SP6 Cap-Scribe (Roche), following the manufacturer's instructions, re-suspended in water and injected at a concentration of 100 ng/µL.
Whole-mount in situ hybridization and immunohistochemistry
Embryos at different stages were collected and pre-treated and fixed as described [57]. Purified plasmids was linearized by selected restriction enzymes and used as templates for in vitro transcription using T7 or Sp6 RNA polymerase to generate DIG-labeled (Roche) sense and anti-sense probes. In situ hybridization was performed as described [62].
F59 antibody was purchased from Developmental Studies Hybridoma Bank (DSHB, University of Iowa, USA), and Prox1 antibody from Abcam (Cambridge, United Kingdom). Zebrafish embryos were fixed overnight in 4% paraformadehyde at 4°C, and then washed in 0.1% Triton in PBS (PBT) and dechorionated. They were then incubated for 1 h in 0.5% Triton in PBS, followed by 5-h incubation in block solution (10% normal goat serum, 1% DMSO, 0.1% Triton in PBS). Embryos were then incubated overnight at 4°C in block solution containing Phalloidin and/or primary antibodies. They were then washed in PBT, and incubated for 5 h at room temperature with secondary antibodies. Antibody and Phalloidin staining of zebrafish embryos were performed as previously described [63].
Transplantation
Embryos, which were used as donors at later stages, were injected between one-cell to two-cell stages with 2.5% fluorescein dextran (Molecular Probes product) alone or in combination with 6 ng morpholino. Donor and host embryos were dechorionated. The cells were sucked from the prospective mesodermal region of a donor at the sphere stage, and transplanted into the same position of wild-type host embryos at the same stage. Locations of transplanted cells in the hosts were determined by fluorescence stereomicroscopy at the shield stage. Distribution of transplanted cells in the hosts were observed and photographed at 24 hpf postfertilization (hpf) using a Leica fluorescence stereomicroscope. We sucked out the embryos with transplanted cell mainly in the somites, which were fixed at 24 hpf for 2 h at room temperature in 4% paraformaldehyde, and later for phalloidin staining.
Statistical analysis
Data are presented as mean±SD. The relative expression level of p-Akt in Mtmr8 morphants and control morpholino groups were compared statistically using one-way analysis of variance (ANOVA), followed by the Tukey's post hoc test for multiple comparisons. A probability (P) of <0.05 was statistically considered significant.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Major Basic Research Program (grant No. 6462004CB117401), the Innovation Project of Chinese Academy of Sciences (grant No.KSCX2-YW-N-020), and the Innovation Project of IHB, CAS (grant No. 075A01-1-301). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wishart MJ, Dixon JE. PTEN and myotubularin phosphatases: from 3-phosphoinositide dephosphorylation to disease. Trends Cell Biol. 2002;12:579–585. doi: 10.1016/s0962-8924(02)02412-1. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Anti-phosphatases take the stage. Nat Genet. 1998;18:303–305. doi: 10.1038/ng0498-303. [DOI] [PubMed] [Google Scholar]
- 3.Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 4.Laporte J, Bedez F, Bolino A, Mandel JL. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 12 Spec No. 2003;2:R285–292. doi: 10.1093/hmg/ddg273. [DOI] [PubMed] [Google Scholar]
- 5.Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science. 1991;253:401–406. doi: 10.1126/science.1650499. [DOI] [PubMed] [Google Scholar]
- 6.Burke TR, Jr, Zhang ZY. Protein-tyrosine phosphatases: structure, mechanism, and inhibitor discovery. Biopolymers. 1998;47:225–241. doi: 10.1002/(SICI)1097-0282(1998)47:3<225::AID-BIP3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Berger P, Schaffitzel C, Berger I, Ban N, Suter U. Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci U S A. 2003;100:12177–12182. doi: 10.1073/pnas.2132732100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzo O, Urbe S, Clague MJ. Systematic analysis of myotubularins: heteromeric interactions, subcellular localisation and endosome related functions. J Cell Sci. 2006;119:2953–2959. doi: 10.1242/jcs.03040. [DOI] [PubMed] [Google Scholar]
- 9.Tsujita K, Itoh T, Ijuin T, Yamamoto A, Shisheva A, et al. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J Biol Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzo O, Urbe S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 11.Galaktionov K, Lee AK, Eckstein J, Draetta G, Meckler J, et al. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Yen C, Liaw D, Podsypanina K, Bose S, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 13.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 14.Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 15.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 16.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 17.Chojnowski A, Ravise N, Bachelin C, Depienne C, Ruberg M, et al. Silencing of the Charcot-Marie-Tooth associated MTMR2 gene decreases proliferation and enhances cell death in primary cultures of Schwann cells. Neurobiol Dis. 2007;26:323–331. doi: 10.1016/j.nbd.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Buj-Bello A, Furling D, Tronchere H, Laporte J, Lerouge T, et al. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum Mol Genet. 2002;11:2297–2307. doi: 10.1093/hmg/11.19.2297. [DOI] [PubMed] [Google Scholar]
- 19.Azzedine H, Bolino A, Taieb T, Birouk N, Di Duca M, et al. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet. 2003;72:1141–1153. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang H, Li Z, Skolnik EY, Fares H. Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol Biol Cell. 2004;15:189–196. doi: 10.1091/mbc.E03-08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Y, Fares H, Grant B, Li Z, Rose AM, et al. Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J Biol Chem. 2003;278:34380–34386. doi: 10.1074/jbc.M303259200. [DOI] [PubMed] [Google Scholar]
- 22.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Deutekom JC, van Ommen GJ. Advances in Duchenne muscular dystrophy gene therapy. Nat Rev Genet. 2003;4:774–783. doi: 10.1038/nrg1180. [DOI] [PubMed] [Google Scholar]
- 24.Gussoni E, Blau HM, Kunkel LM. The fate of individual myoblasts after transplantation into muscles of DMD patients. Nat Med. 1997;3:970–977. doi: 10.1038/nm0997-970. [DOI] [PubMed] [Google Scholar]
- 25.Gussoni E, Soneoka Y, Strickland CD, Buzney EA, Khan MK, et al. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 26.Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D'Antona G, et al. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 27.Guyon JR, Steffen LS, Howell MH, Pusack TJ, Lawrence C, et al. Modeling human muscle disease in zebrafish. Biochim Biophys Acta. 2007;1772:205–215. doi: 10.1016/j.bbadis.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki AT, Firtel RA. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, et al. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 32.Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission 2004 [Google Scholar]
- 33.Carricaburu V, Lamia KA, Lo E, Favereaux L, Payrastre B, et al. The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc Natl Acad Sci U S A. 2003;100:9867–9872. doi: 10.1073/pnas.1734038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faucherre A, Taylor GS, Overvoorde J, Dixon JE, Hertog J. Zebrafish pten genes have overlapping and non-redundant functions in tumorigenesis and embryonic development. Oncogene. 2008;27:1079–1086. doi: 10.1038/sj.onc.1210730. [DOI] [PubMed] [Google Scholar]
- 35.Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, et al. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- 36.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 37.Szeto DP, Kimelman D. The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains. Genes Dev. 2006;20:1923–1932. doi: 10.1101/gad.1435306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blondeau F, Laporte J, Bodin S, Superti-Furga G, Payrastre B, et al. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum Mol Genet. 2000;9:2223–2229. doi: 10.1093/oxfordjournals.hmg.a018913. [DOI] [PubMed] [Google Scholar]
- 39.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 40.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 41.Masai I, Yamaguchi M, Tonou-Fujimori N, Komori A, Okamoto H. The hedgehog-PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development. 2005;132:1539–1553. doi: 10.1242/dev.01714. [DOI] [PubMed] [Google Scholar]
- 42.Whittard JD, Akiyama SK. Positive regulation of cell-cell and cell-substrate adhesion by protein kinase A. J Cell Sci. 2001;114:3265–3272. doi: 10.1242/jcs.114.18.3265. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 44.Chodniewicz D, Zhelev DV. Chemoattractant receptor-stimulated F-actin polymerization in the human neutrophil is signaled by 2 distinct pathways. Blood. 2003;101:1181–1184. doi: 10.1182/blood-2002-05-1435. [DOI] [PubMed] [Google Scholar]
- 45.Qian Y, Corum L, Meng Q, Blenis J, Zheng JZ, et al. PI3K induced actin filament remodeling through Akt and p70S6K1: implication of essential role in cell migration. Am J Physiol Cell Physiol. 2004;286:C153–163. doi: 10.1152/ajpcell.00142.2003. [DOI] [PubMed] [Google Scholar]
- 46.Wang W, Zhang W, Han Y, Chen J, Wang Y, et al. NELIN, a new F-actin associated protein, stimulates HeLa cell migration and adhesion. Biochem Biophys Res Commun. 2005;330:1127–1131. doi: 10.1016/j.bbrc.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 47.Rupper A, Lee K, Knecht D, Cardelli J. Sequential activities of phosphoinositide 3-kinase, PKB/Aakt, and Rab7 during macropinosome formation in Dictyostelium. Mol Biol Cell. 2001;12:2813–2824. doi: 10.1091/mbc.12.9.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elia D, Madhala D, Ardon E, Reshef R, Halevy O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2007;1773:1438–1446. doi: 10.1016/j.bbamcr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edin ML, Howe AK, Juliano RL. Inhibition of PKA blocks fibroblast migration in response to growth factors. Exp Cell Res. 2001;270:214–222. doi: 10.1006/excr.2001.5345. [DOI] [PubMed] [Google Scholar]
- 51.O'Connor KL, Mercurio AM. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J Biol Chem. 2001;276:47895–47900. doi: 10.1074/jbc.M107235200. [DOI] [PubMed] [Google Scholar]
- 52.Howe AK, Hogan BP, Juliano RL. Regulation of vasodilator-stimulated phosphoprotein phosphorylation and interaction with Abl by protein kinase A and cell adhesion. J Biol Chem. 2002;277:38121–38126. doi: 10.1074/jbc.M205379200. [DOI] [PubMed] [Google Scholar]
- 53.Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- 54.Roy S, Wolff C, Ingham PW. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev. 2001;15:1563–1576. doi: 10.1101/gad.195801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons MJ, Campos I, Hirst EM, Stemple DL. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 2002;129:3505–3512. doi: 10.1242/dev.129.14.3505. [DOI] [PubMed] [Google Scholar]
- 56.Henry CA, Amacher SL. Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev Cell. 2004;7:917–923. doi: 10.1016/j.devcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 57.Croushore JA, Blasiole B, Riddle RC, Thisse C, Thisse B, et al. Ptena and ptenb genes play distinct roles in zebrafish embryogenesis. Dev Dyn. 2005;234:911–921. doi: 10.1002/dvdy.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of the Zebrafish (Danio rerio). University of Oregon Press 3rd edn 1995 [Google Scholar]
- 59.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 60.Mei J, Chen B, Yue H, Gui JF. Identification of a C1q family member associated with cortical granules and follicular cell apoptosis in Carassius auratus gibelio. Mol Cell Endocrinol. 2008;289:67–76. doi: 10.1016/j.mce.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Zhu R, Zhang YB, Zhang QY, Gui JF. Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol. 2008;82:6889–6901. doi: 10.1128/JVI.02385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mei J, Zhang QY, Li Z, Lin S, Gui JF. C1q-like inhibits p53-mediated apoptosis and controls normal hematopoiesis during zebrafish embryogenesis. Dev Biol. 2008;319:273–284. doi: 10.1016/j.ydbio.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 63.Koppen M, Fernandez BG, Carvalho L, Jacinto A, Heisenberg CP. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671–2681. doi: 10.1242/dev.02439. [DOI] [PubMed] [Google Scholar]