Abstract
H-Rev107 is a protein that was previously cloned as a negative regulator of proto-oncogene Ras and classified as a class II tumor suppressor. Its structural similarity to lecithin retinol acyltransferase and Ca2+-independent phosphatidylethanolamine (PE) N-acyltransferase led us to analyze H-Rev107 as an enzyme involved in phospholipid metabolism. Here, we show that recombinant H-Rev107s from rat, human, and mouse possess phospholipase (PL) A1 or A2 activity toward phosphatidylcholine (PC). Further examination with purified recombinant protein revealed that H-Rev107 functions as a cytosolic Ca2+-independent PLA1/2 for PC and PE with higher PLA1 activity than PLA2 activity. Dithiothreitol and iodoacetic acid exhibited stimulatory and inhibitory effects, respectively. Histidine-21 and cysteine-111 of rat H-Rev107 were presumed to form a catalytic dyad based on database analysis, and their single mutants were totally inactive. These results suggested that H-Rev107 is a hydrolase of the thiol type. The N-terminal proline-rich and C-terminal hydrophobic domains of H-Rev107 were earlier reported to be responsible for the regulation of cell proliferation. Analysis of deletion mutants indicated that these domains are also catalytically essential, suggesting relevance of the catalytic activity to the anti-proliferative activity.
Keywords: N-acyltransferase, glycerophospholipid, lecithin retinol acyltransferase
Phospholipase (PL) A1 and PLA2 catalyze the esterolytic cleavage at the sn-1 or sn-2 position of glycerophospholipids, respectively, resulting in the formation of FFAs and lysophospholipids (1–5). Arachidonic acid released by PLA2 is further converted into various bioactive eicosanoids (6), whereas lysophospholipids serve as lipid mediators (for example, lysophosphatidic acid, lysophosphatidylserine, and lysophosphatidylinositol) (7–10) or precursors for lipid mediators (platelet-activating factor) (11). These bioactive lipid molecules act principally through G protein-coupled receptors and are involved in a wide range of physiological and pathophysiological events (6–11). PLA1 and PLA2 constitute large families of proteins, respectively. As for PLA2, more than 20 isozymes have been cloned and characterized in mammals, and their physiological functions have been extensively analyzed (1–3). On the other hand, at least 9 isozymes of PLA1 have been cloned (4, 5). Each isozyme is classified into a subgroup, depending on its primary structure, extracellular or intracellular localization, Ca2+ dependency, and specific inhibition by synthetic compounds.
Lecithin retinol acyltransferase (LRAT) is an enzyme-catalyzing transfer of the acyl group at the sn-1 position of phosphatidylcholine (PC) to all-trans-retinol, resulting in the formation of retinyl ester (12). Recently, we found that a rat protein with homology to LRAT is able to transfer an acyl group from the sn-1 and sn-2 positions of PC to the amino group of phosphatidylethanolamine (PE), forming N-acylphosphatidylethanolamine (NAPE) (13). NAPEs are precursors of N-acylethanolamines, including the endocannabinoid anandamide (14, 15). However, several lines of evidence showed that this protein is distinguished from Ca2+-dependent N-acyltransferase, which is generally accepted to be responsible for the NAPE formation in brain and other animal tissues, and we referred to it as Ca2+-independent N-acyltransferase (iNAT) (13). During the study on iNAT, we also noticed that H-Rev107 is another member of the LRAT family. H-Rev107 was originally cloned as a tumor suppressor gene that regulates the activity of proto-oncogene HRAS, although its physiological function remained unclear (16–21). Our preliminary results showed that the cell homogenate containing recombinant H-Rev107 converted [14C]PC and nonradioactive PE to radioactive bands comigrated with authentic FFA and NAPE on the thin-layer plate, suggesting that H-Rev107 has a PLA1/2 activity and a PE N-acylation activity (13). However, we did not perform further characterization.
In the present studies, we examined catalytic properties of recombinant H-Rev107 cloned from rat, human, and mouse, and clarified that H-Rev107 functions principally as a Ca2+-independent PLA1/2 with a higher PLA1 activity. We will also discuss that members of the LRAT family, including LRAT, iNAT, and H-Rev107, have a common property to exert acyltransferase/PLA1/2 activities for glycerophospholipids.
EXPERIMENTAL PROCEDURES
Materials
[1-14C]palmitic acid, 1,2-[1-14C]dipalmitoyl-PC, 1-palmitoyl-2-[1-14C]arachidonoyl-PE, 1-[14C]palmitoyl lyso PC, and [carboxy-14C]triolein were purchased from PerkinElmer Life Science. 1-Palmitoyl-2-[1-14C]palmitoyl-PC, 1-palmitoyl-2-[1-14C]oleoyl-PC, 1-palmitoyl-2-[1-14C]arachidonoyl-PC, 1-palmitoyl-2-[1-14C]linoleoyl-PE, HRP-linked anti-mouse IgG, Hybond P, and an ECL Plus kit were from GE Healthcare. 1,2-Dioleoyl-PE, 1,2-dipalmitoyl-PC, 1-palmitoyl-2-oleoyl-PC, 1-palmitoyl-2-arachidonoyl-PC, 1-palmitoyl-2-linoleoyl-PE, triolein, anti-FLAG monoclonal antibody M2, anti-FLAG M2 affinity gel, FLAG peptide, snake venom PLA2, and Rhizopus arrhizus lipase were from Sigma. DMEM, lipofectamine, fetal calf serum, pcDNA3.1 (+), TRIzol, and Moloney murine leukemia virus RT were from Invitrogen. 1-Palmitoyl-2-arachidonoyl-PE was from Avanti Polar Lipids (Alabaster, AL). Human Testis Marathon-Ready™ cDNA was from Clontech. Nonidet P-40 was from Nacalai Tesque, Inc. (Kyoto, Japan). Random hexamer and Ex Taq DNA polymerase were from TaKaRa Bio, Inc. (Ohtsu, Japan). KOD-Plus DNA polymerase was from TOYOBO (Osaka, Japan). Protein assay dye reagent concentrate was from Bio-Rad, and precoated Silica Gel 60 F254 aluminum sheets (20 × 20 cm, 0.2 mm thick) for TLC were from Merck (Darmstadt, Germany). Bromoenol lactone (BEL) and methyl arachidonyl fluorophosphonate (MAFP) were from Cayman Chemical (Ann Arbor, MI). N-[14C]palmitoyl-PE was prepared from [1-14C]palmitic acid and 1,2-dioleoyl-PE according to the method of Schmid et al. (22). 1-[1-14C]palmitoyl-2-palmitoyl-PC was prepared from 2-palmitoylglycerophosphocholine and [1-14C]palmitic acid (13). 2-[14C]palmitoyl lyso PC was prepared from 1-palmitoyl-2-[1-14C]palmitoyl-PC using Rhizopus arrhizus lipase.
Construction of expression vectors harboring cDNAs of H-Rev107s
The expression vector harboring rat H-Rev107 with a FLAG tag at the N terminus was constructed as described previously (13). The cDNAs encoding C-terminally FLAG-tagged H-Rev107s of human and mouse were amplified by PCR with Human Testis Marathon-Ready™ cDNA or mouse brain cDNA as a template. The mouse brain cDNA was prepared from 5 μg of total RNA of mouse brain using Moloney murine leukemia virus RT and random hexamer. The primers used were the forward primers 5′-CGCAAGCTTGGAAGATGCGTGCGCCCATTCCAG-3′ (human H-Rev107 containing a HindIII site) and 5′-CGCAAGCTTGGAAAATGCTAGCACCCATACCAGAAC-3′ (mouse H-Rev107 containing a HindIII site) and the reverse primers 5′-CGCGAATTCTCACTTATCGTCGTCATCCTTGTAATCTTGCTTTTGTCGCTTGTTTCTTG-3′ (human H-Rev107 containing an in-frame FLAG sequence and an EcoRI site) and 5′-CGCGAATTCTCACTTATCGTCGTCATCCTTGTAATCTTGCTTCTGTTTCTTGTTTC-3′ (mouse H-Rev107 containing an in-frame FLAG sequence and an EcoRI site). PCR was carried out with KOD-Plus DNA polymerase for 30 cycles at 94°C for 20 s, 56°C for 20 s, and 68°C for 60 s in 5% (v/v) Me2SO. The obtained DNA fragments were subcloned into the HindIII and EcoRI sites of pcDNA3.1 (+). All constructs were sequenced in both directions using an ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA).
Overexpression and purification of recombinant proteins
COS-7 cells were grown at 37°C to 60% confluency in 100 mm dishes containing DMEM with 10% fetal calf serum in a humidified 5% CO2 and 95% air incubator. The expression vector harboring H-Rev107 cDNA was introduced into COS-7 cells using lipofectamine according to the manufacturer's instructions. Two days after transfection, cells were harvested and sonicated three times each for 3 s in 20 mM Tris-HCl (pH 7.4), and the homogenates were centrifuged at 105,000 g for 55 min at 4°C. The resultant supernatant was used as the cytosolic fraction, whereas the pellet was suspended in 20 mM Tris-HCl (pH 7.4) and used as the membrane fraction. Purification of recombinant FLAG-tagged rat H-Rev107 was performed as described for iNAT (13). Briefly, cytosolic fractions prepared from the cells grown in ten 100 mm dishes were added to 1 ml of anti-FLAG M2 affinity gel preequilibrated with 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 0.05% Nonidet P-40 (buffer A), and incubated overnight at 4°C under gentle mixing. The gel was packed into a column and washed three times each with 12 ml of buffer A. The FLAG-tagged protein was eluted with buffer A containing 0.1 mg/ml of FLAG peptide and every 0.5 ml fraction was collected. The protein concentration was determined by the method of Bradford with BSA as a standard.
Enzyme assay
For the PLA1/2 assay, the enzyme was incubated with 200 μM 1,2-[1-14C]dipalmitoyl-PC (45,000 cpm) in 100 μl of 50 mM glycine-NaOH (pH 9.0), 2 mM DTT, and 0.05% Nonidet P-40 at 37°C for 30 min. For the PE N-acylation assay, the enzyme was incubated with 40 μM 1,2-[1-14C]dipalmitoyl-PC (45,000 cpm) and 75 μM 1,2-dioleoyl-PE in 100 μl of 50 mM glycine-NaOH (pH 9.0), 2 mM DTT, and 0.05% Nonidet P-40 at 37°C for 30 min. For the lipase assay, the enzyme was incubated with 200 μM [carboxy-14C]triolein (61,000 cpm) in 100 μl of 50 mM potassium phosphate buffer (pH 7.0) containing 2% BSA (23), or in 100 μl of 50 mM Tris-HCl (pH 8.0), 2 mM DTT, and 0.05% Nonidet P-40 at 37°C for 30 min. The reaction was terminated with the addition of 320 μl of a mixture of chloroform-methanol (2:1, v/v) containing 5 mM 3(2)-t-butyl-4-hydroxyanisole. After centrifugation, 100 μl of the lower fraction was spotted on a silica gel thin-layer plate (10 cm height) and developed at 4°C for 25 min either in a mixture of chloroform, methanol, and 28% ammonium hydroxide (80:20:2, v/v) for the PE N-acylation assay, in chloroform-methanol-H2O (65:25:4, v/v) for the PLA1/2 assay, or in chloroform-acetone-acetic acid (96:4:1, v/v) for the lipase assay. The distribution of radioactivity on the plate was quantified using a BAS1500 bioimaging analyzer (FUJIX Ltd., Tokyo, Japan).
Construction and expression of mutants
Mutants were constructed by the PCR method using the expression vector harboring rat H-Rev107 as a template. H21L and C111S were prepared by megaprimer PCR consisting of two sets of PCR reactions. Two oligonucleotides 5′-CTATGTACAGTCTCTGGGCCATCT-3′ (for H21L) and 5′-CTGACCAGTGAGAACAGTGAGCACTTCGTGA-3′ (for C111S) (the underscoring indicates the mismatches) and their complementary oligonucleotides were used as primers. For these mutants, the forward primer was 5′-AAGCTTATGGATTACAAGGATGACGACGATAAGCCCATACCAGAACCCAAGCCT-3′, and the reverse primer was 5′-GAATTCTCACTGCTTCTGTTTCTTGTTTCT-3′. The primers used for deletion mutants of rat H-Rev107 were as follows: the forward primers 5′-CGCAAGCTTGGAAAATGGGAGATCTGATTGAGATTTTC-3′ (for ΔN1), 5′-CGCAAGCTTGGAAAATGCCTATGTACAGTCACTGGGCC-3′(for ΔN2), and 5′-CGCAAGCTTGGAAAATGCCCATACCAGAACCCAAGCC-3′ (for ΔC1), and the reverse primers 5′-CGCGAATTCCTACTTATCGTCGTCATCCTTGTAATCCTGCTTCTGTTTCTTGTTTC-3′ (for ΔN1 and ΔN2) and 5′-CGCGAATTCCTACTTATCGTCGTCATCCTTGTAATCCGCCACCTTGACGGCATCTCTG-3′ (for ΔC1). PCR was carried out with KOD-Plus DNA polymerase for 30 cycles at 94°C for 20 s, 56°C for 20 s, and 68°C for 60 s in 5% (v/v) Me2SO. The resultant DNA fragments were subcloned into the HindIII and EcoRI sites of pcDNA3.1 (+). All constructs were sequenced in both directions. Expression of these mutants was performed as described above for the wild type.
Western blotting
Samples (10 μg protein) were separated by SDS-PAGE on 14% gel and electrotransferred to a hydrophobic polyvinylidene difluoride membrane (Hybond P). The membrane was blocked with PBS containing 5% dried milk and 0.1% Tween 20 (buffer B) and then incubated with anti-FLAG antibody (1:2,000 dilution) in buffer B at room temperature for 1 h, followed by incubation with HRP-labeled secondary antibody (1:4,000 dilution) in buffer B at room temperature for 1 h. FLAG-tagged proteins were visualized using an ECL Plus kit and analyzed using a LAS1000plus lumino-imaging analyzer (FUJIX Ltd.).
PCR
Total RNA was isolated from various organs of Wistar-ST rats (Japan SLC, Inc.) using TRIzol. cDNAs were prepared from 5 μg of total RNA using Moloney murine leukemia virus RT and random hexamer, and subjected to PCR amplification by Ex Taq DNA polymerase. The primers used for rat H-Rev107 were the forward primer 5′-ATGCCCATACCAGAACCCAAGCCTG-3′ and the reverse primer 5′-CTCACTGGTCAGCCTGTACAGCAC-3′ (nucleotides 1–25 and 304–327, respectively, in GenBank™ accession number AB298805), and those for rat GAPDH were the forward primer 5′-AAGGTCGGTGTGAACGGATTTGG-3′ and the reverse primer 5′-ACAAACATGGGGGCATCAGC-3′ (nucleotides 7–29 and 370–389, respectively, in NM_017008). The PCR conditions used were as follows: for H-Rev107, denaturation at 94°C for 20 s, annealing at 54°C for 20 s, and extension at 72°C for 30 s (35 cycles); for GAPDH, denaturation at 95°C for 30 s, annealing at 58°C for 15 s, and extension at 72°C for 30 s (27 cycles).
Real-time quantitative PCR analysis was performed using a LightCycler II (Roche Diagnostics). The primers used for rat H-Rev107 were the forward primer 5′-ATCCAGCGAGCTGAGGAGCTGGTG-3′ and the reverse primer 5′-TGAGAGCATGACTCCAATGAGGCCC-3′ (nucleotides 271–294 and 435–459, respectively, in AB298805), and those for rat GAPDH were the forward primer 5′-CCTGGCCAAGGTCATCCATGACAAC-3′ and the reverse primer 5′-GATGACCTTGCCCACAGCCTTGGC-3′ (nucleotides 546–571 and 710–733, respectively, in NM_017008). The PCR conditions used were as follows: denaturation at 95°C for 6 s, and annealing and extension at 62°C for 20 s (35 cycles).
RESULTS
cDNA cloning and expression of H-Rev107
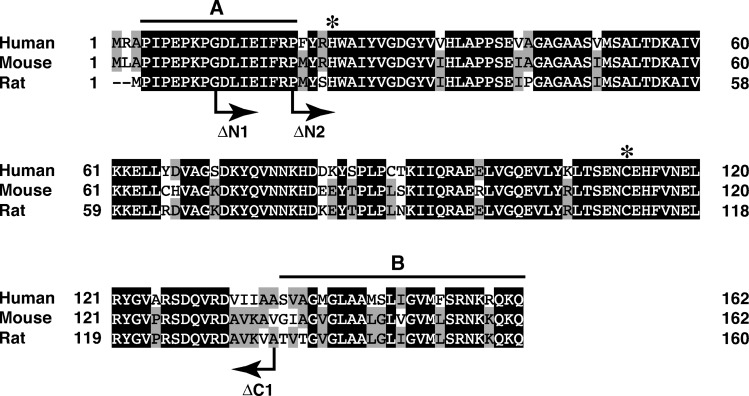
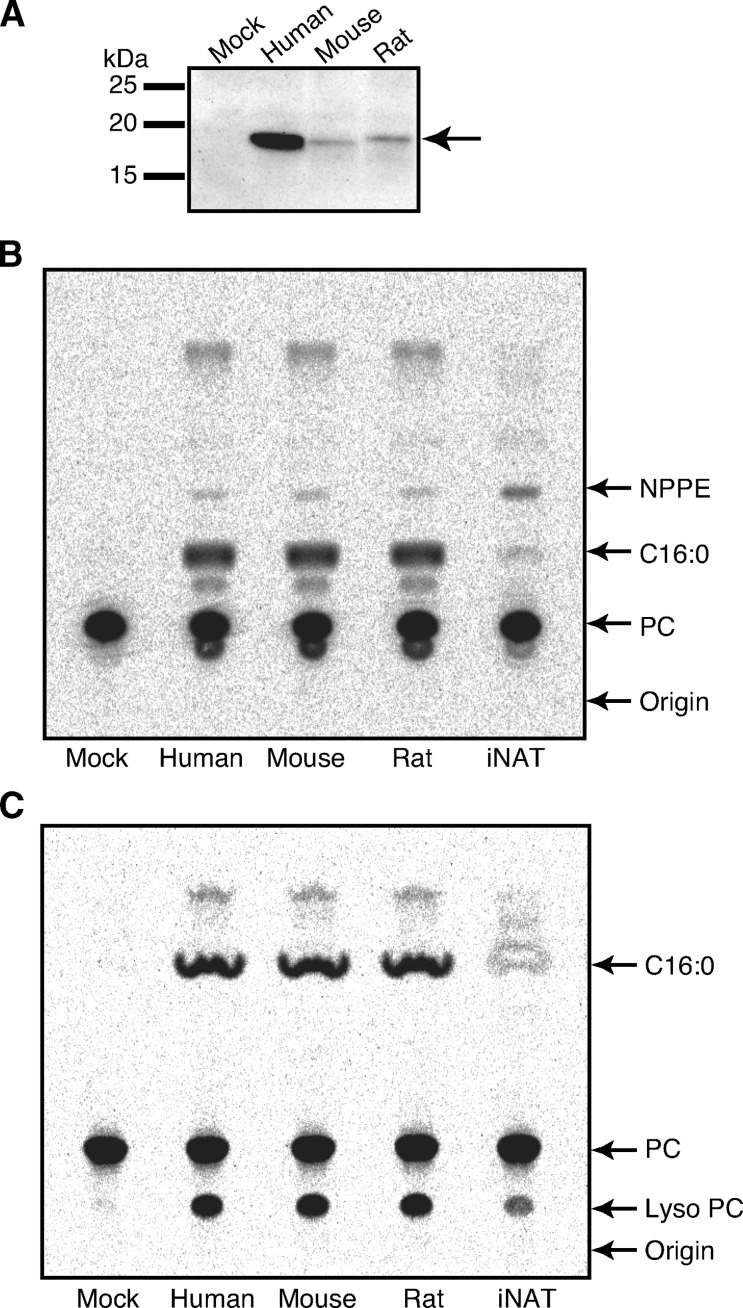
We recently cloned cDNA of rat H-Rev107 (13), which was tentatively termed RLP-3. In addition, we isolated cDNAs of H-Rev107 from human testis and mouse brain based on the reported sequences (GenBank accession numbers NM_007069 and NM_139269). We sequenced the obtained human and mouse cDNAs, which were completely identical to the reported sequences. Figure 1 shows alignment of the amino acid sequences deduced from the cDNAs. The sequences were composed of 162 (human and mouse) and 160 (rat) amino acid residues, and their molecular masses were calculated to be 17,950 (human), 17,872 (mouse), and 17,748 Da (rat). At amino acid level, human and mouse H-Rev107 were 82.1% and 90.1% identical to the rat counterpart, respectively. When the expression vector harboring either human, mouse, or rat H-Rev107 tagged with FLAG was introduced into COS-7 cells, these proteins were successfully expressed with molecular masses around 19 kDa, which were expected for the FLAG-tagged forms (Fig. 2A). The FLAG tag was located either at the N terminus (rat) or at the C terminus (human and mouse). Transient expression of H-Rev107s in COS-7 cells did not appear to influence cell proliferation.
Fig. 1.
Alignment of the deduced amino acid sequences of human, mouse, and rat H-Rev107s. The sequences were aligned using the program GENETYX-MAC (version 10). Closed and shaded boxes indicate identity in all three or any two polypeptides, respectively. Gaps introduced for maximal alignment are indicated by dashes. The highly conserved histidine and cysteine residues in the LRAT family are indicated by asterisks. The N-terminal proline-rich domain (A) and C-terminal hydrophobic domain (B) are indicated by lines. The N-terminal (ΔN1 and ΔN2) or C-terminal (ΔC1) residues of the deletion mutants of rat H-Rev107, which were used in the experiment shown in Fig. 8, are also shown.
Fig. 2.
Transient expression and enzymatic activities of human, mouse, and rat H-Rev107s. COS-7 cells were transfected with the insert-free vector (Mock) or the expression vector harboring FLAG-tagged H-Rev107 of either human, mouse, or rat, or FLAG-tagged Ca2+-independent N-acyltransferase (iNAT) of rat. The cell homogenates (10 μg protein) were analyzed by Western blotting with an anti-FLAG antibody (A). The cell homogenates (10 μg protein) were also assayed for phosphatidylethanolamine (PE) N-acylation activity (B) and for the phospholipase (PL) A1/2 activity (C) as described under Experimental Procedures. The products were separated by TLC. The positions of authentic compounds on the TLC plate are indicated by arrows. NPPE, N-[14C]palmitoyl-PE; C16:0, [14C]palmitic acid; PC, phosphatidylcholine.
Functional analysis of H-Rev107 as an enzyme
First, to examine PE N-acylation activity, we incubated the homogenates from the H-Rev107-expressing cells with radiolabeled dipalmitoyl-PC, in which both 1-O-palmitoyl and 2-O-palmitoyl groups were labeled with 14C as an acyl donor and nonradiolabeled PE as an acyl acceptor. The homogenate of iNAT-expressing cells (13) was used as a control. When the reaction products were separated by TLC, a radioactive band comigrating with authentic N-[14C]palmitoyl-PE was detected with human, mouse, and rat H-Rev107s (Fig. 2B). The activities for PE N-acylation (0.109–0.133 nmol/min/mg of protein) were higher than that of the control COS-7 cells (0.040 nmol/min/mg), but were about half of that of rat iNAT (0.195 nmol/min/mg) (Table 1). Notably, with all the H-Rev107s, the major product was free [14C]palmitic acid, rather than N-[14C]palmitoyl-PE (Fig. 2B), suggesting that the PLA1/2 activity is more potent than the N-acylation activity. In agreement with this finding, in the absence of nonradiolabeled PE, the cell homogenates converted [14C]dipalmitoyl-PC to free [14C]palmitic acid and [14C]lyso PC. The PLA1/2 activities were as high as 12.8–13.8 nmol/min/mg of protein (Table 1). Under the same assay conditions, endogenous activity of the control cells was 0.28 nmol/min/mg, and the PLA1/2 activity of iNAT was 1.2 nmol/min/mg. Thus, although iNAT and H-Rev107s exhibited both N-acylation activity and PLA1/2 activity, H-Rev107s were characterized by a predominant PLA1/2 activity. H-Rev107 without FLAG tag also showed a much higher PLA1/2 activity than PE N-acylation activity, suggesting that the tag does not interfere with PE N-acylation activity (data not shown). We labeled H-Rev107-expressing COS-7 cells and control COS-7 cells with [14C]palmitic acid for 18 h, and after chase with 20 μM palmitic acid for 5 h, measured [14C]palmitic acid released from the cells into the medium. The radioactivity of [14C]palmitic acid released from H-Rev107-expressing COS-7 cells was about 2-fold higher than that released from the control cells. These results suggested that the recombinant H-Rev107 exerts PLA1/2 activity in the whole cells. Western blotting revealed by far the highest expression level of human H-Rev107 (Fig. 2A). However, its activity in the homogenates was similar to those of rat and mouse H-Rev107s, suggesting that most of human H-Rev107 protein is catalytically inactive. Another possibility was that the inherent specific activity of human H-Rev107 is several-fold lower than those of mouse and rat H-Rev107s.
TABLE 1.
PE N-acylation and PLA1/2 activities in the homogenates of COS-7 cells overexpressing recombinant human, mouse, or rat H-Rev107
| PE N-acylation | PLA1/2 | |
|---|---|---|
| nmol/min/mg | ||
| Mock | 0.040 ± 0.004 | 0.28 ± 0.028 |
| Human H-Rev107 | 0.115 ± 0.015 | 13.8 ± 0.38 |
| Mouse H-Rev107 | 0.133 ± 0.013 | 12.8 ± 0.42 |
| Rat H-Rev107 | 0.109 ± 0.021 | 12.9 ± 0.54 |
| Rat iNAT | 0.195 ± 0.014 | 1.2 ± 0.14 |
PE, phosphatidylethanolamine; PLA1/2, phospholipase A1/2; iNAT, Ca2+-independent N-acyltransferase. The homogenates (10 μg protein) of COS-7 cells expressing the indicated proteins were assayed for PE N-acylation activity and PLA1/2 activity as described under Experimental Procedures. Mean values ± SD are shown (n = 3).
Characterization of purified recombinant H-Rev107
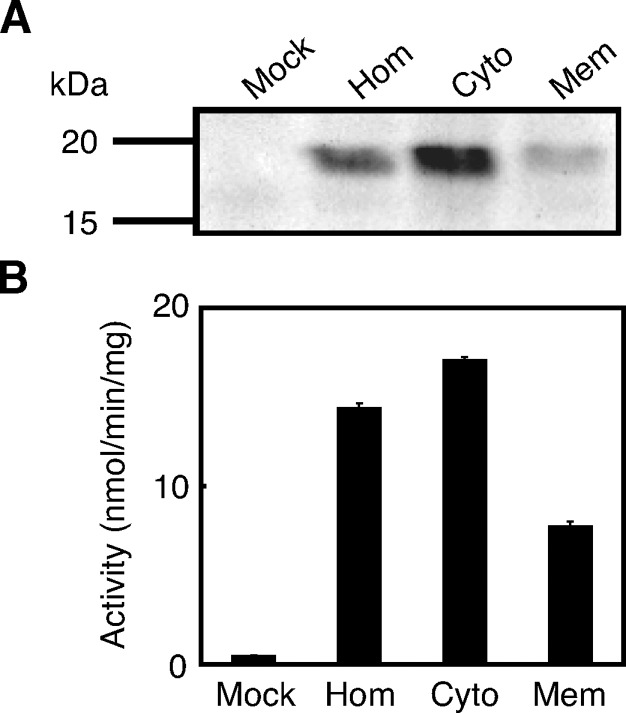
The cell homogenate expressing rat H-Rev107 was subjected to ultracentrifugation at 105,000 g. When analyzed by Western blotting, the content of H-Rev107 per 10 μg of protein in the cytosolic fraction was higher than that in the membrane fraction (Fig. 3A). Corresponding to this finding, PLA1/2 activity in the cytosolic fraction was higher than that in the membrane fraction (Fig. 3B). Moreover, when compared between the total protein contained in the cytosolic fraction and that contained in the membrane fraction prepared from the same homogenate, total PLA1/2 activity in the cytosolic fraction was higher than that in the membrane fraction. The PLA1/2 activity of mouse H-Rev107 without FLAG tag was also detected in both the cytosolic and membrane fractions, with a higher specific activity in the cytosolic fraction (data not shown). The result suggested that the tag does not affect intracellular localization of H-Rev107.
Fig. 3.
Subcellular distribution of H-Rev107. The cell homogenates (Hom), cytosolic fractions (Cyto), and membrane fractions (Mem) (10 μg protein) from COS-7 cells expressing rat H-Rev107 were analyzed by Western blotting (A) or assayed for PLA1/2 activity (B). The homogenates of COS-7 cells transfected with the insert-free vector (Mock) were also analyzed. Enzyme activity is shown as mean values ± SD (n = 3).
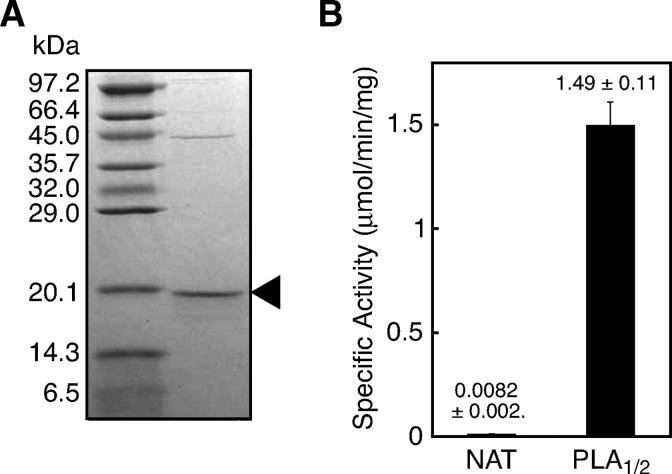
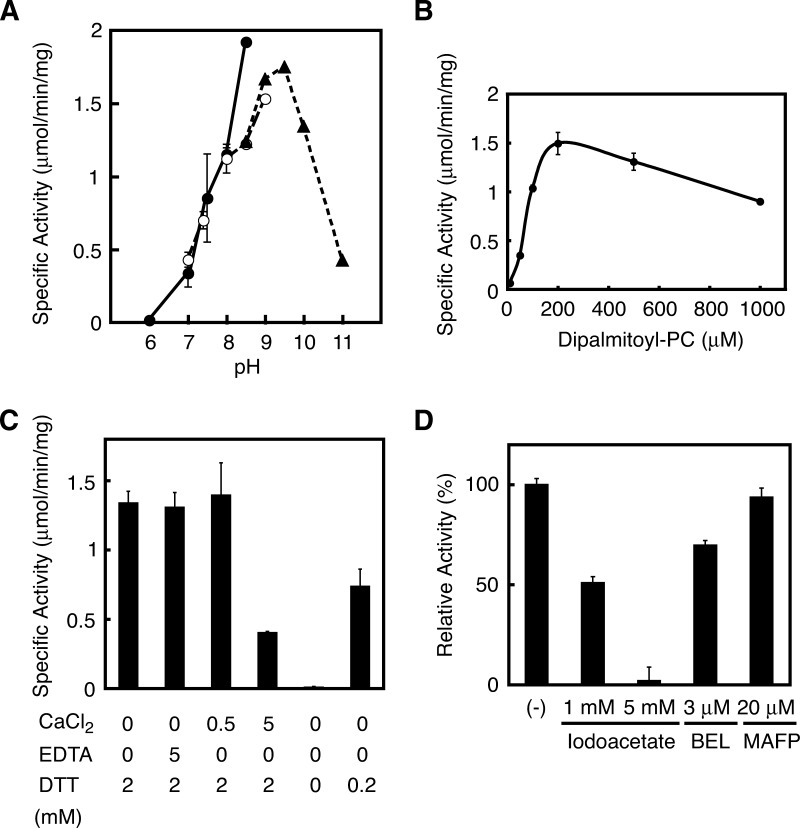
We purified FLAG-tagged rat H-Rev107 from the cytosolic fractions using an anti-FLAG antibody-conjugated column. SDS-PAGE revealed a predominant protein band with a molecular mass of 19 kDa (Fig. 4A). The purified H-Rev107 exhibited an 88-fold higher PLA1/2 activity than the cytosolic fraction (Fig. 4B). A much lower NAT activity was also detected with the purified protein. We also allowed the purified H-Rev107 to react with a mixture of unilamella vesicles derived from 80 μM PC and 40 μM PE in the absence of detergent. The specific PLA1/2 activity and PE N-acylation activity were 215.2 nmol/min/mg and 5.4 nmol/min/mg of protein, respectively. As examined at different pHs, the PLA1/2 activity was detected at pH 7–11. The highest activity was seen at pH 8.0 with HEPES-NaOH or at pH 9.5 with glycine-NaOH (Fig. 5A).
Fig. 4.
SDS-PAGE and enzyme assay with purified H-Rev107. A: Purified FLAG-tagged rat H-Rev107 (2.0 μg protein) was analyzed by SDS-PAGE on 14% gel, followed by Coomassie staining. The band of H-Rev107 is indicated by an arrowhead. The positions of molecular markers are also shown. B: Purified H-Rev107 (0.2 μg protein) was assayed for PE N-acylation activity and PLA1/2 activity. Enzyme activity is shown as mean values ± SD (n = 3).
Fig. 5.
Properties of purified H-Rev107 as PLA1/2. A: Purified rat H-Rev107 (0.2 μg protein) was assayed for PLA1/2 activity at the indicated pH. The buffers used were HEPES-NaOH (closed circles), Tris-HCl (open circles), and glycine-NaOH (closed triangles). B: The assay was performed with different concentrations of 1,2-[14C]dipalmitoyl-PC as a substrate. C, D: The assay was performed in the presence of the indicated substances. In D, H-Rev107 was preincubated with the substances at 4°C for 3 min before the assay. Enzyme activity is shown as mean values ± SD (n = 3). BEL, bromoenol lactone; MAFP, methyl arachidonyl fluorophosphonate.
The PLA1/2 activity increased depending on the concentrations of PC up to 200 μM with an apparent Km at 75 μM (Fig. 5B). The activity was not influenced by either 0.5 mM Ca2+ or 5 mM EDTA, although a high concentration of Ca2+ (5 mM) was inhibitory (Fig. 5C). H-Rev107 required the reducing reagent DTT for full activity, as shown by a DTT-dependent increase in activity (Fig. 5C). The addition of 2 mM DTT increased PLA1/2 activity 167-fold. Nonidet P-40 (0.05%) increased the activity 7-fold (data not shown). Based on these results, the reaction mixture of our standard enzyme assay contained 50 mM glycine-NaOH (pH 9), 200 μM PC, 2 mM DTT, and 0.05% Nonidet P-40. In relation to the requirement of DTT, we examined the inhibitory effect of iodoacetate, an irreversible sulfhydryl (SH) blocker. As shown in Fig. 5D, iodoacetate inhibited the PLA1/2 activity with an IC50 around 1 mM. We also tested BEL, a Ca2+-independent PLA2 (iPLA2) inhibitor (2) and MAFP, an inhibitor of various serine hydrolases, including cytosolic PLA2 and iPLA2 (2). When rat H-Rev107 was incubated with 3 μM BEL, which is sufficient to inhibit iPLA2 completely, only a 30% reduction in activity was observed (Fig. 5D). MAFP at 20 μM did not show any effect on the activity of H-Rev107 (Fig. 5D). Combined with insensitivity to Ca2+ and EDTA (Fig. 5B), these results showed that H-Rev107 is catalytically distinguishable from known PLA2s.
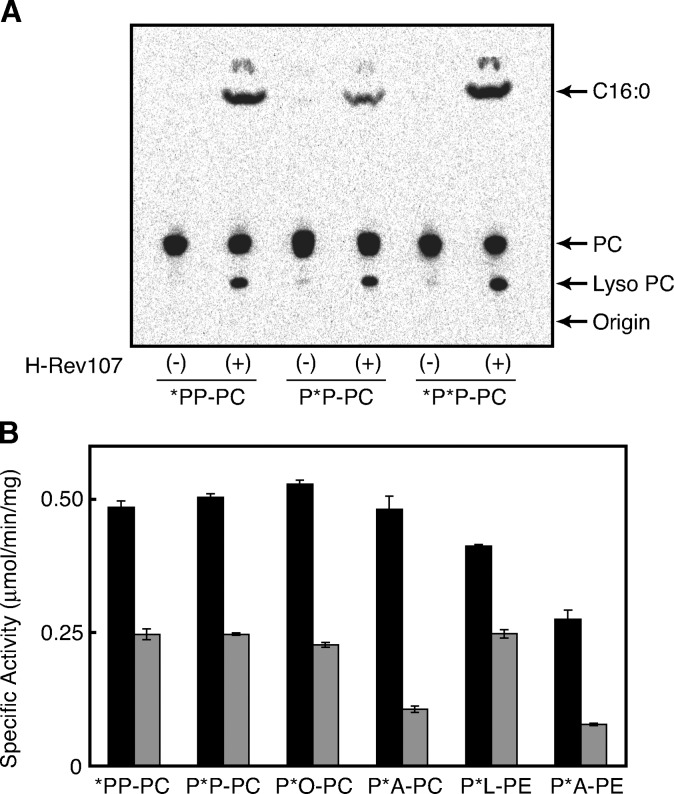
Because dipalmitoyl-PC, which we have used as substrate, was radiolabeled on both the 1-O-palmitoyl and 2-O-palmitoyl groups, we could not determine regioselectivity in terms of the sn-1 and sn-2 positions of PC in the hydrolysis reaction. Therefore, we examined reactivities with 1-[14C]palmitoyl-2-palmitoyl-PC and 1-palmitoyl-2-[14C]palmitoyl-PC. As analyzed by TLC (Fig. 6A), both compounds were converted to [14C]palmitic acid and [14C]lyso PC. [14C]palmitic acid could be produced from 1-[14C]palmitoyl-2-palmitoyl-PC by PLA1 activity and from 1-palmitoyl-2-[14C]palmitoyl-PC by PLA2 activity, whereas [14C]lyso PC could be produced from 1-[14C]palmitoyl-2-palmitoyl-PC by PLA2 activity and from 1-palmitoyl-2-[14C]palmitoyl-PC by PLA1 activity. These results showed that H-Rev107 has both PLA1 and PLA2 activities. The specific activity of PLA1 was higher than that of PLA2 (Fig. 6B). In the presence of nonradioactive PE, [14C]NAPE was formed from both 1-[14C]palmitoyl-2-palmitoyl PC and 1-palmitoyl-2-[14C]palmitoyl PC, showing that both the sn-1 and sn-2 acyl groups can be used for N-acylation as well as hydrolysis (data not shown).
Fig. 6.
PLA1 and PLA2 activities of H-Rev107 toward various glycerophospholipids. The purified rat H-Rev107 (0.2 μg protein) was allowed to react with 200 μM of various PCs or PEs. A: A thin-layer chromatogram is shown. B: PLA1-like activity (black bars) and PLA2-like activity (gray bars) for each substrate are shown. The substrates used were 1,2-[14C]dipalmitoyl-PC (*P*P-PC), 1-[14C]palmitoyl-2-palmitoyl-PC (*PP-PC), 1-palmitoyl-2-[14C]palmitoyl-PC (P*P-PC), 1-palmitoyl-2-[14C]oleoyl-PC (P*O-PC), 1-palmitoyl-2-[14C]arachidonoyl-PC (P*A-PC), 1-palmitoyl-2-[14C]linoleoyl-PE (P*L-PE), and 1-palmitoyl-2-[14C]arachidonoyl-PE (P*A-PE). Enzyme activity is shown as mean values ± SD (n = 3).
We further examined several PC and PE species with different 14C-labeled acyl chains at the sn-2 position. Specific labeling of the radioactive phospholipids at sn-2 was confirmed by digestion with snake venom PLA2. The label in the wrong position was less than 1% with all the substrates. H-Rev107 was active with all the tested phospholipids and revealed both the PLA1 and PLA2 activities (Fig. 6B). It should be noted that PLA1 activity was consistently higher than PLA2 activity. We also tested 1-[14C]palmitoyl lyso PC and 2-[14C]palmitoyl lyso PC for possible lyso PLA activity of the purified H-Rev107. The results showed that H-Rev107 is almost inactive with these lyso PCs (data not shown). Moreover, H-Rev107 did not show any lipase activity toward [14C]triolein (data not shown). These results showed that H-Rev107 functions as a PLA1/2-type enzyme with a preference for PLA1 activity.
Mutagenesis studies on H-Rev107
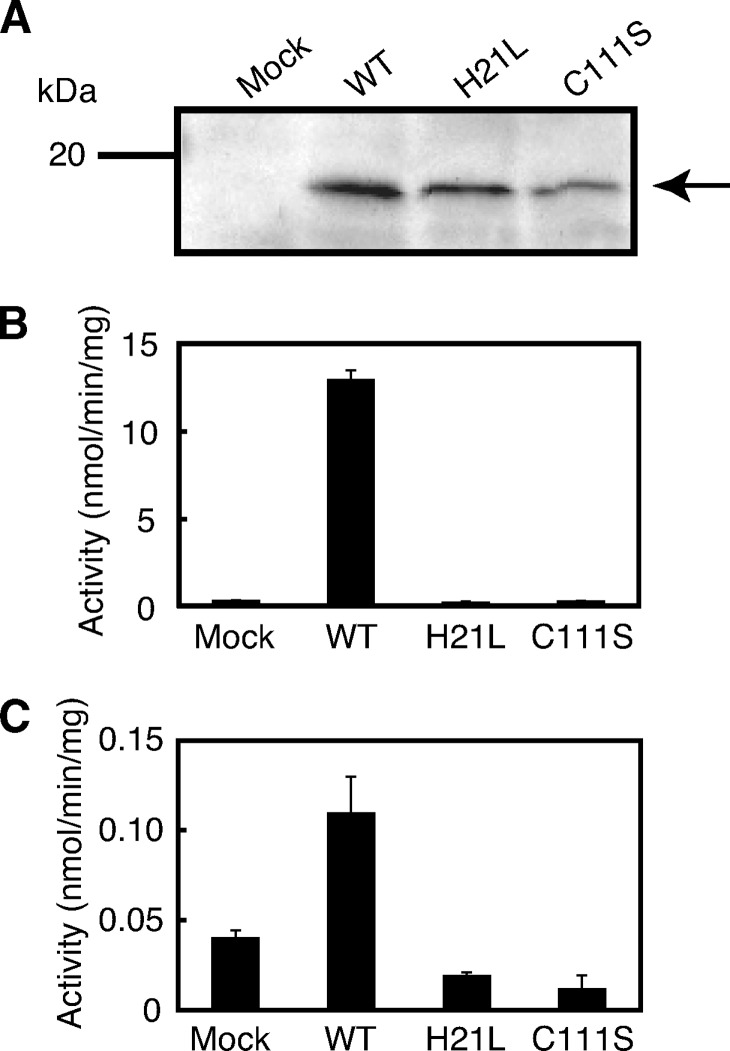
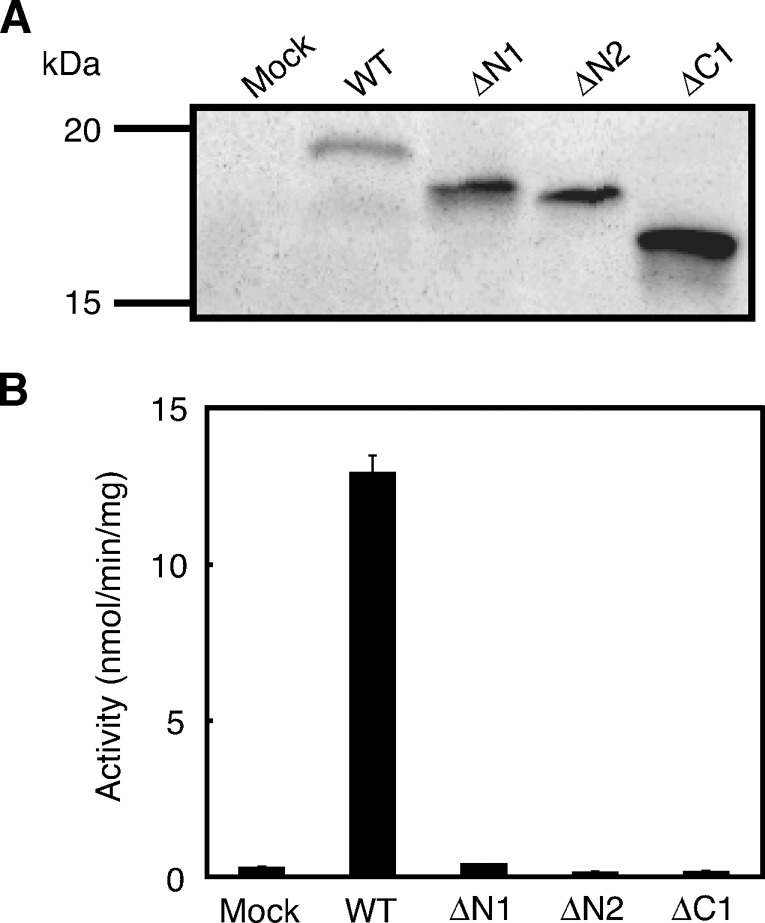
His-21 and Cys-111 of rat H-Rev107 correspond to the histidine and cysteine residues that are highly conserved within the LRAT family and appear to be crucial for catalytic activities (Fig. 1) (24). To examine whether this is the case with H-Rev107, we performed site-directed mutagenesis on His-21 and Cys-111. As shown in Fig. 7A, Western blot analysis revealed that the mutants H21L and C111S were successfully expressed in COS-7 cells. However, neither PLA1/2 nor NAT activity was detected with the mutants (Fig. 7B, C). These results indicated that both residues are absolute requirements for the enzymatic activity and/or proper folding. H-Rev107 possesses a proline-rich domain near the N terminus and a hydrophobic domain near the C terminus (Fig. 1). To understand the functions of these domains, we prepared deletion mutants of rat H-Rev107 lacking either N-terminal 8 or N-terminal 16 amino acids (ΔN1, ΔN2) or C-terminal 25 amino acids (ΔC1) (Fig. 1). Expression of the mutants with the expected molecular masses was confirmed by Western blotting (Fig. 8A). As for PLA1/2 activity, ΔN1 showed very low activity (about 3% that of the wild type), and ΔN2 and ΔC1 were entirely inactive. These results suggested that the N-terminal proline-rich and C-terminal hydrophobic domains are also involved in the catalytic activity and/or proper folding.
Fig. 7.
Analysis of the mutants H21L and C111S of H-Rev107. COS-7 cells were transfected with the insert-free vector (Mock) or expression vectors harboring either the wild type of rat H-Rev107 (WT) or its single mutant H21L or C111S. The cell homogenates (10 μg protein) were analyzed by Western blotting (A) and assayed for PLA1/2 activity (B) and PE N-acylation activity (C). Enzyme activity is shown as mean values ± SD (n = 3).
Fig. 8.
Analysis of deletion mutants of H-Rev107. COS-7 cells were transfected with the insert-free vector (Mock) or the expression vectors harboring either the wild type of rat H-Rev107 (WT) or its deletion mutants ΔN1, ΔN2, or ΔC1. The cell homogenates (10 μg protein) were analyzed by Western blotting (A) or assayed for PLA1/2 activity (B). Enzyme activity is shown as mean values ± SD (n = 3).
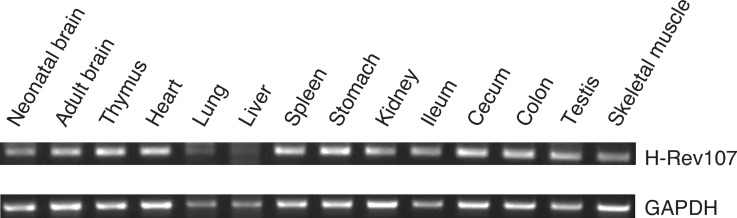
Organ distribution of rat H-Rev107
To examine the organ distribution of rat H-Rev107, RT-PCR was carried out using total RNAs prepared from a series of rat organs. As shown in Fig. 9, mRNA of H-Rev107 was detected in all organs examined, suggesting a universal role for this protein. Very recently, the high expression of H-Rev107 in rat white adipose tissue was reported (25). Therefore, we compared the expression levels of H-Rev107 between white adipose tissue and heart of rat by real-time PCR. The result showed that the ratio of H-Rev107 to GAPDH in white adipose tissues was 10-fold higher than that in heart (data not shown).
Fig. 9.
Tissue distribution of rat H-Rev107 mRNA. Total RNA was isolated from the indicated rat organs and analyzed by RT-PCR using specific primers for H-Rev107 (upper panel) and GAPDH (lower panel).
DISCUSSION
In the present study, we characterized H-Rev107 as a phospholipid-related enzyme. H-Rev107 was originally reported as a negative regulator of proto-oncogene HRAS and categorized as a class II tumor suppressor (16–20). H-Rev107 belongs to the LRAT family in terms of primary structure. This protein family is represented by LRAT, an enzyme that transfers an acyl group of PC to all-trans-retinol, resulting in the formation of retinyl ester, an intracellular storage form of vitamin A (12). However, other members of this family have not been characterized as enzymes until recently. We found that a rat protein that is a member of this family and has been termed HRASLS5 is able to catalyze N-acylation of PE, and we proposed to refer to it as Ca2+-independent N-acyltransferase (iNAT) (13). N-acylation of PE leads to the formation of NAPEs, precursors of bioactive N-acylethanolamines, including anandamide (14, 15).
The alignment of the sequences of human, mouse, and rat H-rev107s revealed 80–90% identity at the amino acid level (Fig. 1). As transiently expressed in COS-7 cells, recombinant proteins of H-rev107s showed a PLA1/2-like hydrolyzing activity toward PC (Fig. 2 and Table 1). In the presence of PE, a weak PE N-acylation activity was also detected. Further analysis with purified protein demonstrated that rat H-Rev107 releases FA from both the sn-1 and sn-2 positions of various PCs and PEs (Fig. 6). For the tested phospholipids as substrates, the PLA1 activity was consistently higher than the PLA2 activity. The reducing reagent DTT was essential for the full activity, whereas the activity was not affected by Ca2+ or EDTA (Fig. 5C). The SH-blocker iodoacetate inhibited the activity (Fig. 5D). Furthermore, replacement of His-21 and Cys-111 with leucine and serine, respectively, resulted in complete loss of activity (Fig. 7). These two amino acid residues are highly conserved within the LRAT family and are considered to constitute a catalytic dyad, with the cysteine functioning as the active site nucleophile (24). These results clarified that H-Rev107 is a Ca2+-independent PLA1/2 of the thiol esterase type. Our results also showed that three members of the LRAT family (LRAT, iNAT, and H-Rev107) have a common ability to abstract an acyl group from glycerophospholipids, but use different acyl acceptors, i.e., retinol is used by LRAT, the amino group of PE by iNAT, and water by H-Rev107. However, the sharp pH profile at basic pH in the PLA1/2 reaction (Fig. 5A) may be due to the hydrolysis of a metastable acyl enzyme intermediate through a thioester bond. We cannot rule out the possibility that we missed a right acceptor substrate different from PE. Thus, the dominant PLA1/2 activity over transacylase activity of H-Rev107 may be caused by our assay conditions without the right acceptor.
A previous study reported that the N-terminal proline-rich domain of H-Rev107 is responsible for its binding to PR65α, which is one of major serine/threonine phosphatases in eukaryotic cells (26). Because a deletion mutant of this domain failed to cause apoptosis, it was shown that the binding of H-Rev107 to PR65α through this domain is required for the induction of programmed cell death in ovarian carcinoma cells (26). Another report indicated that the C-terminal hydrophobic domain, consisting of 25 amino acids, is not only essential for the association with plasma membrane, but also required for the inhibitory effect on proliferation of rat hepatoma cells and HRAS-transformed rat fibroblasts in vitro and in vivo (19). We showed that deletion of the proline-rich domain (the mutants ΔN1 and ΔN2) and the C-terminal hydrophobic domain (the mutant ΔC1) resulted in the abrogation of PLA1/2 activity (Fig. 8). These results suggest that enzymatic activity of H-Rev107 is involved in the induction of programmed cell death through the association with PR65α and in the suppression of cell proliferation by H-Rev107. By the PLA1/2-type reaction, FFAs and lysophospholipids are produced. These products or their further metabolites, including eicosanoids, lysophosphatidic acid, and other lipid mediators, may be directly or indirectly responsible for the anti-proliferating activity of H-Rev107 as a negative regulator of HRAS. Further studies will be needed to elucidate the physiological role of H-Rev107.
During the preparation of this manuscript, Duncan et al. (25) reported that H-Rev107 encodes white adipose tissue-specific PLA2, and designated it AdPLA. However, we noticed several obvious differences between their results and ours. First, they identified H-Rev107 as a PLA2 and classified it into a novel group (group XVI) of PLA2, although a coexisting minor PLA1 activity was also mentioned. As we discussed above, our results showed that the PLA1 activity is consistently higher than the PLA2 activity. This discrepancy appeared to be due to the difference in the assay systems used; we used regiospecifically radiolabeled substrates in the assays shown in Fig. 6, and quantified the radioactivity of the produced FFAs and lysophospholipids on the TLC plates. This method enabled us to measure PLA1 activity and PLA2 activity simultaneously, but separately. On the other hand, Duncan et al. measured enzymatic activity principally by lipoxygenase-coupled spectrometry. By this method, they detected linoleic acid released from dilinoleoyl-PC and 1-palmitoyl-2-linoleoyl-PC. Because the phospholipid in which linoleic acid was specifically bound to the sn-1 position was not used, they did not measure PLA1 activity directly. They also suggested the involvement of H-Rev107 in the production of eicosanoids because arachidonic acid is mostly bound to the sn-2 position of glycerophospholipids. However, considering the positional specificity of H-Rev107 that we revealed in the present study, its contribution to the eicosanoid formation should be minor, if any. Second, they did not refer to iNAT or discuss a common property of the members of the LRAT family (LRAT, iNAT, and H-Rev107) as thiol esterases with acyltransferase/PLA1/2 activity. Indeed, they classified H-Rev107 as belonging to group XVI of PLA2. Third, in our assay, DTT was an essential factor for the full PLA1/2 activity of H-Rev107, whereas they did not use DTT or its equivalent in the reaction mix. Finally, in agreement with their results, we observed a remarkably high expression of H-Rev107 in white adipose tissue. However, the significant expression of H-Rev107 in all of the tissues examined (Fig. 9) suggests its physiological role in various tissues.
In summary, we demonstrated that the class II tumor suppressor H-Rev107 is able to act as a Ca2+-independent PLA1/2 of the thiol hydrolase type. Our mutagenesis studies suggested the involvement of enzyme activity in its anti-proliferating activity. The present results also extended our understanding of the LRAT family, including LRAT, iNAT, and H-Rev107, all of which were capable of catalyzing transacylation and/or PLA1/2-type hydrolysis of glycerophospholipids.
Abbreviations
BEL, bromoenol lactone
iNAT, Ca2+-independent N-acyltransferase
iPLA2, Ca2+-independent PLA2
LRAT, lecithin retinol acyltransferase
MAFP, methyl arachidonyl fluorophosphonate
NAPE, N-acylphosphatidylethanolamine
PC, phosphatidylcholine
PE, phosphatidylethanolamine
PL, phospholipase
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and Japan Society for the Promotion of Science, grants-in-aid from Medical Institution Union Foundation and the Japan Foundation for Applied Enzymology, Kagawa University Specially Promoted Research Fund 2008, and the Fund for Kagawa University Young Scientists 2008.
Published, JLR Papers in Press, December 1, 2008.
References
- 1.Kudo I., and M. Murakami. 2002. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 68–69 3–58. [DOI] [PubMed] [Google Scholar]
- 2.Schaloske R. H., and E. A. Dennis. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 1761 1246–1259. [DOI] [PubMed] [Google Scholar]
- 3.Lambeau G., and M. H. Gelb. 2008. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu. Rev. Biochem. 77 495–520. [DOI] [PubMed] [Google Scholar]
- 4.Aoki J., Y. Nagai, H. Hosono, K. Inoue, and H. Arai. 2002. Structure and function of phosphatidylserine-specific phospholipase A1. Biochim. Biophys. Acta. 1582 26–32. [DOI] [PubMed] [Google Scholar]
- 5.Aoki J., A. Inoue, K. Makide, N. Saiki, and H. Arai. 2007. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 89 197–204. [DOI] [PubMed] [Google Scholar]
- 6.Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294 1871–1875. [DOI] [PubMed] [Google Scholar]
- 7.Sugo T., H. Tachimoto, T. Chikatsu, Y. Murakami, Y. Kikukawa, S. Sato, K. Kikuchi, T. Nagi, M. Harada, K. Ogi, et al. 2006. Identification of a lysophosphatidylserine receptor on mast cells. Biochem. Biophys. Res. Commun. 341 1078–1087. [DOI] [PubMed] [Google Scholar]
- 8.Oka S., K. Nakajima, A. Yamashita, S. Kishimoto, and T. Sugiura. 2007. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem. Biophys. Res. Commun. 362 928–934. [DOI] [PubMed] [Google Scholar]
- 9.Meyer zu Heringdorf D., and K. H. Jakobs. 2007. Lysophospholipid receptors: signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim. Biophys. Acta. 1768 923–940. [DOI] [PubMed] [Google Scholar]
- 10.Moolenaar W. H., L. A. van Meeteren, and B. N. G. Giepmans. 2004. The ins and outs of lysophosphatidic acid signaling. Bioessays. 26 870–881. [DOI] [PubMed] [Google Scholar]
- 11.Prescott S. M., G. A. Zimmerman, D. M. Stafforini, and T. M. McIntyre. 2000. Platelet-activating factor and related lipid mediators. Annu. Rev. Biochem. 69 419–445. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz A., A. Winston, Y-H. Lim, B. A. Gilbert, R. R. Rando, and D. Bok. 1999. Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem. 274 3834–3841. [DOI] [PubMed] [Google Scholar]
- 13.Jin X-H., Y. Okamoto, J. Morishita, K. Tsuboi, T. Tonai, and N. Ueda. 2007. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J. Biol. Chem. 282 3614–3623. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto Y., J. Morishita, K. Tsuboi, T. Tonai, and N. Ueda. 2004. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 279 5298–5305. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto Y., J. Wang, J. Morishita, and N. Ueda. 2007. Biosynthetic pathways of the endocannabinoid anandamide. Chem. Biodivers. 4 1842–1857. [DOI] [PubMed] [Google Scholar]
- 16.Hajnal A., R. Klemenz, and R. Schäfer. 1993. Up-regulation of lysyl oxidase in spontaneous revertants of H-ras-transformed rat fibroblasts. Cancer Res. 53 4670–4675. [PubMed] [Google Scholar]
- 17.Hajnal A., R. Klemenz, and R. Schäfer. 1994. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 9 479–490. [PubMed] [Google Scholar]
- 18.Kiess M., B. Scharm, A. Aguzzi, A. Hajnal, R. Klemenz, I. Schwarte-Waldhoff, and R. Schäfer. 1995. Expression of ril, a novel LIM domain gene, is down-regulated in Hras-transformed cells and restored in phenotypic revertants. Oncogene. 10 61–68. [PubMed] [Google Scholar]
- 19.Sers C., U. Emmenegger, K. Husmann, K. Bucher, A. C. Andres, and R. Schäfer. 1997. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J. Cell Biol. 136 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roder K., K. H. Kim, and H. S. Sul. 2002. Induction of murine H-rev107 gene expression by growth arrest and histone acetylation: involvement of an Sp1/Sp3-binding GC-box. Biochem. Biophys. Res. Commun. 294 63–70. [DOI] [PubMed] [Google Scholar]
- 21.Anantharaman V., and L. Aravind. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4 R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid P. C., P. V. Reddy, V. Natarajan, and H. O. O. Schmid. 1983. Metabolism of N-acylethanolamine phospholipids by a mammalian phosphodiesterase of the phospholipase D type. J. Biol. Chem. 258 9302–9306. [PubMed] [Google Scholar]
- 23.Zimmermann R., J. G. Strauss, G. Haemmerle, G. Schoiswohl, R. Birner-Gruenberger, M. Riederer, A. Lass, G. Neuberger, F. Eisenhaber, A. Hermetter, et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306 1383–1386. [DOI] [PubMed] [Google Scholar]
- 24.Jahng W. J., L. Xue, and R. R. Rando. 2003. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 42 12805–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan R. E., E. Sarkadi-Nagy, K. Jaworski, M. Ahmadian, and H. S. Sul. 2008. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J. Biol. Chem. 283 25428–25436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazarenko I., R. Schäfer, and C. Sers. 2007. Mechanisms of the HRSL3 tumor suppressor function in ovarian carcinoma cells. J. Cell Sci. 120 1393–1404. [DOI] [PubMed] [Google Scholar]