Abstract
In response to a variety of cell stresses, e.g. endoplasmic reticulum (ER) stress, expression of REDD1 (regulated in development and DNA damage responses) is transcriptionally upregulated. However, the mechanism through which ER stress acts to upregulate REDD1 expression is unknown. In the present study, REDD1 expression was found to be upregulated by ER stress in several cell lines. However, in MEF cells lacking the eIF2α kinase PERK, ER stress failed to upregulate REDD1 expression, demonstrating that phosphorylation of eIF2α was necessary for the effect. Moreover, ER stress led to upregulated expression of the transcription factor ATF4, but in MEF cells lacking ATF4, REDD1 mRNA expression was not increased by ER stress. In contrast, exogenous expression of ATF4 was sufficient to induce REDD1 expression. Overall, the results suggest that REDD1 expression is upregulated during ER stress through a mechanism involving activation of PERK, phosphorylation of eIF2α, and increased ATF4 expression.
Keywords: REDD1, Rtp801, mTOR, ER stress, PERK, ATF4
Introduction
The protein referred to as regulated in development and DNA damage responses (REDD1; aka Rtp801, Ddit4, dig2) was originally identified during a screen for genes whose expression was altered in response to hypoxia in cells in culture [1]. In that, and subsequent studies, REDD1 expression was shown to be upregulated in response to a variety of cell stresses, including treatment with hydrogen peroxide [1], exposure to conditions promoting DNA damage [2], during starvation of Drosophila [3], dexamethasone treatment [4; 5], and treatment with chemicals that induce stress in the endoplasmic reticulum [ER, 4]. These early studies suggested that REDD1 acted to regulate apoptosis, and could either promote [1] or repress [4] it depending on cell type and experimental context. For example, in MCF7 and undifferentiated PC12 cells, exogenous REDD1 expression protected cells from glucocorticoid- [4], hydrogen peroxide-, hypoxia-, and serum deprivation-induced apoptosis [1], suggesting that REDD1 does not promote apoptosis but instead acts to protect cells from apoptosis. In contrast, REDD1 overexpression increased the sensitivity of differentiated PC12 cells to hydrogen peroxide- and hypoxia-induced, but not UV irradiation-induced, apoptosis. The basis for the disparate response observed in various studies is unknown. However, exogenous expression of the REDD1 orthologs scylla and/or charybdis in Drosophila extended mean life span up to two-fold [6], suggesting that moderate overexpression of REDD1 may be beneficial, rather than detrimental to cell function and organism survival.
More recent studies have shown that REDD1, and a related protein, REDD2, act to repress signaling through the mammalian target of rapamycin complex 1 (mTORC1) in mammalian cells and in Drosophila [6; 7; 8; 9; 10]. For example, induced expression of REDD1 in a tetracycline regulatable system led to a rapid decline in phosphorylation of two well-characterized substrates of mTORC1, eukaryotic initiation factor 4E binding protein 1 (4E-BP1) and the ribosomal protein S6 kinase, S6K1 [9; 10]. Although the mechanism through which REDD1 acts to repress mTORC1 signaling is incompletely defined, it is clear that it functions upstream of the tuberous sclerosis complex (TSC) in a pathway that appears to be parallel to that involving the AMP-activated protein kinase, AMPK [7; 10]. The TSC1•TSC2 complex acts as a GTPase activator for the small GTPase, ras homolog enriched in brain (Rheb) [11; 12; 13]. The GTP-bound form of Rheb binds to and activates mTORC1, whereas the GDP-bound form does not [14]. Thus, under conditions where TSC1•TSC2 activity is low, Rheb is predominantly present in the GTP-bound form, and mTORC1 signaling is high.
Loss of TSC1 or TSC2 leads to constitutive activation of mTORC1 kinase that can result in the development of benign tumors and neurological disorders. Moreover, the loss of these tumor suppressors causes ER stress and triggers the unfolded protein response (UPR) [15]. This is likely due to overloading of the protein folding machinery and chaperones of the ER as a result of dysregulated protein synthesis. In mammals, the UPR is a complex signaling network mediated by three ER-resident factors: the type-I transmembrane kinase, inositol-requiring enzyme-1 (IRE1), the PKR like ER kinase (PERK), and the activating transcription factor 6 (ATF6). The ER lumenal domains of each of these factors are normally bound to glucose-regulated protein (GRP)78. Upon the accumulation of unfolded proteins in the lumen of the ER, GRP78 dissociates from these three sensors permitting their signaling capabilities. Activated IRE1 acts as both a kinase and an endoribonuclease. IRE1 alternatively splices the mRNA of the X-box DNA-binding protein 1 (XBP1) removing a 26 nucleotide intron that allows sXBP1 to act as a transcriptional activator for several ER chaperones. Activated PERK phosphorylates eukaryotic initiation factor 2 on its α-subunit (eIF2α) resulting in a reduction in translation of most mRNAs, although translation of a few mRNAs, e.g. those encoding the transcription factors ATF4 and ATF5, paradoxically is upregulated. Upon dissociating from GRP78, ATF6 travels to the Golgi and is cleaved. The cleaved ATF6 (nATF6) relocalizes to the nucleus where it functions in complex with the transcription factor NF-Y and induces several ER stress responsive genes and chaperones [reviewed in 16; 17].
Recent evidence suggests that ER stress and activation of the UPR pathways contribute to a negative-feedback regulation of mTOR. For example, activation of JNK by IRE1 resulted in an inhibition of IRS1 activity leading to insulin resistance [15]. Furthermore, the mTOR repressor REDD1 was induced during ER stress [4; 18]. Here, we examine the mechanism responsible for the increased expression of REDD1 in response to ER stress. The results demonstrate that the increase in REDD1 expression requires PERK and a downstream transcription factor ATF4. They also show that overexpression of ATF4 is sufficient to induce REDD1 expression.
Materials and Methods
Cell culture, transfections and plasmids
Human HepG2 cells and HEK293T cells were cultured in high glucose Dulbecco’s modified Eagle’s Medium (DMEM, Invitrogen) with 10% fetal bovine serum (Atlas Biologicals) and 1% penicillin-streptomycin (Invitrogen) at 37°C. PERK+/+, PERK−/−, ATF4+/+, and ATF4−/− MEFs (kindly provided by Drs. David Ron and Heather Harding, NY University School of Medicine) were similarly maintained with the addition of 1.5 μg/ml puromycin to the medium of the PERK MEFs. ATF4−/− cells medium was supplemented with 55 μM β-mercaptoethanol (Invitrogen) and 1x nonessential amino acids (Invitrogen). Cells were seeded onto 6-well plates and grown to ~60–80 % confluency. On the day of the experiment, cells were treated with vehicle (DMSO or ethanol), 10 μg/ml tunicamycin in DMSO, or 100 nM thapsigargin in ethanol, for 4 h. Transfections were carried out using a calcium phosphate-based method. Mouse ATF4 plasmid was kindly provided by Dr. David Ron.
Mouse ATF4-myc plasmid was kindly provided by Dr. Jawed Alam (He et. al., 2001)
RNA isolation and quantitative real-time PCR analysis
RNA was extracted from cells with TRIzol reagent according to the manufacturer’s protocol (Invitrogen). RNA (1 μg) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and subjected to quantitative real-time PCR as described previously (Wang et. al., 2006). Mouse specific primers for GAPDH and REDD1 used for amplification were previously described (Kimball et. al., 2007). Human specific primers used were as follows: GAPDH forward, 5′-GGTGGTCTCCTCTGACTTCA ACA-3′; GAPDH reverse, 5′-GTTGCTGTAGCCAAATTCGTTGT-3′; REDD1 forward, 5′-TGGTGCCCACCTTCCAGCTG-3′; and REDD1 reverse 5′-GTCAGGGACTGGCTGAAGCC-3′. REDD1 mRNA expression level were normalized to GAPDH mRNA expression levels.
Western blot analysis
Cells were lysed and harvested in SDS sample buffer, boiled for 5 min, and an equal volume (30 μl) of each sample was subjected to SDS-PAGE. Resolved samples were transferred onto 0.45 μm PVDF membrane and blocked for 1 h with 5% nonfat dry milk. Membranes were incubated overnight at 4°C with one of the following primary antibodies: anti-REDD1 (ProteinTech Group Inc.), anti-GAPDH (Santa Cruz), anti-phosphorylated eIF2α (Ser51) (Biosource), anti-eIF2α-Total [19], or anti-ATF4 (Santa Cruz). Membranes were incubated with appropriate secondary antibody (Bethyl) at room temperature for 1 h. Blots were developed with ECL Plus reagents (Amersham Biosciences). Images were acquired using a GeneGnome HR Bioimager (SynGene) and GeneSnap software (SynGene).
Results
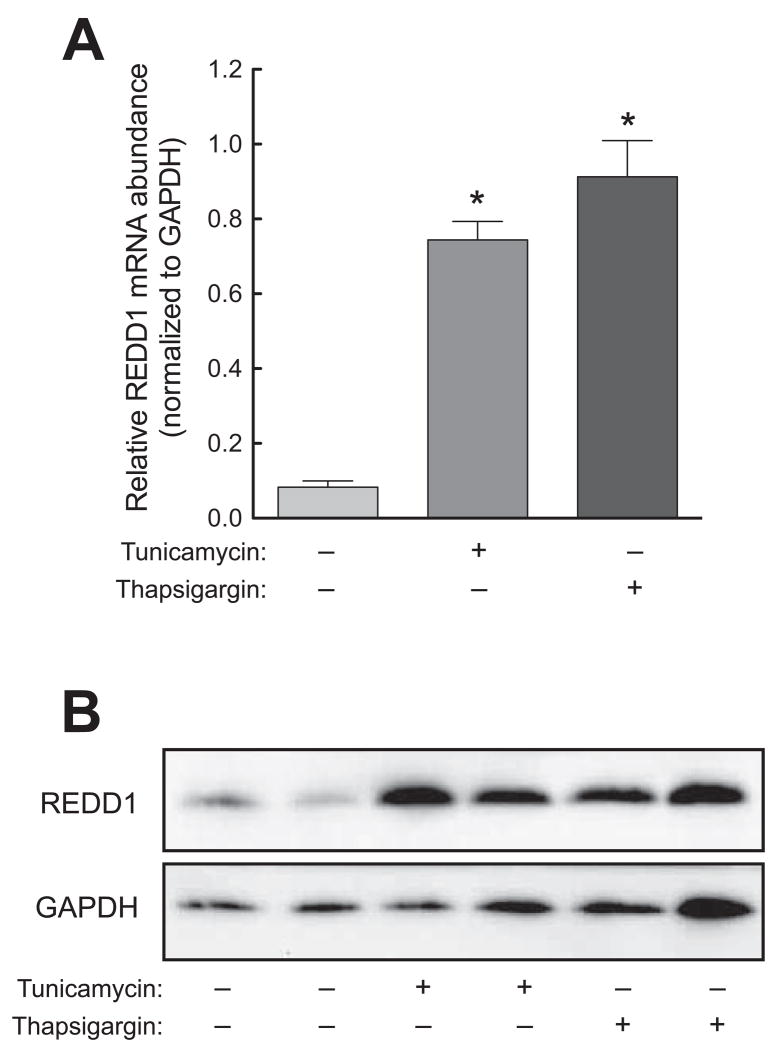
In two previous reports [4; 18], REDD1 mRNA expression was shown to be induced in response to ER stress. Herein, we confirm the results of the previous reports using HepG2 hepatoma cells subjected to ER stress induced by tunicamycin or thapsigargin. Consistent with the previous reports, ER stress induced by either agent resulted in upregulated REDD1 mRNA expression (Fig. 1A). Moreover, the upregulated mRNA expression corresponded to an elevation in REDD1 protein (Fig. 1B).
Fig. 1.
ER stress induces REDD1 expression in a liver-derived cell line. HepG2 cells were exposed to 10 μg/ml tunicamycin, 100 nM thapsigargin, or vehicle control in normal growth medium for 4 h. In separate experiments, RNA or protein was prepared from cell lysates and analyzed by qRT-PCR or Western blot respectively. (A) RNA was analyzed for the REDD1 and GAPDH mRNA expression. The results were normalized to GAPDH mRNA and are expressed as means ± SEM (n=3). *p<0.05 vs. control. (B) Western blot analysis using anti-REDD1 (top panel) and anti-GAPDH (bottom panel) antibodies. The results are representative of 3 studies that were performed.
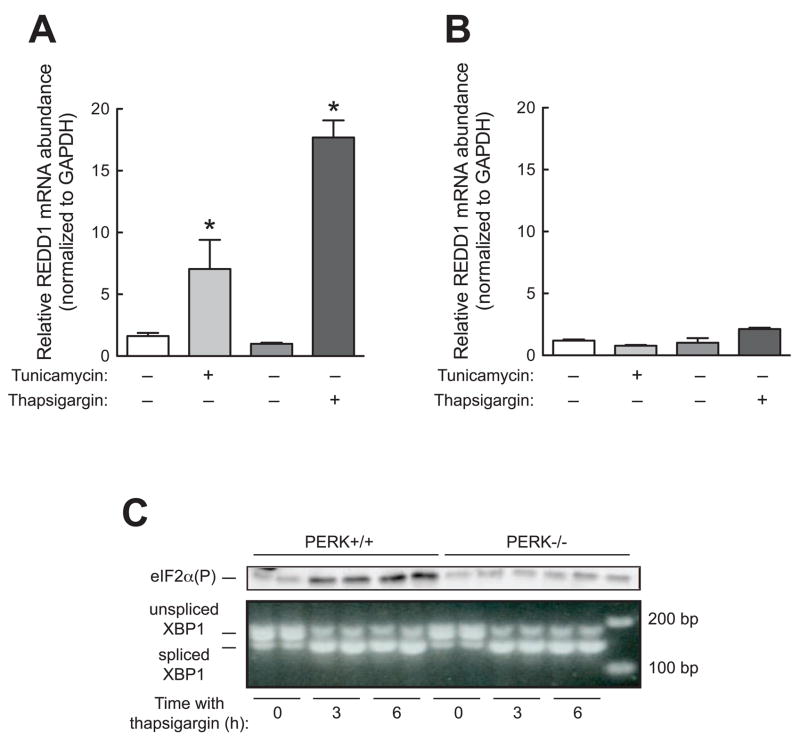
We next examined the mechanism through which REDD1 mRNA expression is upregulated during ER stress. When mammalian cells are subjected to ER stress, three distinct ER-resident stress sensors, PERK1, IRE1 and ATF6, become activated. Activation of these signaling pathways leads to a change in gene expression due to increased expression of various transcription factors including ATF4, ATF6, and XBP1 [17]. We hypothesized that one or more of these signaling networks was responsible for the enhanced expression of REDD1 during ER stress. Initially, we employed PERK−/− MEFs to assess whether PERK activity was necessary for increased REDD1 expression in response to stress induced by tunicamycin or thapsigargin. REDD1 expression increased during ER stress in wild type cells, but not PERK−/− cells (Fig. 2A and 2B). A possible explanation for the ineffectiveness of thapsigargin and tunicamycin in upregulating REDD1 expression in PERK−/− cells is that the cells might be resistant to induction of ER stress. PERK functions as an eIF2α kinase [20; 21], and as expected, wild type (PERK+/+) cells exhibited increased phosphorylation of eIF2α in response to treatment with thapsigargin (Fig. 2C, top panel). However, such treatment did not lead to eIF2α phosphorylation in PERK−/− cells. Since eIF2α phosphorylation was not increased in PERK−/− cells during ER stress, another index of ER stress, XBP1 processing was assessed. In unstressed PERK+/+ and PERK−/− cells, the majority of XBP1 was present in the unspliced form (Fig. 2C, bottom panel). However, at three and six hours after treatment with thapsigargin, the amount of XPB1 present in the spliced form was increased in both cell types. These data suggest that both wild type and PERK−/− cells are equally sensitive to thapsigargin-induced ER stress and that REDD1 expression was increased during ER stress through a mechanism requiring eIF2α phosphorylation.
Fig. 2.
Induction of REDD1 expression upon ER stress requires PERK. Wild type (PERK+/+) and PERK−/− MEFs were treated with DMSO (vehicle for tunicamycin), 10 μg/ml tunicamycin, ethanol (vehicle for thapsigargin), or 100 nM thapsigargin for 4 h. After treatment, cells were harvested for qRT-PCR analysis as described under Materials and Methods. The results represent mean ± SEM (n=3). *p<0.05 vs. control. (A) PERK+/+ MEFs. (B) PERK−/− MEFs. The relative amount of REDD1 mRNA was normalized to GAPDH mRNA. (C) PERK+/+ and PERK−/− MEFs were incubated for 0, 3, or 6 h with thapsigargin and then harvested for Western blot analysis or PCR analysis of XBP1 splicing as described under Materials and Methods.
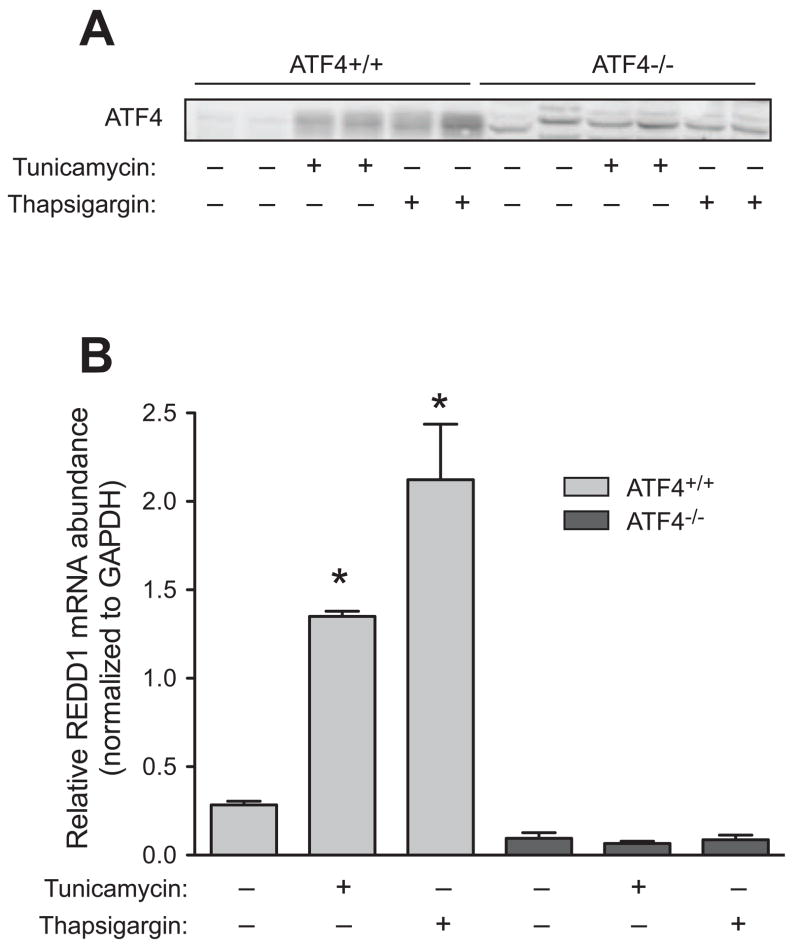
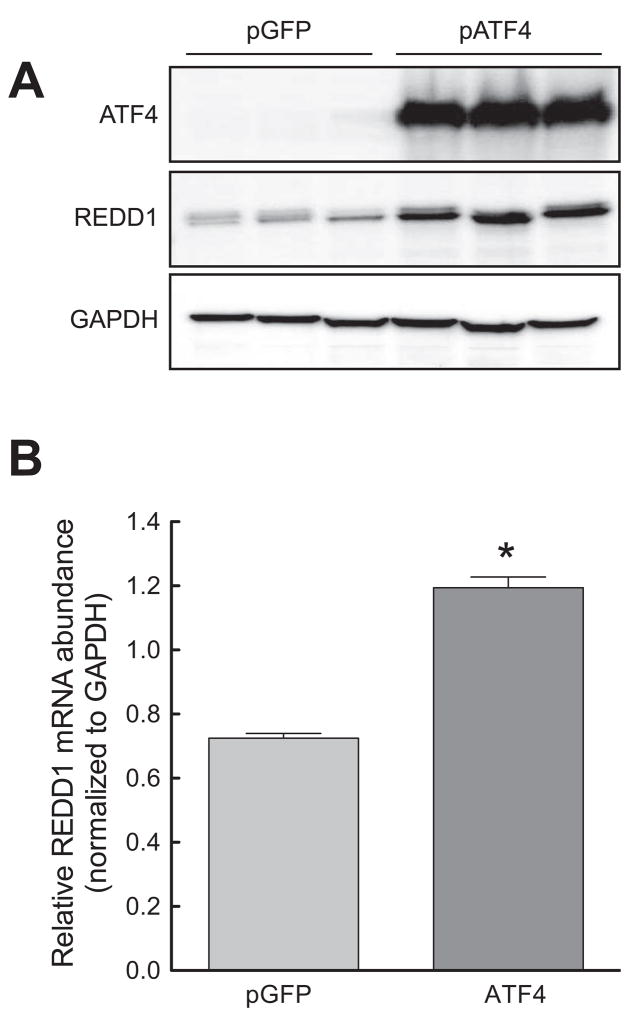
Phosphorylation of eIF2α leads to a global decrease in translation [22]. Paradoxically, the translation of a small subset of mRNAs becomes increased. The transcription factor ATF4 is a well-characterized member of this subset of mRNAs [23; 24; 25; 26]. As expected, ATF4 protein expression increased during conditions that resulted in eIF2α phosphorylation (Fig. 3A). Interestingly, ATF4−/− MEFs exposed to either tunicamycin or thapsigargin lacked the enhanced REDD1 expression displayed by their wild type counterparts (Fig. 3B). This finding shows that ATF4 was necessary for ER stress-induced upregulation of REDD1 expression, but did not demonstrate sufficiency. To determine whether increased ATF4 was sufficient to induce REDD1 expression, ATF4 was constitutively overexpressed in HEK293T cells, and the effect on REDD1 assessed. Transfection of cells with a plasmid expressing ATF4 resulted in an increase in REDD1 protein levels (Fig. 4A) and mRNA expression (Fig. 4B) relative to cells transfected with a plasmid expressing GFP, demonstrating that increased ATF4 expression was sufficient to upregulate REDD1 expression.
Fig. 3.
Induction of REDD1 expression upon ER stress requires ATF4. ATF4+/+ and ATF4−/− MEFs were treated with vehicle, 10 μg/ml tunicamycin, or 100 nM thapsigargin for 4 h. The results represent mean ± SEM (n=3). (A) Western blot analysis using anti-ATF4 antibodies. (B) qRT-PCR analysis was performed on REDD1 and GAPDH mRNA extracted from cells. The relative amount of REDD1 mRNA was normalized to GAPDH mRNA. The results represent mean ± SEM (n=3). *p<0.05 vs. control.
Fig. 4.
Overexpression of ATF4 leads to induction of REDD1 expression. HEK293T cells were transfected with pMAX-GFP (control) or a plasmid containing ATF4. (A) Western blot analysis was performed on cell homogenates using anti-REDD1, anti-ATF4, and anti-GAPDH antibodies. (B) RNA was extracted from cells and analyzed by qRT-PCR. The relative amount of REDD1 mRNA was normalized to GAPDH mRNA. The results represent mean ± SEM (n=3). *p<0.05 vs. control.
Discussion
Previous reports have shown that expression of the mRNA encoding REDD1 is upregulated during ER stress [4; 18]. In the present study, we show that ER stress-induced upregulation of REDD1 expression is dependent on PERK, suggesting that the effect requires eIF2α phosphorylation. This suggestion is supported by the finding that in PERK−/− MEFs, histidine deprivation, which activates the eIF2α kinase GCN2 [27], increased both eIF2α phosphorylation and REDD1 expression (data not shown). In one of the first steps in translation initiation, eIF2 binds GTP and the initiator form of methionyl-tRNA (met-tRNAi), and the ternary complex then binds to the 40S ribosomal subunit [reviewed in 28]. During a subsequent step, the GTP bound to eIF2 is hydrolyzed to GDP, resulting in release of the eIF2•GDP binary complex from the 40S ribosomal subunit. The GDP bound to eIF2 then must be exchanged for GTP in order for eIF2 to bind met-tRNAi to reform the ternary complex. The GDP-GTP exchange reaction that is catalyzed by another initiation factor, eIF2B. Phosphorylation of eIF2α converts the protein from a substrate into a competitive inhibitor of eIF2B, thereby repressing ternary complex formation. Because the translation of all eukaryotic mRNAs is initiated by the eIF2 ternary complex, inhibition of eIF2B results in downregulated translation of most messages. However, a few mRNAs, such as those encoding ATF4 and ATF5, continue to be translated, even when eIF2B is inhibited. Unlike most mRNAs, the mRNAs encoding these proteins have multiple upstream open reading frames in their 5′-untranslated regions. When eIF2α phosphorylation is low, and ternary complex is abundant, the presence of upstream open reading frames acts to repress translation of the authentic coding region. However, through a complex mechanism explained in detail elsewhere [29; 30], when ternary complex availability is low, translation of mRNAs bearing multiple upstream open reading frames is maintained, or even enhanced. Thus, the finding that ER stress upregulates ATF4 expression supports the idea that eIF2α phosphorylation is involved in the observed increase in REDD1 expression in cells treated with tunicamycin or thapsigargin.
ATF4 is a basic leucine zipper transcription factor that regulates the transcription of genes encoding proteins such as the system A sodium-dependent neutral amino acid transporter 2 [SNAT2, 31], asparagine synthetase [ASNS, 32], and growth arrest and DNA damage 34 [GADD34, 33]. ATF4-mediated upregulation of such transcripts is thought to play a key role in the recovery of the cell from the stress that induced eIF2α phosphorylation. For example, the amino acid deprivation-induced increase in expression of SNAT2, ASNS, and other genes encoding proteins that function to increase amino acid uptake or biosynthesis would help increase intracellular amino acid concentrations for use in mRNA translation and metabolism. Similarly, GADD34 targets protein phosphatase 1 to eIF2, resulting in dephosphorylation of the protein [34; 35]. By promoting eIF2α dephosphorylation, GADD34 derepresses eIF2B activity resulting in increased ternary complex formation, and resumption of translation of most mRNAs. Simultaneously, the translation of mRNAs with multiple upstream open reading frames, such as that encoding ATF4, is repressed.
In the present study, we show that increased ATF4 expression is both necessary for upregulation of REDD1 expression in response to ER stress and sufficient to upregulate expression of the protein. Since many of the genes induced by ATF4 function in recovery processes, this finding implies that REDD1 might act to help the cell recover from ER stress. The best characterized REDD1 function is to repress signaling through mTORC1. In this regard, a recent report [15] showed that loss of either TSC1 or TSC2 leads to a rapamycin-sensitive increase in XBP1 processing and S6K1 and PERK phosphorylation. Because TSC1 and TSC2 function as a complex to repress signaling through mTORC1, activation of the ER stress response in cells lacking these proteins suggests that mTORC1 activation promotes ER stress. Thus, it is tempting to speculate that the ER stress-induced upregulation of REDD1 expression observed in the present study might provide a feedback mechanism to reduce mTORC1 signaling and thereby attenuate ER stress.
Overall, the results of the present study are consistent with a model in which ER stress leads to activation of PERK which subsequently phosphorylates eIF2. Phosphorylation of eIF2 leads to increased expression of the transcription factor ATF4, and ATF4 is both necessary and sufficient for increased expression of REDD1. Future studies will be required to identify the mechanism through which ATF4 upregulates REDD1 expression.
Acknowledgments
The authors would like to thank Holly Lacko for performing the qRT-PCR assays described herein. The studies described in this manuscript were supported by a grant from the National Institutes of Health (DK13499).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel Hypoxia-Inducible Factor 1-responsive gene, RTP801, involved in apoptosis. Molecular and Cellular Biology. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Molecular Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 3.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO Journal. 2002;21:6162–73. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Malone MH, Thomenius MJ, Zhong F, Xu F, Distelhorst CW. Dexamethasone-induced Gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. The Journal of Biological Chemistry. 2003;278:27053–27058. doi: 10.1074/jbc.M303723200. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. The Journal of Biological Chemistry. 2006;281:39128–39134. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 6.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes & Development. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & Development. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801l are negative regulators of the mammalian Target of Rapamycin pathway. The Journal of Biological Chemistry. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 9.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2 mTOR signaling and tumor suppression through REDD1-mediated 14 3 3 shuttling. Genes & Development. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Molecular and Cellular Biology. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garami A, Zwartkruis FJT, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP1 signaling, is inhibited by TSC1 and 2. Molecular Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 12.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes and Development. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR by acting as a GTPase-activating protein complex toward Rheb. Current Biology. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 14.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Current Biology. 2005;15:702. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 15.Ozcan U, Ozcan L, Yilmaz E, D¸vel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Molecular Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji C, Kaplowitz N. ER stress: Can the liver cope? Journal of Hepatology. 2006;45:321–333. doi: 10.1016/j.jhep.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiological Reviews. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 18.Protiva P, Hopkins ME, Baggett S, Yang H, Lipkin M, Holt PR, Kennelly EJ, Bernard WI. Growth inhibition of colon cancer cells by polyisoprenylated benzophenones is associated with induction of the endoplasmic reticulum response. International Journal of Cancer. 2008;123:687–694. doi: 10.1002/ijc.23515. [DOI] [PubMed] [Google Scholar]
- 19.Scorsone KA, Panniers R, Rowlands AG, Henshaw EC. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. The Journal of Biological Chemistry. 1987;262:14538–14543. [PubMed] [Google Scholar]
- 20.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Molecular and Cellular Biology. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pain VM. Initiation of protein synthesis in mammalian cells. Biochemical Journal. 1986;235:625–637. doi: 10.1042/bj2350625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding HP, Novoa I, Zhang Y, Zeng H, Wek RC, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 24.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Molecular Cell. 2001;7:1165–76. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 25.Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS. MEK signaling Is required for phosphorylation of eIF2α following amino acid limitation of HepG2 human hepatoma cells. Journal of Biological Chemistry. 2008;283:10848–10857. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, Cavener DR. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Molecular & Cellular Biology. 2002;22:6681–8. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Current Opinion in Structural Biology. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 29.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annual Review of Microbiology. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 30.Wek RC, Cavener DR. Translational Control and the Unfolded Protein Response. Antioxidants & Redox Signaling. 2007;9:2357–2372. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 31.Gjymishka A, Palii SS, Shan J, Kilberg MS. Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity Is repressed in HepG2 cells. Journal of Biological Chemistry. 2008;283:27736–27747. doi: 10.1074/jbc.M803781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 Is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. Journal of Biological Chemistry. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 33.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. Journal of Biological Chemistry. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 34.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Molecular & Cellular Biology. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. Journal of Cell Biology. 2001;153:1011–1021. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]