Abstract
The complexity of the adult brain is the result of an integrated series of developmental events that depends on appropriate timing of differentiation. The importance of transcriptional regulatory networks and epigenetic mechanisms of regulation of gene expression is becoming increasingly evident. Among these mechanisms, previous work has revealed the importance of histone deacetylation in oligodendrocyte differentiation. In this manuscript we define the region of interaction between transcription factor Yin-Yang 1 (YY1) and histone deacetylase 1, and characterize the functional consequences of YY1 overexpression on the differentiation of oligodendrocyte progenitors.
Keywords: chromatin, transcription, development, brain, myelin, epigenetic
INTRODUCTION
Oligodendrocytes, the myelin forming cells of the CNS, are generated from oligodendrocyte progenitor cells (OPCs) during early postnatal days (Sauvageot and Stiles, 2002). During this developmental window OPCs that reside throughout the neuraxis exit from the cell cycle and differentiate throughout the CNS (Baumann and Pham-Dinh, 2001). Although the mechanisms regulating proliferation in these cells have been studied extensively (Casaccia-Bonnefil et al., 1996; Casaccia-Bonnefil et al., 1999; Durand and Raff, 2000), the characterization of the molecular events at the interface between cell-cycle exit and the initiation of the complex program of differentiation remain elusive.
During the past decade, the role of epigenetic factors (post-translational modifications of the histone tails, histone variants switching, ATP-remodeling complexes and microRNAs) in the regulation of brain development (Seo et al., 2005; Matsumoto et al., 2006; Cao et al., 2006), has been investigated extensively. Studies from our group (Marin-Hustegge et al. 2002; Shen et al. 2005) and others (Hsieh et al., 2004; Kondo and Raff, 2004; Lyssiotis et al., 2007) have identified global deacetylation of histone H3 as one of the first epigenetic events occurring in oligodendrocyte progenitors during the transition between cell-cycle exit and initiation of the transcriptional program of differentiation.
The removal of acetyl groups from lysine residues in the histone tails is performed by specific enzymes called histone deacetylases (HDACs). However these enzymes do not have the ability to directly bind to DNA. A recent study in our laboratory has identified the transcription factor Yin Yang 1 (YY1) as a recruiter of HDAC1 on the promoter of the transcriptional inhibitors Tcf4 and Id4 in order to decrease their transcription levels during oligodendrocyte progenitor differentiation (He et al., 2007).
The transcription factor Yin Yang 1 (YY1) is a multi-functional protein that can either activate or repress gene expression depending on the binding partner, including HDAC1, p300, TBP, Rb and E1A. It has been shown that YY1 regulates the expression of diverse genes (e.g. p53, c-myc, IFN-β and CREB) that are important for cellular activity. However different studies describe distinct roles of YY1 in different aspects of cellular activities, including proliferation, apoptosis and differentiation (reviewed in Shi et al., 1997). The role of yy1 in nervous system development, was suggested in studies in Xenopus embryo in which yy1 deletion caused neuronal defects (Kwon and Chung, 2003; Morgan et al., 2004). The role of YY1 in oligodendrocyte differentiation has been indicated by in silico-analysis of the promoter sequences of genes that are downregulated by histone deacetylation during oligodendrocyte differentiation. It was noted that almost one third of the downregulated genes have YY1-binding sites on their promoter and conditional ablation of YY1 in the oligodendrocyte lineage significantly impairs myelination (He et al., 2007). In this manuscript we have explored the role of YY1 as recruiter of HDAC activity by studying the consequences of complex disruption and investigated the effects of overexpressing YY1 in immortalized mouse OPCs (Jung et al., 1995).
OBJECTIVES
METHODS
Animals
All the mice and rats used in this study were handled according to protocols approved by the Institutional IACUC committee. Yy1flox/flox mice were previously generated by Dr Y. Shi (Affar et al., 2006). Cnp1-cre mice in C57BL were provided by Dr K. Nave (Max Planck Institute, 2003). Yy1 conditional knockout mice and control siblings were obtained from breeding yy1flox/+; cnp-cre+/− pairs or breeding yy1flox/flox with yy1flox/+; cnp-cre−/− mice. Genotyping was performed as reported previously (He et al., 2007).
Antibodies and plasmids
Acetylated H3 (rabbit polyclonal; 1:1,000; Upstate Biotechnology), CC1/APC (mouse monoclonal; 1:50; Oncogene Research Products), Flag (mouse monoclonal; 1:1,000 for IHC and 1:2,000 for WB; Sigma) HDAC1 (rabbit polyclonal; 1:6,000; Affinity BioReagents, Inc.), NG2 chondroitin sulfate proteoglycan (mice monoclonal; 1:200; CHEMICON International), O4 (mouse monoclonal; 1:10, gift from Dr Bansal, University of Farmington), Sox2 (rabbit polyclonal; 1:1,000; CHEMICON International), Tcf4 (rabbit polyclonal; 1:100; Santa Cruz), and YY1 (mouse monoclonal sc-7341; 1:100; Santa Cruz).
PCEP4F-yy1 encoding full-length, human YY1 fused with flag at the N terminal was a gift from Dr Seto of University of South Florida. To better visualize the transfected cells, the coding sequence of flag-YY1 was subcloned into pCX vector which expresses EGFP under the IRES element. To create a YY1 mutant that is defective for binding to HDAC1, six lysine residues within the central domain of the molecule (amino acids 173,174,178,179,182 and 183) were changed to arginine by mutating the AAG codon into AGG using the QuikChange II Site Directed Mutagenesis Kit (Stratagene). The mutations were confirmed by DNA sequencing.
Oli-Neu cell culture and transfections
Mouse oligodendrocyte progenitors immortalized with the neu antigen (Oli-Neu cell line) were a gift of Dr J. Trotter (University of Mainz, Germany) (Jung et al., 1995). Cells were grown on poly-ornithine coated culture dishes and maintained proliferating in growth medium ODM plus 1% horse serum (HS) as described previously (He et al., 2007). Proliferating Olineu cells were transfected with different plasmids using FuGENE 6 (Roche) at a ratio of 3 µl FuGENE6: 1 µg DNA according to the manufacturer’s manual. Two days later cells were immunostained for AcH3.
Adenoviral infection of mouse OPCs
OPCs from yy1flox/flox neonatal mice were isolated and infected with adenovirus-CMV-Cre as described previously (He et al., 2007). Two days later the cells were processed for YY1 and AcH3 immunocytochemistry.
Immunohistochemistry and immunocytochemistry
Immunocytochemistry of cultured cells with O4 antibodies was performed live. Cells were rinsed gently in phosphate-buffered saline (PBS; 10 mM sodium phosphate, pH 7.4, and 150 mM NaCl) and incubated live with O4 hybridoma supernatant (1:10) for 30 min at 37°C. Cells were then fixed with 4% PFA for 20 min at room temperature and stained with secondary antibodies as described. For immunohisto-chemistry, animals were anesthetized and perfused intracardially with 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2. The brains were removed, post-fixed overnight in the same solution at 4°C, and then cryopreserved in 30% sucrose in 0.1 M phosphate buffer and embedded in OTC. Frozen sections (20 µm) were cut and incubated in blocking buffer (0.1 M phosphate buffer, 0.1% gelatin, 1% bovine serum albumin, 0.002% sodium azide) containing 10% normal goat serum (0.5% TritonX-100 was added for cytoplasmic and nuclear antigens) for 30 min and then incubated overnight with the primary antibody diluted in the same blocking buffer. After rinsing in PBS sections were incubated with the appropriate secondary antibodies conjugated to either fluorescein or rhodamine (Southern Biotechnologies, Amersham Biosciences, Jackson ImmunoResearch and Vector Laboratories). DAPI (1:1,000; Molecular Probes, Inc.) was used as nuclear counterstain.
Image acquisition, processing and quantification
Immunostained sections were analyzed using fluorescence microscopy (DM RA; Leica) followed by confocal microscopy analysis (LSM 510 Meta confocal laser scanning microscope; Carl Zeiss MicroImaging, Inc.). Confocal images of mouse brain sections were captured at 1 µm intervals and stacks of six slices were typically used for the generation of the projections. The intensity of AcH3 fluorescence staining was measured in arbitrary units using NIH ImageJ software. For the quantification of the in vitro experiments on transfected immortalized progenitors, a minimum 40 GFP+ cells were measured in each duplicate repeat of two independent experiments. For the quantification of the AcH3 immunostaining in the cerebellum, 20 randomly selected CC1+ cells were measured in the similar area of each section, a minimum of three sections (corresponding to the levels of plates 127–136 from (Sidman et al., 1997) per mouse and three mice each genotype were evaluated. Results were expressed as mean ± SD and statistically analyzed using two-tailed Student’s t test, P<0.05 was considered statistically significant.
Immunoprecipitation and Western blot
Proteins from the transfected Oli-Neu cells were extracted using a buffer containing 50 mM Hepes, pH 7.0, 250 mM NaCl, 0.5% NP-40, 1 mM DTT, 1 mM EDTA, 0.01% PMSF, 1 mM aprotinin and 1 mM leupeptin for 15 min on ice. Equal amounts of protein (1 mg) were immunoprecipitated for 16–18 hours at 4°C using 2 µg anti-flag antibody. Pcx-yy1 transfected cell lysate immunoprecipitated with normal mouse IgG (Santa Cruz) was used as negative control. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane, followed by Western blot probed with anti-flag and anti-HDAC1 antibody. Immunoreactive bands were visualized using HRP-conjugated secondary antibodies followed by chemiluminescence using ECL Plus Western Blotting Detection System (Amersham Biosciences).
HDAC activity assay
HDAC activity was measured by using HDAC Activity Assay/Drug Discovery Kit (BIOMOL Research Laboratories, Inc.). Experimental procedures were designed and performed according to the kit protocol provided. In brief, tissue lysates from transfected Oli-Neu cells (prepared according to the same procedure described in Western blot analysis section) were used as sources for HDAC activity. Protein lysates (25 µg) were added to a 96-well plate in 25 µl HDAC assay buffer (BIOMOL Research Laboratories, Inc.). A fluorimetric acetylated substrate was added and the reaction allowed to proceed at room temperature for 1 hour, followed by incubation with the developer for 10–15 min. Enzymatic activity was evaluated in Perseptive Biosystems CytoFlour multi-well plate reader-4000 (excitation, 360 nm; detection of emitted light, 460 nm). The results were normalized to the amount of the protein.
Quantitative and semi-quantitative reverse transcriptase PCR
Mouse tissues of each age (embryonic day 18, postnatal days 2, 4, 9, 12, 15 and 22) or cell pellets were homogenized in Trizol® Reagent and RNA isolated following the manufacturer’s instruction and cleaned using RNeasy Mini kit (Qiagen). Total RNA (2 µg) was used in a 40 µl reverse transcription (RT) reaction using the SuperScript RT-PCR kit (Invitrogen). Semi-quantitative PCR was performed in a 20 µl reaction mixture containing 2 µl cDNA as template and 0.1 µM specific oligonucleotide primer pairs using program denaturation at 94°C for 60 sec, annealing at 55°C for 30 sec and extension at 72°C for 45 sec for 25 cycles. Actin was used as loading control. Quantitative RT-PCR was performed using Applied Biosystems SYBR green PCR master mix in 7900HT Sequence Detection PCR System. The melting curve of each sample was measured to ensure the specificity of the products. Data were normalized to the internal control GAPDH and analyzed using Pfaffl ΔΔCt method. Primers sequences are given in Table 1. All the primers were designed for mouse genes except primers for yy1 and β-actin, which recognize mouse, rat and human homology.
Table 1.
PCR primers Semi-quantitative PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Semi-quantitative PCR primers | ||
| β-actin | 5′-TGGAATCCTGTGGCATCC-3′ | 5′-TCGTACTCCTGCTTGCTG-3′ |
| CGT | 5′-TGCCAACGTATCCTTCTTCC-3′ | 5′-CATTGTCCCATGTCAAGCAC-3′ |
| Hes5 | 5′-CAAGGAGAAAAACCGACTGC – 3′ | 5′-GCTGGAAGTGGTAAAGCAGC-3′ |
| Id4 | 5′-GATGAAGGCGGTGAGCCCGGTG-3′ | 5′-GACAGCATTCTCTGCCGCTGA-3′ |
| Mash1 | 5′-CTTCCTTAAGGCCTCTGGCT-3′ | 5′-GAACCCGCCATAGAGTTCAA-3′ |
| MBP | 5′-ATGGCATCACAGAAGAGACC – 3′ | 5′-CATGGGAGATCCAGAGCGGC -3′ |
| NG2 | 5′-GAACGCATCAGCCACCGTAA -3′ | 5′-GGACGCTTCTTCCTGGTTTC-3′ |
| Olig1 | 5′-ATGAGCTGGTGGGTTACAGG -3′ | 5′-CACCAGCTGGGAGAGAGAAC-3′ |
| Sox2 | 5′-ATGATGGAGACGGAGCTGAA -3′ | 5′-CTCCGGGAAGCGTGTACTTA-3′ |
| Sox10 | 5′-GACCAGTACCCTCACCTCCA-3′ | 5′-CCCCTCTAAGGTCGGGATAG-3′ |
| Tcf4 | 5′-CTCACGCCTCTCATCACGTA-3′ | 5′-TGAATGCATTAAGGGGCTTC-3′ |
| yy1 | 5′-GGTGCAGATCAAGACCCTGGA-3′ | 5′-GTGTGCGCAAATTGAAGTCCA-3′ |
| Quantitative PCR primers | ||
| GAPDH | 5′-ACCCAGAAGACTGTGGATGG-3′ | 5′-CACATTGGGGGTAGGAACAC-3′ |
| plp | 5′-CCCACCCCTATCCGCTAGTT -3′ | 5′-CAGGAAAAAAAGCACCATTGTG-3′ |
| Id4 | 5′- GCCCAACAAGAAAGTCAGCAA -3′ | 5′-CCAGCTGCAGGTCCAGGAT-3′ |
| Sox11 | 5′-GACGACCTCATGTTCGACCT -3′ | 5′-TCCAGGTCCTTATCCACCAG-3′ |
| Tcf4 | 5′-GTCCTCGCTGGTCAATGAAT -3′ | 5′-CCCTTAAAGAGCCCTCCATC-3′ |
RESULTS
Because expression of the encoding myelin is regulated by a complex network of activators and inhibitors we reasoned that differentiation timing might be affected by the equilibrium between inhibitors and activators (Gokhan et al., 2005). In support of this hypothesis, quantitative real-time PCR analysis of RNA isolated from the developing nervous system of mice between embryonic day 18 and postnatal day 22 revealed that myelin gene expression is preceded by downregulation of transcriptional inhibitors Id4, Tcf4 and Sox11 Fig. 1. Our previous studies indicate that YY1 acts as lineage-specific repressor of these genes during oligodendrocyte progenitor differentiation by forming a protein complex with HDAC1 (He et al., 2007). We now demonstrate that the activating function of YY1 depends on its ability to directly bind HDAC1. Briefly, either full-length or mutant YY1 containing six aminoacid substitutions (KR) in the central HDAC binding domain (Yao et al., 2001), were subcloned into a pCX vector containing EGFP under the IRES element (Fig. 2A). The inability of mutant YY1 (KR) to bind to HDAC1 was confirmed by immunoprecipitation followed by Western blot analysis (Fig. 2B) of protein extracts from transfected Oli-neu cells (Jung et al., 1995). The effect of YY1 overexpression on the enzymatic activity of HDAC1 was inferred by the decrease in global levels of acetylation of histone H3 detected in transfected OPCs (Fig. 2). Transfected oligodendrocyte progenitors were identified by green fluorescence and NG2 immunoreactivity (Fig. 2C). The relative levels of acetylation of histone H3 (AcH3) in transfected cells were calculated by measuring the average fluorescence intensity of the AcH3+ nuclei using NIH ImageJ (Fig. 2C,D). Although the AcH3 intensity measured within each group varied (possibly reflecting either different levels of ectopic gene expression or different stages of the cells), the average pixel intensity in the cells transfected with full-length YY1 was significantly lower than vector transfected controls (Fig. 2D). In contrast, the average intensity measured in cells expressing the mutant form of YY1 was equivalent to the intensity measured in vector transfected controls (Fig. 2D).
Figure 1. Timing of oligodendrocyte differentiation is characterized by a decrease in inhibitor levels before activation of myelin gene expression.
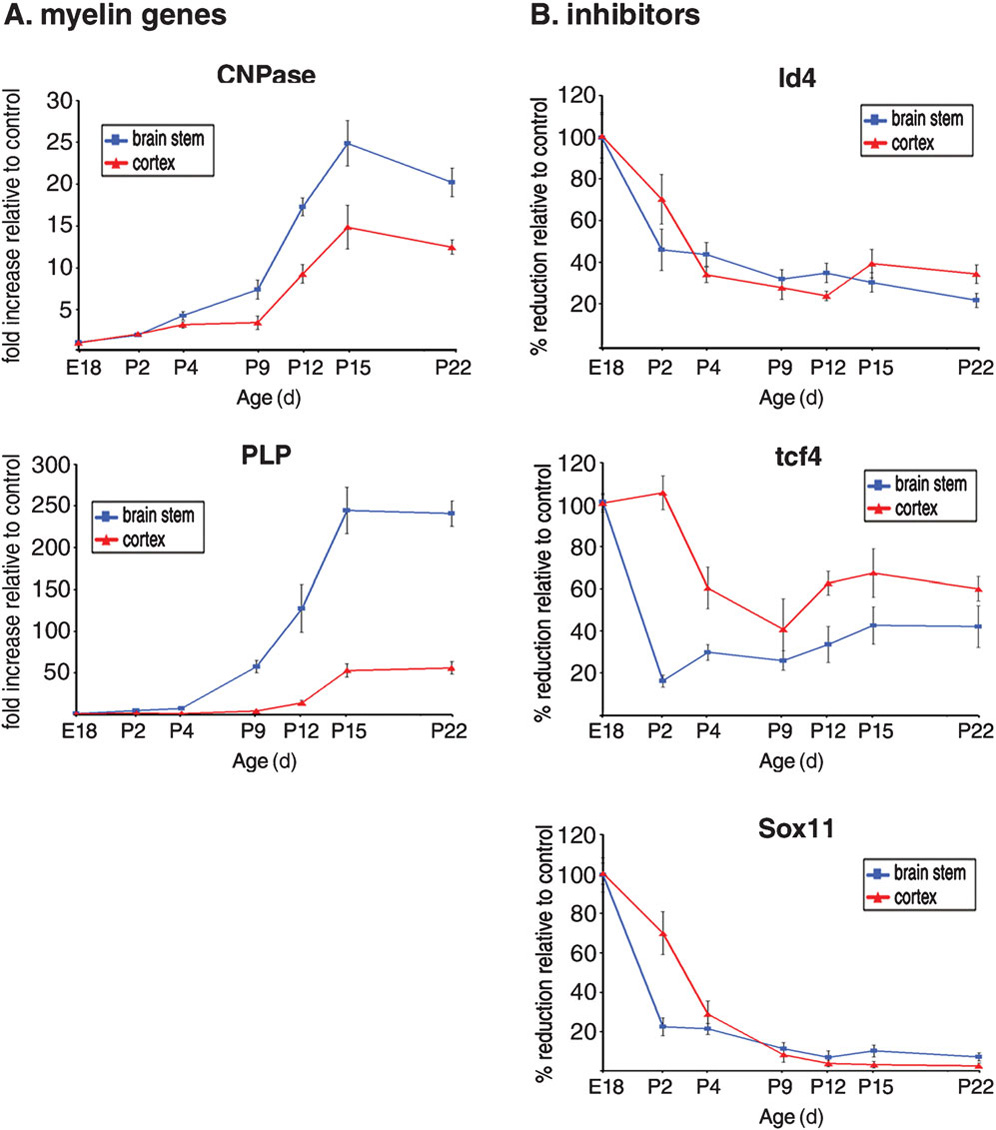
(A) Time course of myelin gene expression during development from embryonic day 18 (E18) to post-natal day 22 (P22). RNA levels of myelin genes CNPase and plp were measured by quantitative PCR of brain stem and cortical areas of individual mice, harvested at the indicated time points (n = 3 mice at each time point). The values were normalized to GAPDH and referred as relative fold increase of the values measured in mice at E18. Data are mean ± SD. (B) Transcript levels of oligodendrocyte differentiation inhibitors (Id4, tcf4 and sox11) were analyzed by qPCR during development in the brain stem and cortex. The values were normalized to GAPDH and referred as relative fold increase of the values measured in mice at E18.
Figure 2. YY1 modulates global histone acetylation through interacting with HDAC1.
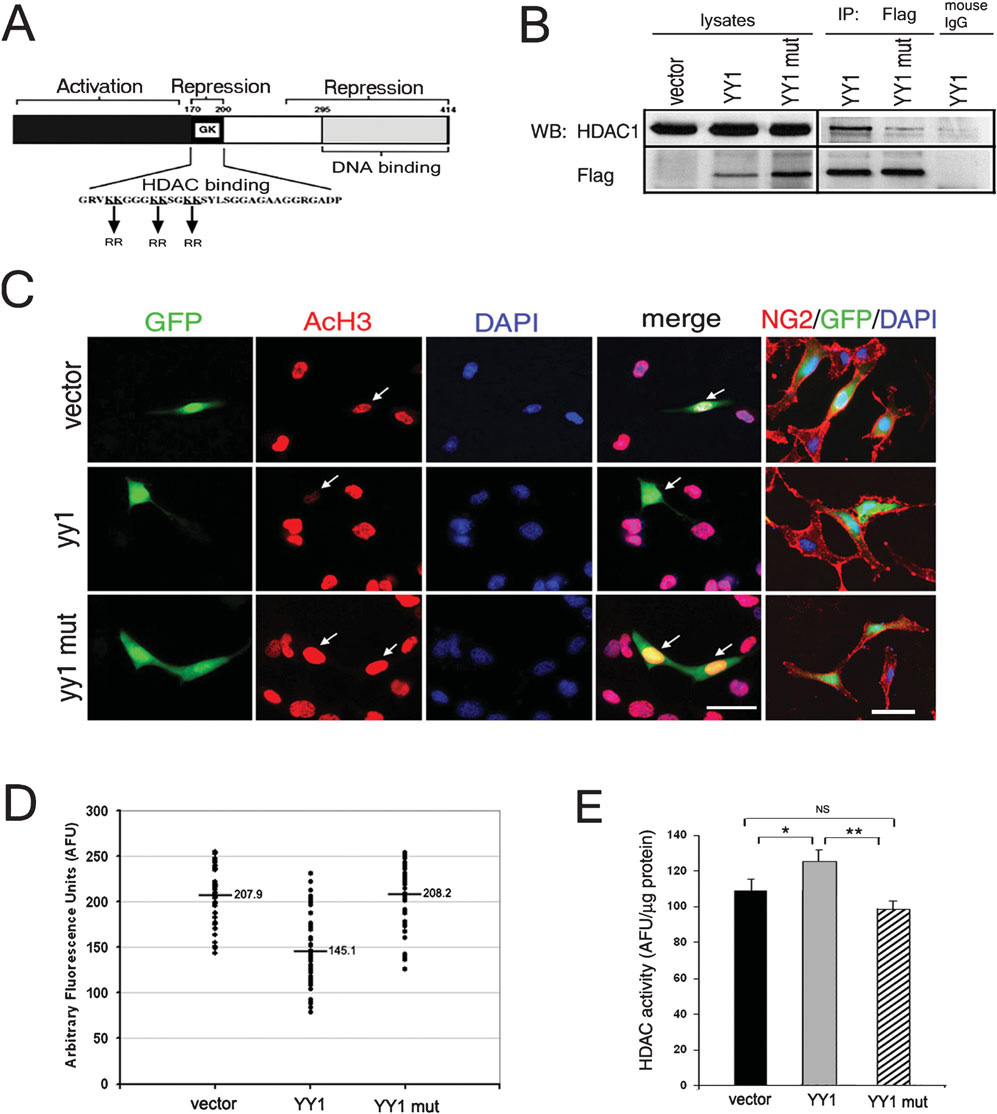
(A) The functional domains of YY1, including the DNA binding domain and the HDAC-interacting region. The arrows indicate the position of the amino acid substitutions in the HDAC1 binding-deficient mutant of YY1, in which the six lysine residues in the central HDAC-binding domain are replaced by arginines. (B) Oli-Neu cells were transfected with empty vector, Flag-full-length yy1 (YY1) or Flag-mutant yy1 (YY1mut). Protein lysates were immunoprecipitated with anti-Flag mouse monoclonal antibody. After SDS-PAGE separation and transfer, the blots were processed for Western blot analysis using antibodies specific for HDAC1 and Flag. Immunoprecipitation with normal mouse IgG was used as negative control. Note that HDAC1 co-immunoprecipitates wild-type but not mutant YY1. (C) Immortalized murine oligodendrocyte progenitors (i.e. Oli-neu cells) were transfected (green) with pCX-GFP (vector), full-length yy1-GFP (yy1) or mutant yy1-GFP (yy1mut) and stained for acetylated histone H3 (AcH3, red). Note the decreased AcH3 immunoreactivity in cells transfected with full-length YY1 but not with the mutant YY1 compared to the vector. Cells were stained for progenitor marker NG2 to define their morphology. Scale bars, 25 µm. (D) Scatter plot of fluorescence intensity of AcH3 in the nuclei of GFP+ transfected cells, measured by NIH ImageJ. The average values from 40, randomly chosen cells in each group are presented. Note that even though the AcH3 level varies in individual cells, only the full-length YY1, but not the mutant form of YY1 had the overall effect of decreasing AcH3 level. (E) Total HDAC enzymatic activity in whole cell extracts of control (vector) and YY1-transfected cultures, measured by a fluorimetric assay, reveals that increased levels of wild-type YY1 but not the mutant form increases HDAC activity. Data are mean ± SD. *P<0.05, *P<0.01 (Student’s t-test).
Finally, we used a fluorimetric assay to compare the levels of HDAC enzymatic activity in cultured progenitors transfected with full-length YY1 (pCX-yy1-EGFP) with those in progenitors transfected with the control vector (pCX- EGFP) (Fig. 2E). The overall enzymatic activity of HDAC in cells that overexpress full-length YY1 (125.5 ± 6.1 AFU/µg protein) was 25% greater than the activity in cells transfected with vector alone (109.2 ± 6.2 AFU/µg protein). Together these results indicate that the ability of YY1 to modulate the acetylation of histone H3 depends on the formation of a protein complex with HDAC1.
To confirm these data independently we tested the consequences of in vitro and in vivo ablation of yy1 on the global levels of acetylated nuclear histone H3 in oligodendrocyte progenitors. Ablation of yy1 in vitro was achieved by infecting yy1 flox/flox oligodendrocyte progenitors with adenoviral vectors expressing the recombinase Cre as described previously (He et al., 2007). Consistent with a crucial role of YY1 in modulating HDAC activity, the impaired removal of acetyl groups from the histones of YY1− cells resulted in increased acetyl H3 levels compared to YY1+ cells (Fig. 3A). In yy1 conditional knockout mice, a similar deficit of HDAC activity was inferred by the detection of higher levels (~1.5 fold increase) of acetylated histone H3 in CC1+ oligodendrocyte lineage cells within white matter tracts of conditional mutants compared to wild type mice (Fig. 3B,C).
Figure 3. Defective oligodendrocyte differentiation in yy1 conditional-knockout (cko) mice is associated with persistent global histone acetylation.
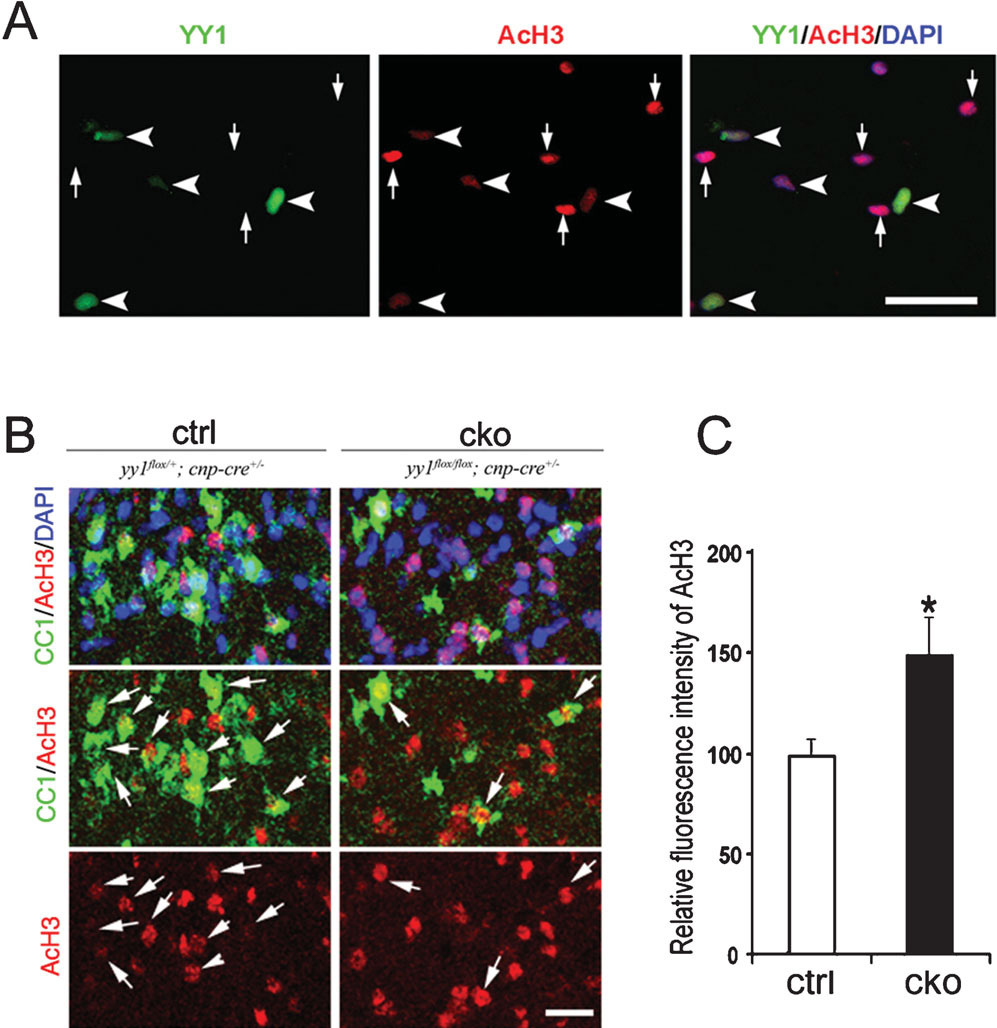
(A) Oligodendrocyte progenitors generated from neonatal yy1flox/flox mice were infected with adenovirus-CMV-Cre and 48-hours later stained for YY1 (green) and AcH3 (red). Note that YY1-deleted cells (arrowhead) have higher AcH3 immunoreactivity compared to uninfected cells (arrow) in the same field. Scale bar, 20 µm. (B) Sagittal sections of the cerebellum of control and yy1-cko mice stained for oligodendrocyte marker CC1 (green) AcH3 (red) shows a higher level of AcH3 in yy1-cko mice compared to sibling controls at P18. Scale bar, 20 µm. (C) Fluorescence intensity of AcH3 in CC1+ cells in control mice (n = 3) and yy1-cko mice (n = 3) was calculated using NIH ImageJ on acquired confocal images. The average fluorescence intensity in the control siblings is set arbitrarily at 100. Data are mean ± SD. *P<0.05, Student’s t-test.
Our previous work indicates that loss of yy1 prevents the progression of OPCs to terminal differentiation and arrests the cells at a stage characterized by simple morphology and O4 immunoreactivity (He et al., 2007). We therefore tested the effects of yy1 overexpression on the progression of NG2+ cells along the lineage. Whereas transfection of the pCEP4F vector did not affect the overall expression of NG2 in Oli-Neu cells, transfection of pCEP4F-yy1 decreased NG2 expression and increased O4 immunoreactivity (Fig. 4A). A possible explanation for these results is that YY1 has a repressive role on the expression of differentiation inhibitors, but does not directly affect activators or modulate the expression of myelin genes. In agreement with this interpretation, RT-PCR of RNA samples isolated from yy1-overexpressing cultures revealed decreased levels of Id4, Tcf4, Hes5 and Sox2 and unchanged levels of the transcription activators (i.e. Sox10, Olig1 and Mash1) and of myelin gene products (i.e. myelin basic protein and ceramide galactosyltransferase) compared to controls (Fig. 4B). The effect of YY1 on Sox2 and TCf4 expression was further validated by immunocytochemistry (Fig. 4C). Together these results indicate that YY1 is necessary but not sufficient to drive NG2 progenitors to a terminally differentiated phenotype.
Figure 4. Overexpression of yy1 decreases the concentration of oligodendrocyte differentiation inhibitors but does not promote the differentiation.
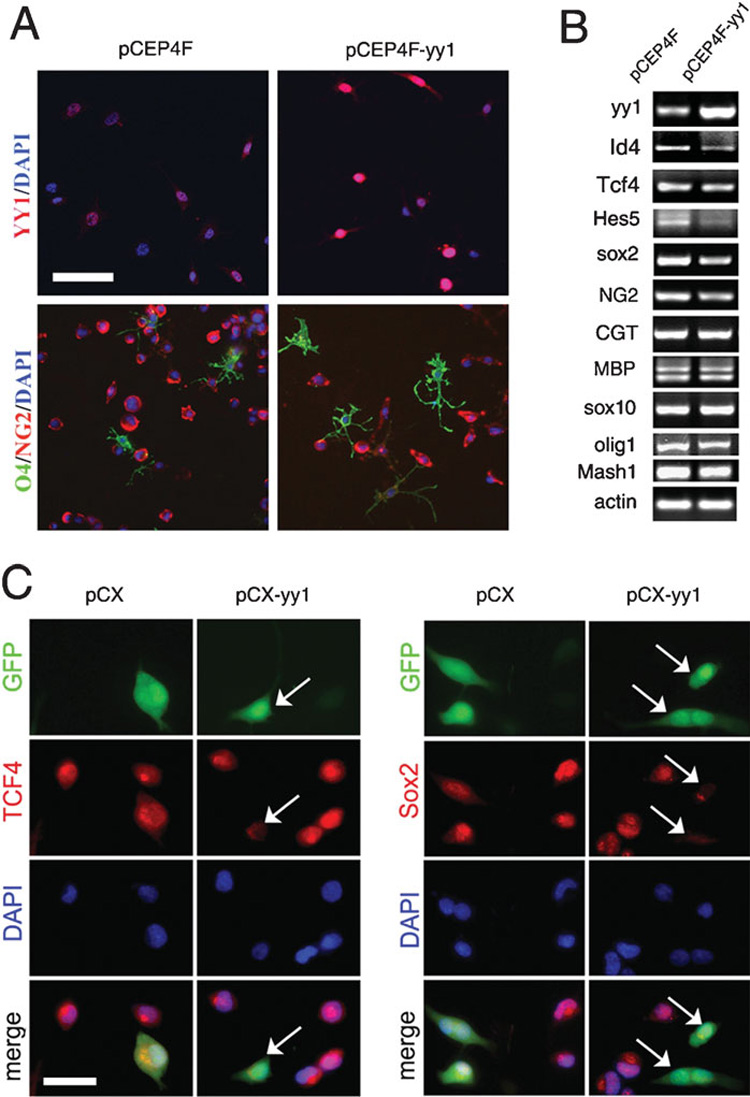
(A) Oli-Neu cells were transfected with either pCEP4F vector or pCEP4F-yy1 plasmid in proliferating medium. After 24 hours the cells were induced to differentiate by adding cAMP. At 1 day later, the cultures were stained for YY1 to confirm overexpression and were double immunostained for progenitor marker NG2 and early differented marker O4. Scale bar, 50 µm. (B) Oli-Neu cells were transfected with either pCEP4F vector or pCEP4F-yy1 plasmid in proliferating medium. After 24 hours the cells were induced to differentiate and 48 hours later the total mRNA was extracted and semi-quantitative PCR performed. Note the decreased levels of NG2, TCF4, Id4, Hes5 and Sox2. However the concentration of myelin genes did not increase, probably because the levels of transcriptional activators such as Sox10, Olig1 and Mash1 is not upregulated. (C) Oli-Neu cells were transfected with either pCX vector or pCX-yy1 plasmid and maintained in proliferating medium. After 2 days the cells were fixed and subjected to immunostaining for TCF4 and Sox2. The concentration of both proteins decreased in cells that overexpress yy1 compared to either untransfected cells in the same field or vector-transfected cells. Scale bar, 20 µm.
CONCLUSIONS
Although the transition between exit from the cell cycle and the onset of oligodendrocyte differentiation remains poorly characterized, the timing of oligodendrocyte progenitor differentiation is regulated by extrinsic and intrinsic signals.
Crucial intrinsic factors in this transition are HDACs and YY1, which both initiate differentiation by repressing transcriptional inhibitors of myelin gene expression.
However, extrinsic signals that regulate HDACs and YY1 in oligodendrocyte development are unknown and deserve further investigation.
YY1 is part of the polycomb complex that catalyzes the methylation of histone tails, experiments are in progress to define whether either this modification or other epigenetic events contributes to the repression of inhibitory molecules.
DISCUSSION
Three major cell types define the structure and function of the brain in both adult and developing animals: neurons, astrocytes and oligodendrocytes. This complex architecture is achieved by precisely orchestrating timing of differentiation of distinct cell types in overlapping, but temporally distinct, waves, with the neurogenic phase occurring during early embryogenesis, followed by astrogliogenesis and, finally, by oligodendrogliogenesis (Sauvageot and Stiles, 2002). This sequential order of lineages is determined by the integration of extracellular signals and cell-intrinsic changes (Qian et al., 2000; Shen et al., 2006). During the last decade identification of the intrinsic mechanisms that underlie cellular differentiation in the developing CNS has been the subject of intense investigation. One line of investigation has focused on the role of chromatin modifiers as intrinsic modulators of fate choice during brain development (Ballas et al., 2001; Marin-Husstege et al., 2002; Hsieh and Gage, 2004; Ballas et al., 2005; Fan et al., 2005; Colvis et al., 2005; Miller and Gauthier, 2007; Lessard et al., 2007; He et al., 2007; Liu et al., 2007; Shen and Casaccia-Bonnefil, 2007). Among the epigenetic factors that modulate brain development, different groups have focused on post-translational modification of the histone tails, switching histone variants, ATP-remodeling complexes and microRNAs (Seo et al., 2005; Matsumoto et al., 2006; Cao et al., 2006).
The concept of the existence of an intrinsic timer in oligodendrocyte progenitors was suggested in a series of elegant experiments in clonal cultures of cells isolated from the optic nerve and studied using time-lapse video microscopy (Temple and Raff, 1986). It was proposed that in the presence of mitogens such as platelet-derived growth factor, progenitors undergo a certain number of cell divisions until they growth arrest. Differentiation would result from the progressive accumulation of intrinsic signals modulating the responsiveness of the cells to the differentiative effect of retinoic acid and thyroid hormone (Barres et al., 1994). Because of the intriguing relationship between cell-cycle exit and differentiation, it has been proposed that the progressive accumulation of cell-cycle inhibitors such as p27Kip1, p18Ink4c and p57Kip2 (Durand and Raff, 2000; Tokumoto et al., 2002; Dugas et al., 2007) might be responsible for the timing of differentiation onset. However, it has been difficult to determine whether cell-cycle inhibitors modulate differentiation directly or indirectly, by affected the proliferative state of the cell (Casaccia-Bonnefil et al., 1997; Casaccia-Bonnefil et al., 1999; Ghiani and Gallo, 2001). Because overexpression of the cell-cycle inhibitors p27Kip1 or p18Ink4c did not induce differentiation of oligodendrocyte progenitors, even though cells were arrested at the G1/S transition, it has been proposed that cell-cycle exit is necessary but not sufficient to induce differentiation (Tikoo et al., 1998; Tang et al., 1999; Tokumoto et al., 2002). An alternative possibility is that some cell-cycle genes might affect the transcriptional machinery after the cells have exited from the cell cycle. This hypothesis is supported by evidence that links p27Kip to transcriptional activation of myelin genes (Miskimins et al., 2002) and p21Cip to oligodendrocyte differentiation independent of its role in cell-cycle regulation (Zezula et al., 2001). More recent studies further support an alternative role for the cell-cycle related molecule p57Kip2 as a modulator of the transcriptional network regulating oligodendrocyte differentiation (Dugas et al., 2007). Together these data indicate the existence of a complex series of events that links exit from the cell cycle to differentiation of oligodendrocyte progenitors.
Timing of differentiation caused by disinhibition of late differentiation genes: examples from the astrocytic and neuronal lineages
In the neuronal lineage, timing of differentiation of neuronal precursors has been linked to the expression of neuronal genes following a de-repression event linked to the activity of HDACs (Ballas et al., 2001; Hsieh and Gage, 2004; Ballas et al., 2005). It has been shown that activation of neuronal gene expression is achieved by the removal of multi-protein complexes containing co-repressors, HDACs and repressor element 1 (RE1)-silencing transcription factor (REST) from the promoters of differentiation-asociated genes (Roopra et al., 2000; Ballas et al., 2005). In agreement with these findings, treating cells with HDAC inhibitors (Kondo, 2006; Balasubramaniyan et al., 2006; Lyssiotis et al., 2007; Siebzehnrubl et al., 2007), and systemic administration of HDAC inhibitors to either neonatal (Liu et al., 2007) or adult animals (Hsieh et al., 2004) increases neuronal gene expression and favors neurogenesis. More recently, it has also been suggested that the assembly of specific SWI/SNF complexes participates in defining the timing of neurogenesis (Lessard et al., 2007). Thus, these studies identify histone acetylation of neuronal gene promoters as a crucial event for differentiation.
Similarly, studies of astrocytic cell differentiation have reported the gradual epigenetic de-repression of chromatin in specific promoter regions of the astrocytic lineage marker glial fibrillary acidic protein (gfap). This gene is repressed in neural precursor cell because DNA methylation of CpG islands in its promoter precludes the access of the crucial transcription factor signal transducer and activator of transcription (STAT) (Bonni et al., 1997; Feng et al., 2005). The increase in concentration of gfap transcripts correlates with a gradual decrease in DNA demethylation and concomitant increase in acetylation and methylation of crucial lysine residues on the histone tails of the chromatin region around the promoter, and with the appearance of astrocytes during development (Nakashima et al., 1999; Takizawa et al., 2001; Song and Gosh 2004). Thus astrogliogenesis is favored by histone acetylation and requires DNA demethylation.
In the oligodendrocyte lineage myelin genes, such as mbp, proteolipid protein (plp) and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase), define the differentiated state of oligodendrocytes. Previous studies have reported the presence of repressive complexes containing HDACs and the transcriptional factors NKX2.2 or HES5 on the mbp promoter (Wei and Miskimins, 2005; Liu et al., 2006). Similar repressive complexes containing HDAC and the co-repressor SIN3 have been identified on the proximal region of plp promoter (Romm et al., 2005). By analogy to the astrocytic and neuronal lineages it has been suggested that timing of differentiation might be achieved by counteracting the enzymatic activities in the repressive complexes. However, pharmacological inhibition of HDACs in vitro prevents rather than facilitates the expression of myelin genes (Marin-Husstege et al., 2002). In addition, systemic administration of HDAC inhibitors to developing rat pups alters the timing of oligodendrocyte differentiation and delays myelination (Shen et al., 2005). Therefore, inhibition of repressive complexes containing HDAC on myelin genes is not sufficient to initiate oligodendrocyte differentiation.
Because myelin gene expression is regulated by a complex network of activators and inhibitors, we reasoned that differentiation timing might be affected by the equilibrium between inhibitors and activators (Gokhan et al., 2005). In this study we present evidence of a temporal correlation between decreased levels of transcriptional inhibitors and the onset of myelin gene expression. It is important to underline the role of these inhibitory basic-loop-helix factors such as members of the ID and HES family, and additional inhibitory molecules such as ATF5 for preventing oligodendrocyte differentiation (Kondo and Raff, 2000a; Kondo and Raff, 2000b; Samanta and Kessler, 2004; Mason et al., 2005; Marin-Husstege et al., 2006) and inhibiting myelin gene expression (Liu et al., 2006). Because decreased expression of transcriptional inhibitors is important for the timing of progenitor differentiation and for myelin gene expression, we searched for a mechanism that regulates the expression levels of the inhibitors themselves. Studies from our group have identified that global deacetylation of histone H3 is crucial for the decrease in transcriptional inhibitors of differentiation and that this occurs during the transition between cell-cycle exit and differentiation onset (Marin-Husstege et al.., 2002; Shen et al., 2005). Therefore we proposed that global histone deacetylation is responsible for the decreased transcription of differentiation inhibitors.
Histone deacetylation is mediated by the catalytic activity of HDACs, enzymes that do not have the ability to bind directly to DNA. We therefore sought to identify DNA-binding proteins that bind to the promoter of the differentiation inhibitors and recruit HDACs in order to decrease their transcript levels. The transcription factor YY1 is a recruiter of HDAC1 on the promoter of the transcriptional inhibitors Tcf4 and Id4 (He et al., 2007). The transcription factor Yin Yang 1 (YY1) is a multifunctional protein that can either activate or repress gene expression depending on the binding partner. Its role in the developing nervous system was originally suggested from studies in Xenopus embryos (Kwon and Chung, 2003; Morgan et al., 2004). However the analysis of HDAC1 function in mammals was limited by the fact that yy1-null mice do not reach gastrulation (Donohoe et al., 1999). To overcome this, we have generated conditional-knockout mice by crossing yy1flox/flox with the cnp-cre line to specifically delete yy1 in the oligodendrocytic lineage.
The yy1 conditional-knockout mice have a severe hypomyelinating phenotype that is characterized by tremor, ataxia and a shaking. This dramatic phenotype is consequent to impaired differentiation of oligodendrocyte progenitors, which are arrested at the transition between cell cycle exit and initiation of the differentiation program due to high levels of inhibitory molecules such as Id4, Tcf4 and Sox11 (He et al., 2007). These phenotypes resemble the one detected in animals treated with HDAC inhibitors (Shen et al., 2005; Liu et al., 2007).
YY1 binds to the promoter region of oligodendrocyte differentiation inhibitors (i.e. Tcf4 and Id4) and recruits HDAC1 into repressive complexes. In this manuscript, we provide additional experimental evidence to support the role of YY1 as crucial activator of HDAC activity in differentiating oligodendrocyte progenitors, because mutation in the interaction domain of HDAC1 caused severe consequences on the global levels of histone deacetylation. We also report that overexpressing YY1 in immortalized OPCs is sufficient to decrease the levels of multiple inhibitory molecules and to promote the progression of NG2 cells to the O4+ stage. However, we did not detect precocious myelin gene expression, possibly because YY1 did not increase the levels of transcriptional activators that are necessary to reach a specific threshold in order to activate myelin gene expression.
A better understanding of the mechanisms that underlie the differentiation of oligodendrocyte progenitors will provide the basis for the development of potential therapies of demyelinating disorders by stimulating either endogenous or transplanted oligodendrocyte progenitors.
ACKNOWLEDGEMENTS
This work was supported in part by grants from NIH-NINDS (RO1NS042925), the National Multiple Sclerosis Society (NMSS3957) and the Christopher Reeve Foundation to PCB.
REFERENCES
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Molecular and Cellular Biology. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniyan V, Boddeke E, Bakels R, Küst B, Kooistra S, Veneman A, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiological Reviews. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annual Review of Neuroscience. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Carter BD, Dobrowsky RT, Chao MV. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature. 1996;383:716–719. doi: 10.1038/383716a0. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedich V, Jr, Chao MV, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes and Development. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- Colvis CM, Pollock JD, Goodman RH, Impey S, Dunn J, Mandel G, et al. Epigenetic mechanisms and gene networks in the nervous system. Journal of Neuroscience. 2005;25:10379–10389. doi: 10.1523/JNEUROSCI.4119-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Molecular and Cellular Biology. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Raff MC. A cell-intrinsic timer that operates during oligodendrocyte development. Bioessays. 2000;22:64–71. doi: 10.1002/(SICI)1521-1878(200001)22:1<64::AID-BIES11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. Journal of Neuroscience. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. Journal of Neuroscience Research. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Ghiani C, Gallo V. Inhibition of cyclin E-cyclin-dependent kinase 2 complex formation and activity is associated with cell cycle arrest and withdrawal in oligodendrocyte progenitor cells. Journal of Neuroscience. 2001;21:1274–1282. doi: 10.1523/JNEUROSCI.21-04-01274.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fintanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. Journal of Neuroscience. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Science of the USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Current Opinion in Genetics and Development. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Jung M, Krämer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. European Journal of Neuroscience. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff MC. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO Journal. 2000a;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff MC. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000b;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes and Development. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T. Epigenetic alchemy for cell fate conversion. Current Opinion in Genetics and Development. 2006;16:502–507. doi: 10.1016/j.gde.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Chung HM. Yin Yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochemical Biophysical Research Communications. 2003;306:1008–1013. doi: 10.1016/s0006-291x(03)01071-4. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, Casaccia-Bonnefil P. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. Journal of Neuroscience. 2007;27:7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO Journal. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proceedings of the National Academy Science of the USA. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ, Woltering JM, In der Rieden PM, Durston AJ, Thiery JP. YY1 regulates the neural crest-associated slug gene in Xenopus laevis. Journal of Biological Chemistry. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Li J, Kondo T, Sablitzsky F, Casaccia-Bonnefil P. Multiples roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Marin-Hustegge M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. Journal of Neuroscience. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Angelastro JM, Ignatova TN, Kukekov VG, Lin G, Greene LA, et al. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Molecular and Cellular Neuroscience. 2005;29:372–380. doi: 10.1016/j.mcn.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Developmental Biology. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Miskimins R, Srinivasan R, Marin-Husstege M, Miskimins WK, Casaccia-Bonnefil P. p27(Kip1) enhances myelin basic protein gene promoter activity. Journal of Neuroscience Research. 2002;67:100–105. doi: 10.1002/jnr.10080. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, et al. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Molecular and Cellular Biology. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylases to regulate neural transcription. Journal of Neurochemistry. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Current Opinion in Neurobiology. 2002;12:244–249. doi: 10.1016/s0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nature Neuroscience. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. Journal of Cellular Biology. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte and disease. Journal of Molecular Neuroscience. 2007 doi: 10.1007/s12031-007-9014-x. PMID: 17999198 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochimica Biophysica Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Angevine JB, Pierce ET. Atlas of the Mouse Brain and Spinal Cord. Harvard University Press; 1971. pp. 202–206. [Google Scholar]
- Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Experimental Brain Research. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- Song MR, Gosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nature Neuroscience. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Ochiai W, Uemura V, Yanagisawa M, Fujita N, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Developmental Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tang XM, Beesley JS, Grinspan JB, Seth P, Kamholz J, Cambi F. Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. Journal of Cellular Biochemistry. 1999;76:270–279. doi: 10.1002/(sici)1097-4644(20000201)76:2<270::aid-jcb10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Temple S, Raff MC. Clonal analysis of oligodendrocyte development in culture: evidence for a developmental clock that counts for divisions. Cell. 1986;44:773–779. doi: 10.1016/0092-8674(86)90843-3. [DOI] [PubMed] [Google Scholar]
- Tikoo R, Osterhout DJ, Casaccia-Bonnefil P, Seth P, Koff A, Chao MV. Ectopic expression of p27Kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. Journal of Neurobiology. 1998;36:431–440. [PubMed] [Google Scholar]
- Tokumoto YM, Apperly JA, Gao FB, Raff MC. Posttranscriptional regulation of p18 and p27 Cdk inhibitor proteins and the timing of oligodendrocyte differentiation. Developmental Biology. 2002;245:224–234. doi: 10.1006/dbio.2002.0626. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. Journal of Biological Chemistry. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and Deacetylation. Molecular and Cellular Biology. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, Osterhout DJ, Levine JM, Dowdy SF, et al. p21cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal. EMBO reports. 2001;2:27–34. doi: 10.1093/embo-reports/kve008. [DOI] [PMC free article] [PubMed] [Google Scholar]


