Abstract
Summary
The distributed annotation system (DAS) defines a communication protocol used to exchange biological annotations. It is motivated by the idea that annotations should not be provided by single centralized databases but instead be spread over multiple sites. Data distribution, performed by DAS servers, is separated from visualization, which is carried out by DAS clients. The original DAS protocol was designed to serve annotation of genomic sequences. We have extended the protocol to be applicable to macromolecular structures. Here we present SPICE, a new DAS client that can be used to visualize protein sequence and structure annotations.
Availability
1 INTRODUCTION
A variety of manual, computational and experimental annotations of biological data, such as genome and protein sequences, are being developed by different groups all around the world. The distributed annotation system (DAS) protocol allows data producers to share their results with the community without requiring aggregation into a central database (Dowell et al., 2001). Resources such as the EnsEMBL genome browser (Hubbard et al., 2005) use the DAS protocol to link new data to the browser and visualize it. Although originally designed to serve annotations of genomes, in the last year DAS has also received some interest from the protein-bioinformatics community, largely because of the BioSapiens (http://www.biosapiens.info/) and eFamily (http://www.efamily.org.uk/) projects.
DAS provides a simple convention to encode a DNA or protein sequence and its annotated features into simple XML documents that are exchanged via the Internet (http:// www.biodas.org/). To make the DAS protocol applicable for protein structures we developed two extensions that allow alignments and 3D structure information (http://www. sanger.ac.uk/Users/ap3/DAS/) to be transmitted. Using these extensions a variety of new DAS clients become possible. For example, pairwise or multiple alignments of chromosomes or protein sequences could be visualized. Here we are using these extensions to support a new DAS client, SPICE, that can be used to visualize annotations of protein sequences and protein structures.
2 THE SPICE DAS CLIENT
SPICE is a Java program that can be started using Java Web Start simply by following a link from a web page. It accepts either a PDB (Berman et al., 2000) or a UniProt code (Appweiler et al., 2004) as an argument. When the application is started for the first time, Web Start will download the program automatically. Once SPICE is running, it connects to the DAS registration service (http://das.sanger.ac.uk/registry/) (manuscript in preparation) to retrieve a list of available DAS servers.
Four different types of DAS servers contribute to a complete SPICE display:
(1) A protein sequence server that provides the sequence. Typically this will be a UniProt sequence, but because of the data independence achieved by using the DAS protocol, it is possible to use any other sequence source if annotations are provided using the other types of DAS servers.
(2) An alignment server that provides the alignment between the protein sequence and its structure. At the time of submission of this manuscript the UniProt to PDB alignment used in SPICE is based on the MSD mapping (Boutselakis et al., 2003). If other alignments are made available the user can choose between alternative servers and compare the alignments provided. The 3D structure of a protein may have been resolved several times, and a structure can consist of several sequences. To deal with this many to many relationship, a window is available that allows the user to choose which of the alignments to display.
(3) A structure server that serves the 3D coordinates displayed. If a local PDB installation is available, SPICE can be configured to check there first when looking for structure data. If a required structure is not found, it is automatically retrieved using the macromolecular structure DAS extension.
(4) Several feature servers that provide pre-calculated annotations. These can be served in either protein sequence position or PDB residue number coordinates. The registration server keeps track of which coordinate system is used by a server to avoid confusion. At the time of submission of this manuscript the DAS registration server contains 16 DAS servers provided by 8 different institutions serving either UniProt or PDB annotations.
The available annotations include active site definitions, domain assignments and secondary structure assignments. One of the DAS sources provided is a mapping of genomic features including SNPs and intron/exon borders onto UniProt sequences. Using SPICE it is therefore possible to visualize the location of DNA features on the protein structure.
The SPICE viewer window consists of three main panels, as illustrated in Figure 1:
(1) The structure panel provides a 3D visualization of the molecule using the open source Jmol library (http://www.jmol.org/). Jmol provides a powerful 3D visualization tool and can interpret RASMOL-like (Sayle and Milner-White, 1995) scripting commands entered at a command line.
(2) Annotations provided by the distributed servers are displayed in a 2D feature panel, regardless of whether they were originally provided in sequence positions or PDB residue coordinates, since SPICE can use alignment data to project features between coordinate systems. This is required for the interaction between the sequence and structure panels. If an annotation is selected, its location is shown in both the 3D structure and the sequence panel. The feature panel contains a listing of available annotations, making it easy to compare the annotations made by different methods. If the user finds a particular source unreliable it is possible to disable it from the display.
(3) A sequence panel that displays the amino acid sequence of the protein. For a selected region the corresponding positions in the other panels are shown. The sequence can be searched for motifs.
Fig. 1.
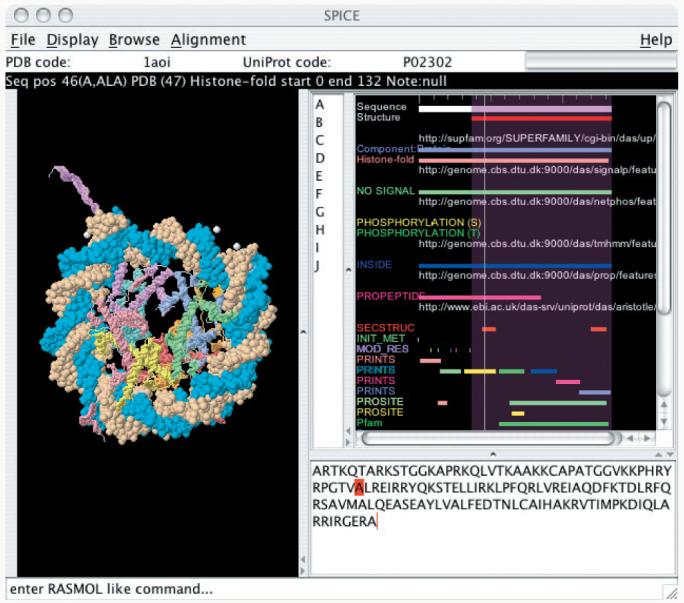
The SPICE client allows users to browse through annotations of protein sequences and structures. It retreives data from different sites across the Internet via the DAS protocol.
It is possible to add new DAS sources to SPICE. The SPICE configuration allows access to local DAS sources that are still under development or have not been registered with the DAS registration server. In this way SPICE can be used to evaluate new methods by comparing new results with the information that can be obtained from other sources. Since features can contain links back to the original data, providing data with DAS can also be used as a way to advertise and draw attention to new methods. Future developments include integrating SPICE into the EnsEMBL website. We will set up DAS sources that provide alignments of UniProt sequences and PDB structures to the EnsEMBL predicted peptides, through which it will be possible to launch SPICE.
3 CONCLUSION
SPICE is a tool to visualize protein sequence and protein structure annotations. It utilizes the DAS protocol to retrieve its data from separate sites on the Internet. It can be used to browse and compare the available annotations for a particular protein as well as to compare the results of newly developed methods with the pre-existing data.
4 AVAILABILITY
The SPICE source code is available under the Lesser General Public Licence (LGPL) from http://www.derkholm.net/svn/repos/spice/. Some modules are being made available through BioJava, which is also available under the LGPL from http://www.biojava.org/.
ACKNOWLEDGEMENTS
This work has been supported by the Medical Research Council. Thanks to Rob Finn for suggesting the name SPICE and to Andreas Kähäri for many feature suggestions. Thanks to everybody who is setting up DAS servers—the system would not work without you.
REFERENCES
- Appweiler R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutselakis H, et al. E-MSD: The European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res. 2003;31:458–462. doi: 10.1093/nar/gkg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, et al. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, et al. ENSEMBL 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle R, Milner-White J. RasMol: Biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]


