Abstract
Ever shorter telomeres 3 (Est3) is an essential telomerase regulatory subunit thought to be unique to budding yeasts. Here we use multiple sequence alignment and hidden Markov model–hidden Markov model (HMM-HMM) comparison to uncover potential similarities between Est3 and the mammalian telomeric protein Tpp1. Analysis of site-specific mutants of Candida albicans Est3 revealed functional distinctions between residues that are conserved between Est3 and Tpp1 and those that are unique to Est3. Although both types of residues are important for telomere maintenance in vivo, only the former contributes to telomerase activity in vitro and facilitates the association of Est3 with telomerase core components. Consistent with a function in protein-protein interaction, the residues common to Est3 and Tpp1 map to one face of an OB-fold model structure, away from the canonical nucleic acid binding surface. We propose that Est3 and the OB-fold domain of Tpp1 mediate a conserved function in telomerase regulation.
Telomeres are specialized nucleoprotein structures that maintain the integrity of eukaryotic chromosomal termini by protecting them from fusion and recombination, and promoting their replication (for reviews, see refs. 1–4). In most organisms, telomeric DNA consists of short repetitive sequences that are rich in G residues on the 3′ end–containing strand. These repeats are maintained by a ribonucleo-protein (RNP) known as telomerase, which acts as an unusual reverse transcriptase (for reviews, see refs. 3,5–7). Both telomere binding proteins and telomerase are crucial for the maintenance of telomere integrity through multiple cell divisions, which in turn is pivotal in supporting genome stability and promoting cellular life span.
Remarkably, components of both the telomeric protein complex and the telomerase complex have been observed to be evolutionarily malleable. For example, the terminal G-strand overhangs of mammalian telomeres are bound by Pot1 and Tpp1, which are subunits of a larger complex known as shelterin1,8. In contrast, the budding yeast telomere overhangs evidently interact with a Replication protein A–like heterotrimeric complex consisting of Cdc13, Stn1 and Ten1 (refs. 9,10). Although each of the mammalian and yeast proteins consists of a variable number of OB-fold domains, no orthologous relationship has been established between any protein pairs from either sequence or functional comparison. Similarly, no telomerase subunit except the catalytic protein (telomerase reverse transcriptase (TERT)) and template RNA (TER) is universally conserved. Instead, both mammals and yeast possess species-specific factors that are crucial for telomerase assembly and function5,6. Thus, the fundamental importance of the telomere-maintenance machinery is in striking contrast to its evolutionary plasticity.
Est3 is a small but functionally essential subunit of the yeast telomerase complex11. Deletion of EST3 leads to a progressive telomere-attrition phenotype that mimics closely deletion of TERT or TER12,13. Yet the protein is not essential for the catalytic activity of telomerase in vitro12,14,15. Recent experiments, however, point to a role for Candida albicans Est3 in promoting holoenzyme assembly and stimulating the polymerization activity of telomerase in a primer substrate–dependent fashion16. To gain further insights into Est3 mechanisms, we used a combination of bioinformatic analysis and site-specific mutagenesis to identify structural attributes of the protein necessary for its biochemical functions. Our results reveal unexpected similarities between Est3 and mammalian Tpp1.
RESULTS
Similarities between Est3 and Tpp1
To delineate further the structure and function of Est3, we first undertook a computational analysis of Est3 homologs, which can be readily identified in many Saccharomyces and Candida species (Fig. 1). A multiple sequence alignment was generated from 12 Est3 homologs and used to query the HHpred server17, which exploits HMM-HMM comparisons for the identification of distantly related proteins. Remarkably, the top-scoring hit from this analysis was the mammalian telomeric protein Tpp1 (E-value = 0.71, P-value = 91.5). The plausibility of the Est3 and Tpp1 connection is reinforced by two recent studies that revealed an interaction between Tpp1 and telomerase as well as a stimulatory effect of Tpp1 on telomerase activity and processivity18,19.
Figure 1.
Sequence and structural similarity between Est3 and Tpp1 homologs. (a) Sequence comparison between Est3, Tpp1 and TEBPβ. The secondary-structural elements derived from the human Tpp1 crystal structure are illustrated as follows: α-helices, green cylinders; β-strands, blue arrows. Blue and cyan letters highlight mutated residues that are conserved between Est3 and Tpp1. Note that these residues are also conserved in the recently identified S. pombe homolog Tpz1 (ref. 24 and data not shown). Green dots highlight mutated residues that are conserved only in Est3. (b) A hypothetical structure of Est3 based on the Tpp1 crystal structure. Residues predicted to be important for telomerase interaction based on structure-function analysis are shown in van der Waals sphere representations in blue (Asp91) and cyan (Trp36 and Asp169).
Effects of EST3 mutations on telomeres and telomerase assembly
We proceeded to generate and analyze a series of site-specific C. albicans EST3 mutants designed on the basis of a composite sequence alignment between Est3 and Tpp1 homologs (Fig. 1a). This sequence alignment highlights three C. albicans residues that are well conserved in all Est3 and Tpp1 family members (Trp36, Asp91 and Asp/Glu169), as well as residues that are conserved only in Est3 homologs (Arg72, Ser92, Phe109, Arg116, Thr121 and Gln170). These residues (and a nonconserved Asp59) were replaced with alanine, and the resulting mutants were reintegrated into an est3Δ/est3Δ strain for functional analysis. To facilitate biochemical studies, the mutant alleles were each fused C-terminally to a protein-A tag, which has no discernable effect on telomere maintenance16 (data not shown).
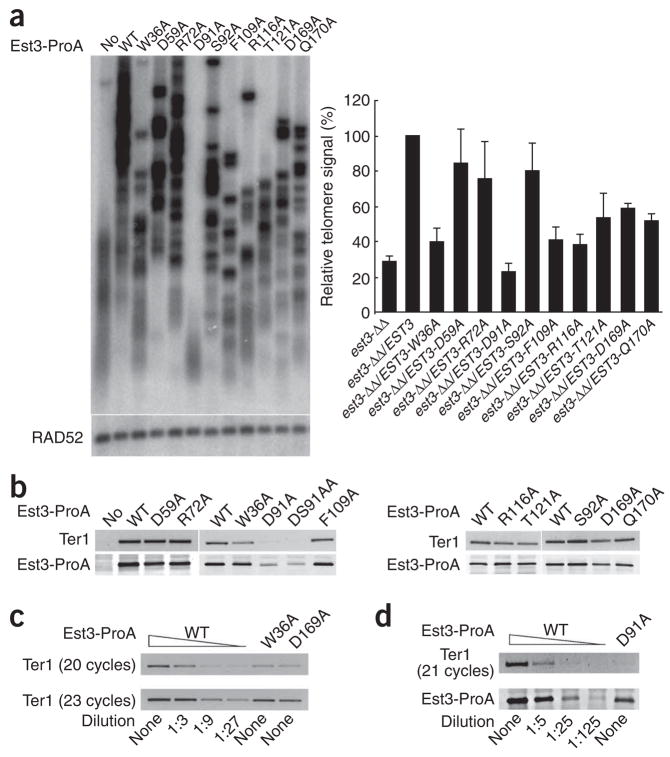
We first examined the impact of mutations on telomere lengths and contents by telomere Southern (Fig. 2a and Supplementary Fig. 1 online). All of the mutants, with the exception of D59A, R72A and S92A, manifested substantial defects in telomere maintenance. Notably, both the D91A and DS91AA mutants behaved as null alleles with regard to telomere maintenance, whereas the others (W36A, F109A, R116A, T121A and Q170A) retained partial function. We then investigated the levels of mutant Est3 proteins and their association with telomerase RNA (Ter1) by IgG-Sepharose pull-down, western blotting and reverse-transcription PCR (RT-PCR). All of the mutant proteins were present at near wild-type levels, except for D91A and DS91AA, which were reduced by approximately five-fold (Fig. 2b). On the other hand, quantitative RT-PCR revealed no detectable Ter1 in the D91A and DS91AA pull-down samples, and Ter1 levels were reduced to ~20% of the normal Ter1 levels in the W36A and D169A samples (Fig. 2c, d). Titration of the wild-type extract indicates that the absence of Ter1 in the D91A pull-down sample cannot be explained by reduced protein level. In particular, the D91A protein level is reduced approximately 5-fold, whereas the associated Ter1 is reduced more than 25-fold (Fig. 2d). Thus, Asp91 enhances the stability of Est3 and is essential for telomerase association, whereas Trp36 and Asp169 are not required for protein stability but promote telomerase association. The other mutated residues seem dispensable for both functions. Notably, the null-like phenotype of the D91A and DS91AA mutants with regard to telomere maintenance argues that telomerase binding is essential for Est3 function. Specifically, we found that the W36A and D169A mutations each resulted in a five-fold reduction in the level of Est3-associated telomerase complex (Fig. 2c), yet this reduced level can still prevent the null-like phenotype. Thus, the five-fold reductions observed for the D91A and DS91AA mutant proteins by themselves cannot account for the complete loss of function. Rather, the effect of the mutations on telomerase association is likely to be responsible.
Figure 2.
The effects of C. albicans Est3 mutations on telomere maintenance and telomerase association. (a) Left, est3Δ/est3Δ strains containing the wild-type and mutant alleles of protein A (ProA)-tagged EST3 were passaged on plates by successively streaking for single colonies. Chromosomal DNAs were prepared from either streaks 16 or 20 of each indicated strain and subjected to telomere Southern blotting (above). The blot was then stripped and reprobed using a labeled RAD52 fragment (below). Right, the telomere-hybridization signals and the RAD52 signals obtained from the Southern blots shown and from three other independent blots were quantified using a PhosphorImager. The relative ratios of the telomere to RAD52 signals were calculated and plotted for the wild-type and all mutant samples. Shown are means ± s.d. from four independent experiments. (b) Est3 and associated Ter1 in wild-type and mutant strains were analyzed by IgG-Sepharose pull-down followed by western analysis (right) and RT-PCR (20 cycles) (left), respectively. (c) Extracts from strains containing ProA-tagged wild-type EST3 (WT), the W36A mutant and the D169A mutant were subjected to IgG-Sepharose pull-down and RT-PCR to detect Est3-associated Ter1. To facilitate quantitative comparison, the WT sample was subjected to three-fold serial dilution before RT-PCR. Thermocycling was performed for 20 or 23 cycles to ensure the linearity of the signals. (d) The Est3 protein and associated Ter1 in the wild-type and D91A mutant strains were analyzed by IgG-Sepharose pull-down followed by western blotting and RT-PCR (below and above, respectively). The WT sample was subjected to five-fold serial dilution before the pull-down.
Effects of EST3 mutations on telomerase activity
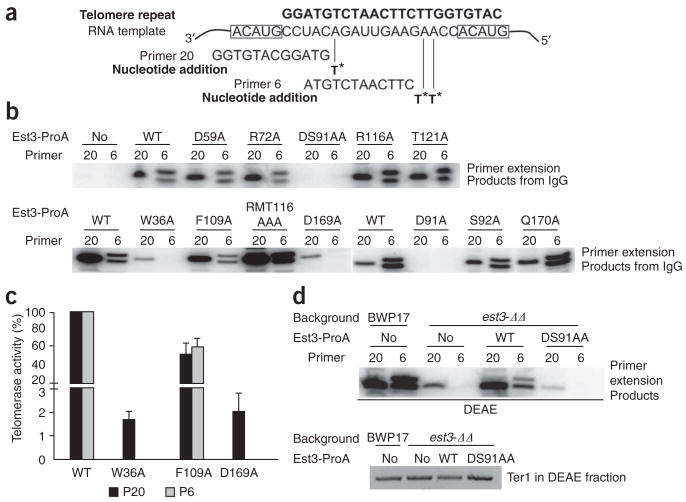
To determine the effect of EST3 mutations on telomerase activity in vitro, we first measured primer-extension activity using the IgG-Sepharose precipitates and two different primers (Fig. 3a). Our previous study demonstrated that C. albicans Est3 is essential for the extension of some primers (for example, P6 in Figure 3a) by telomerase but not others (for example, P20 in Figure 3a). Notably, mutations in the residues conserved only in Est3 generally had little effect on telomerase activity on either primer (Fig. 3b). An exception is the F109A mutant, which showed an approximately two-fold reduction in activity on both P6 and P20 (Fig. 3b, c). Telomerase from the W36A and D169A mutants showed greatly reduced activity on P20 and no detectable activity on P6 (Fig. 3b). For both mutants, the magnitude of reduction in activity on P20 (approximately 50-fold) was far greater than that of telomerase RNA (approximately 5-fold) (compare Figs. 2c and 3c), indicating a dramatic reduction in specific activity. Thus, Trp36 and Asp169 must have a function in stimulating telomerase activity that goes beyond their role in promoting telomerase association of Est3, possibly through an allosteric effect.
Figure 3.
The effects of C. albicans Est3 mutations on telomerase primer extension activity in vitro. (a) All primer extension assays were performed using two different 12-nt primers, P6 and P20, which correspond to different regions of the C. albicans telomere repeat and support the synthesis of primer+1 or primer+2 products in the presence of labeled dTTP (T*). (b) Telomerase isolated from the indicated strains by IgG-Sepharose pull-down were assayed for primer extension activity using primers P6 and P20 in the presence of labeled dTTP. (c) The activities for the W36A, F109A and D169A mutants relative to the wild-type enzyme on the P6 and P20 primer were quantified and plotted; shown are means ± s.d. from three independent experiments. (d) Telomerase isolated by DEAE chromatography from the indicated strains was tested for enzyme activity using primers P6 and P20 in the presence of labeled dTTP (above). The levels of Ter1 in the fractions were measured by RT-PCR (below).
As expected, no telomerase activity was detected in the D91A and DS91AA IgG-Sepharose precipitates as a result of their lack of telomerase association (Fig. 3b). To assess the enzymatic defect of these mutants, we isolated telomerase by diethylaminoethyl (DEAE) chromatography, which does not depend on the association of Est3 with telomerase core components. Notably, telomerase obtained by this method from the D91A and DS91AA mutants behaved identically to that from the est3Δ/est3Δ strain; that is, the telomerase showed reduced activity on P20 and no activity on P6 (Fig. 3d and Supplementary Fig. 2 online). These results are consistent with the in vivo null-like phenotype of these mutants and highlight the importance of telomerase association for Est3 function. We also purified several additional mutant telomerases by DEAE chromatography in cases where the mutant Est3 was capable of telomerase association. Comparison of the DEAE-derived and IgG-Sepharose–derived telomerase revealed no substantial differences16 (Supplementary Fig. 2).
Our previous study uncovered, in addition to the nucleotide-addition defect, an elongation or processivity defect for telomerase derived from the est3Δ/est3Δ strain16. We therefore investigated the elongation properties of the Est3 mutant enzymes (derived by IgG-Sepharose pull-down) using an 8-nt primer to which 6 nt can be added in the presence of dCTP and dTTP (Supplementary Fig. 3 online). Notably, even though total telomerase activity was reduced for the W36A, F109A and D169A mutants, each mutant enzyme generated a similar banding pattern, adding all 6 nt to the primer. This result indicates that none of the mutants is grossly defective in processivity and suggests that the stimulatory effect of Est3 on nucleotide addition can be uncoupled from its effect on enzyme processivity.
Contributions of Est3 residues to holoenzyme assembly
Our earlier studies also suggested that in C. albicans, the association of Est1 and Est3 with the telomerase core complex is mutually dependent. If this were case, then the binding of Est1 to the telomerase core should be impaired by specific EST3 mutations such as W36A, D91A and D169A. To test this prediction, we used DEAE chromatography to monitor the association of Est1 with telomerase in mutant extracts (Supplementary Fig. 4a, b online). As expected, wild-type Est1 and Est3 coeluted in high-salt fractions from a DEAE column. In contrast, the level of Est1 in the high-salt elution fractions was reduced by the Est3 W36A mutation and abolished by the DS91AA mutation. We tested the effect of EST3 mutations on Est1 association using an alternative method that involves affinity pull-down of tagged Est1 by Streptavidin agarose and obtained similar results (Supplementary Fig. 4c). Thus, the extent of Est3 association with telomerase correlates with that of Est1 association, supporting the notion that their assembly is mutually dependent.
DISCUSSION
Altogether, our functional analysis of Est3 mutants has revealed three classes of important residues (class I to III in Table 1). Class I, represented by Asp91, is essential for both the in vivo and in vitro function of Est3. Substitution of Asp91 results in the concurrent dissociation of Est3 and Est1 from the telomerase complex and primer-dependent loss of telomerase activity, thus leading to progressive telomere attrition. Class II residues, including Trp36 and Asp169, are less important functionally. Substitution of these residues results in partial dissociation of Est3 and Est1 from telomerase and partial loss of telomerase activity. The extent of telomere loss in these mutants is accordingly less severe than in the D91A mutant. Class III residues, including Phe109, Arg116, Thr121 and Gln170, are also required for normal telomere maintenance. However, mutations in these residues have little effect on the telomerase association of Est3 and telomerase activity. The existence of class III residues points to an additional molecular function for Est3 that is not revealed by the current in vitro assay.
Table 1.
Phenotypes of the EST3 mutants
| Class | Mutated residues | Telomere maintenance | Telomerase association | Telomerase activity |
|---|---|---|---|---|
| I | Asp91 | Similar to deletion mutant | No | No |
| II | Trp36, Asp169 | Moderate and progressive loss | Reduced (B53) | Reduced (B503) |
| III | Phe109 | Moderate and progressive loss | Normal | Reduced (B23) |
| Arg116, Thr121 | Moderate and progressive loss | Normal | Normal | |
| Gln170 | Short and stable | Normal | Normal | |
| IV | Asp59, Arg72, Ser92 | Similar to wild type | Normal | Normal |
Notably, class I and II residues are conserved between Est3 and Tpp1, whereas class III residues are shared only by Est3 family members. The availability of a Tpp1 crystal structure allowed us to map the location of the Tpp1 residues that are equivalent to the Est3 class I and II residues (Fig. 1b). In Tpp1, the side chain of Asp146 (equivalent to Asp91 of C. albicans Est3) makes four hydrogen-bonding interactions with the backbone amino groups, hence stabilizing helix αA at one end of the β-barrel of the OB-fold (Supplementary Fig. 5a online). Notably, Asp91 is conserved in many OB-fold–containing proteins other than Est3 and Tpp1 and has a similar structural role as in Tpp1 (Supplementary Fig. 5b and data not shown). In contrast, class II residues (Trp36 and Asp169) are shared only by Est3 and Tpp1 (Fig. 1a). All three residues map to one face of the OB-fold that is away from the typical DNA-interacting surface, suggesting that these residues are required for protein binding rather than DNA binding. Consistent with this notion, we have been unable to detect DNA binding by Est3 using various substrates (data not shown). A plausible protein target of Est3 may be the N-terminal domain of TERT, which is conserved between yeast and human telomerase, and has been linked to Est3 genetically20.
The structural and functional similarities uncovered in the current study suggest that Est3 could be orthologous to the OB-fold domain of Tpp1. This then begs the question as to how orthologous proteins can be components of different macromolecular complexes in different organisms. A speculative evolutionary scenario is as follows (Supplementary Fig. 6 online): the ancestral Tpp1/Est3 was a component of the telomeric protein complex and, as such, a stimulatory factor for telomerase action. It has been suggested that, during budding yeast evolution, Pot1 might have been lost from telomeres as a result of mutations in the telomere repeat sequence21, thus resulting in the concomitant dissociation of Tpp1. The yeast mutant would be expected to face profound selection pressures for alternative mechanisms of telomere protection and telomerase stimulation. The telomere-protective function was apparently assumed by the Cdc13–Stn1–Ten1 complex. The telomerase-stimulatory function, on the other hand, was rescued by new interactions between Est3 and the telomerase protein Est1, which stabilized the association between Est3 and TERT16,22. In this scenario, Est3 is orthologous to only the telomerase-stimulatory domain of Tpp1, having lost the region(s) required for telomere localization (for example, the Tin2 and Pot1 binding domain). The fact that budding yeast has retained Est3 despite the drastic remodeling of its telomere-maintenance machinery suggests that this telomerase-stimulatory function may be universally required. In this regard, it would be interesting to determine whether the ciliate TEBPβ and S. pombe Tpz1, which are homologous to Tpp1 (refs. 18,19,23,24), also mediate telomerase stimulation. Moreover, an apparent homolog of Est3 and Tpp1 has not been identified in plants. Instead, a Pot1 homolog is reportedly associated with the telomerase RNP25. Understanding the shared and distinctive properties of these factors would surely provide valuable insights on telomere evolution.
METHODS
Strains
The parental C. albicans strain BWP17 and the est3Δ/est3Δ strain have been described previously26. The deletion strain reconstituted with untagged EST3 or protein A (ProA)–tagged EST3 have also been described13,16. To generate an est3Δ/est3Δ strain containing both EST3-ProA and EST1-ProA, we transformed SnaBI-digested pBS-CaEST1-ProA-URA into the est3Δ/est3Δ EST3-ProA strain. An EST1 gene tagged with Glycine8-Streptavidin binding peptide–Calmodulin binding peptide (GSC) was engineered to allow co-immunopreciptation with EST3-ProA. To generate a strain that contains both EST3-ProA and EST1-GSC, we transformed SnaBI-digested pBS-CaEST1-GSC-URA into the est3Δ/est3Δ EST3-ProA strain. Proper integration of the tagged genes at the respective native chromosomal loci was confirmed by Southern blot analysis. Each tagged strain contains one copy of the tagged gene.
Plasmids and site-directed mutagenesis
The plasmid pBS-CaEST3-ProA-HIS containing tagged Est3 has been described previously16. We constructed plasmids containing mutated EST3 genes using the QuikChange site-directed mutagenesis method (Stratagene). To generate pBS-CaEST1-ProA-URA, we introduced a PCR product from pGEM3-URA3 containing the URA3 marker between ApaI and HindIII sites of pBS-CaEST1-ProA-HIS16, resulting in the replacement of the HIS1 marker with a URA3 marker. The protein-A tag of pBS-CaEST1-ProA-URA was then replaced with GSC (consisting of a Gly8 linker, Streptavidin binding peptide (SBP) and Calmodulin binding peptide (CBP)) to yield pBS-CaEST1-GSC-URA (the complete sequence of the GSC fragment is available on request).
Sequence alignment and structure prediction
The sequences of 12 Est3 homologs from budding yeast were culled from the National Center for Biotechnology Information (NCBI) and Broad Institute databases and aligned using T-coffee (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi). The alignment was used to query the HHpred server, resulting in the identification of human TPP1 as a potential homolog17. A composite alignment of the 12 Est3 and 3 Tpp1 homologs (from humans, mice and Xenopus laevis) was then generated using the ProbCons server27.
Telomere-length analysis
Chromosomal DNAs were isolated from 4–5 ml of saturated C. albicans culture by the smash-and-grab method, digested with AluI and NlaIII, and fractionated in 0.8% agarose gels. Following transfer to nylon membranes, the telomere-restriction fragments were detected as previously described using an oligonucleotide probe that contains two copies of the C. albicans telomere repeat15. We performed the hybridization at 50 °C.
IgG-Sepharose pull-down for the analysis of Est3-associated Ter1 and telomerase activity
TMG(n) buffer (10 mM Tris-HCl, pH 8.0, 1.2 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 10% (v/v) glycerol (n refers to the millimolar concentration of sodium acetate)) was used throughout the study. We pretreated 45 μl IgG-Sepharose beads with 100 μg tRNA in 1 ml TMG(0) at 4 °C for 30 min to minimize nonspecific binding. After one wash in TMG(0), the beads were incubated with extracts (5 mg) in 1.2 ml TMG(400) and subjected to gentle rotation at 4 °C for 2 h. The beads were then washed three times with TMG(600) and twice with TMG(0), and divided into four equal aliquots. One aliquot was subjected to western blot analysis and another aliquot was treated with proteinase K, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and subjected to ethanol precipitation to recover RNA. We then measured the level of Ter1 (C. albicans telomerase RNA) by semiquantitative RT-PCR (2× RT-PCR master mix; USB Corporation), using primer pairs designed to amplify a 350-bp fragment (forward, 5′-CCCATATTCAATGCTCTTGGAGTGTG-3′; reverse, 5′-CTCCACAAGGTATCATACAAATTATGG-3′). The linearity of the assay was confirmed by titrating the samples and by using different cycle numbers. The last two aliquots were used for telomerase primer-extension assays using 12-nt primers (P6 and P20) and a single-labeled nucleotide triphosphate that can be incorporated at the primer +1 (or primer +1 and primer +2) position. The pull-down assays for wild-type and mutant Est3-ProA were each performed three or four times, and the results were highly reproducible.
DEAE chromatography for purification of telomerase complex and analysis of telomerase activity
Telomerase-containing DEAE column fractions were prepared and subjected to primer extension assays with 12-nt primers (P6 and P20) and a single-labeled nucleotide triphosphate as described above15,16. To analyze telomerase association of Est1 in various EST3 mutant backgrounds, extracts from strains containing ProA–tagged EST1 and EST3 were fractionated by DEAE chromatography, and the levels of ProA–tagged Est1 and Est3 in column fractions were measured by western blotting using antibodies directed against ProA.
Streptavidin-agarose pull-down assay for testing the association of Est1 with telomerase in strains containing mutant EST3
We carried out Streptavidin-agarose pull-down following the same protocol as for IgG-Sepharose pull-down with a few modifications. Briefly, 45 μl of Streptavidin-agarose beads were pretreated with 100 μg tRNA at 4 °C for 30 min to minimize nonspecific binding. After one wash in TMG(0), the beads were incubated with extracts (15 mg) in 3.6 ml TMG(400) with rotation at 4 °C for 2 h to precipitate Est1-GSC–associated telomerase complex. The beads were then washed three times with TMG(600) and twice with TMG(0). One-sixth of the pull-down sample was used for RNA isolation. We then determined the level of Ter1 by semiquantitative RT-PCR.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank A. Den and J. Kang for technical assistance. This work was supported by grants from the US National Institutes of Health (GM069507 to N.F.L. and GM083015 to M.L.).
Footnotes
AUTHOR CONTRIBUTIONS
E.Y.Y. performed the experiments and analyzed the data. F.W. and M.L. contributed reagents and helped interpret the data. N.F.L. conceived the project, analyzed the experiments and wrote the paper.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira MG, Miller KM, Cooper JP. Indecent exposure: when telomeres become uncapped. Mol Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 3.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 5.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 6.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Mammalian telomeres. In: de Lange T, Lundblad V, Blackburn E, editors. Telomeres and Telomerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 387–431. [Google Scholar]
- 9.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 10.Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Lundblad V. Budding yeast telomeres. In: de Lange T, Lundblad V, Blackburn E, editors. Telomeres and Telomerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 345–386. [Google Scholar]
- 12.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg-Neifach O, Lue NF. Modulation of telomere terminal structure by telomerase components in Candida albicans. Nucleic Acids Res. 2006;34:2710–2722. doi: 10.1093/nar/gkl345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SM, Steinberg-Neifach O, Mian IS, Lue NF. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot Cell. 2002;1:967–977. doi: 10.1128/EC.1.6.967-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu M, Yu EY, Singh SM, Lue NF. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, et al. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 19.Xin H, et al. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 20.Friedman KL, Heit JJ, Long DM, Cech TR. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol Biol Cell. 2003;14:1–13. doi: 10.1091/mbc.E02-06-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 22.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 23.Paeschke K, et al. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1–Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 25.Surovtseva YV, et al. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.