Summary
The RNA helicase eIF4A plays a key role in unwinding of mRNA and scanning during translation initiation. Free eIF4A is a poor helicase and requires the accessory proteins eIF4G and eIF4H. However, the structure of the helicase complex and the mechanisms of stimulation of eIF4A activity have remained elusive. Here we report the topology of the eIF4A/4G/4H helicase complex, which is built from multiple experimentally observed domain-domain contacts. Remarkably, some of the interactions are continuously rearranged during the ATP binding/hydrolysis cycle of the helicase. We show that the accessory proteins modulate the affinity of eIF4A for ATP by interacting simultaneously with both helicase domains and promoting either the closed, ATP-bound conformation or the open, nucleotide-free conformation. The topology of the complex and the spatial arrangement of the RNA-binding surfaces offer insights into their roles in stimulation of helicase activity and the mechanisms of mRNA unwinding and scanning.
Introduction
Control of translation is vital for cell proliferation and differentiation. Translation initiation entails the process of locating the correct translation start codon on the mRNA and the assembly of an active ribosome. It requires a number of eukaryotic translation initiation factors (eIFs) and for the majority of mRNAs consists of several steps: 1) preinitiation complex assembly by binding of several factors to the small ribosomal subunit; 2) recruitment of the preinitiation complex to the 5′-cap of the mRNA; 3) scanning along the mRNA in the 3′ direction in search of the start codon; 4) start codon recognition; and 5) joining of the large ribosomal subunit to form a ribosome with a bound initiator Met-tRNAi ready to translate the mRNA. Initiation of translation is typically the rate-limiting step and is the main target of regulation (reviewed in (Marintchev and Wagner, 2004)).
The eukaryotic translation initiation factor eIF4A is an ATP-dependent RNA helicase, which unwinds RNA secondary structures in the 5′-untranslated region (5′-UTR) of mRNA and promotes scanning by the translation pre-initiation complex toward the start codon. Unwinding secondary structures in the 5′-cap proximal region of mRNA also serves to clear the “landing pad” for recruitment of the small ribosomal subunit. eIF4A (Figure 1A) has two domains, both of which are involved in binding RNA and ATP. In the “closed”, active conformation, the two domains form a contiguous RNA-binding surface (see Figure 1D, left), and the ATP-binding site is at the interface between the two domains (Andersen et al., 2006). ATP and RNA bind to eIF4A cooperatively (Lorsch and Herschlag, 1998a). The ATPase and helicase activity of free eIF4A is low; however it is greatly stimulated when eIF4A is part of a multiprotein complex that includes eIF4G, eIF4E, eIF4B and eIF4H (Grifo et al., 1984; Pause et al., 1994; Richter-Cook et al., 1998). At the center of this complex is eIF4G (Figure 1C), a large protein that provides the platform for binding several initiation factors. eIF4G recruits the small ribosomal subunit to mRNA through interactions with the cap-binding protein eIF4E and eIF3. It also promotes circularization of mRNA through interactions with the 3′-poly-A tail binding protein PABP.
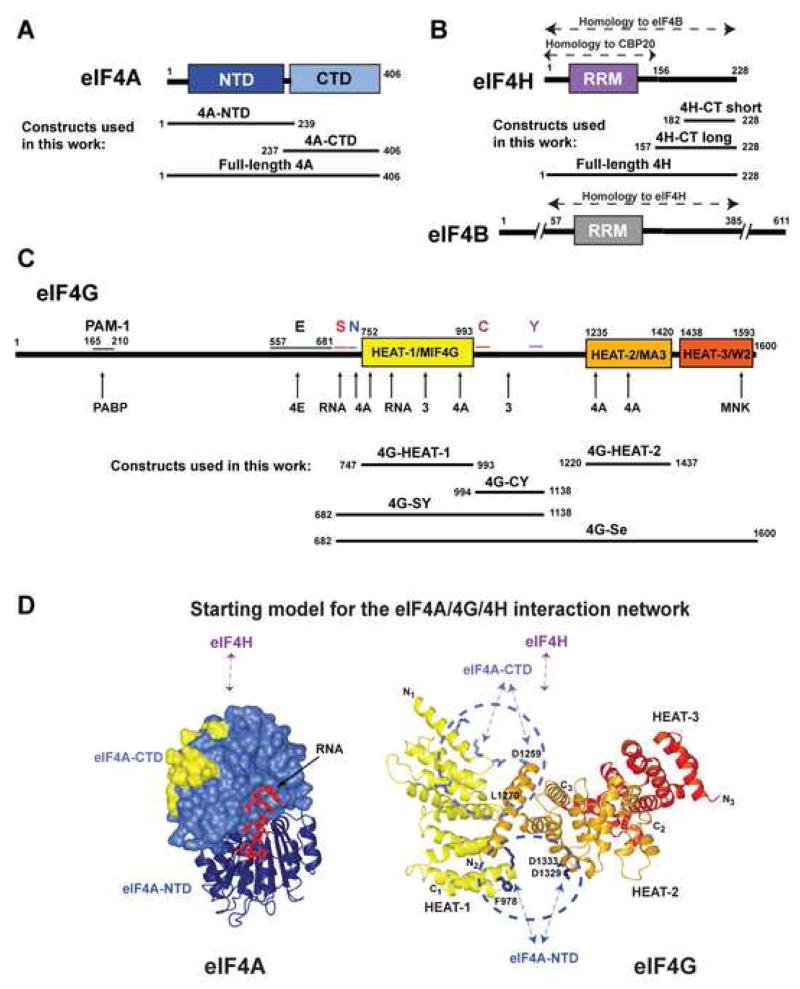
Figure 1. Domain organization of eIF4G, eIF4A and eIF4H.
(A-C) Domain organization of eIF4A (A), eIF4H and eIF4B (B), and eIF4G (C) and constructs used in this work. In panel C, conserved sequence segments in eIF4G are marked above the diagram. Sites of interaction with other proteins and RNA are marked with arrows below the diagram. Abbreviations: NTD, N-terminal domain; CTD, C-terminal domain; RRM, RNA recognition motif domain; 4H-CT-long and 4H-CT-short, long and short C-terminal fragments of eIF4H; PAM-1, PABP-binding motif-1; E, predicted extended eIF4E-binding region; S, RNA-binding region important for scanning, aa. 682–721 (Prevot et al., 2003); N, H1-NT motif, aa. 722–741; C, H1-CT motif, aa. 994–1027; Y, y4G-CT motif, aa. 1118–1136 (Marintchev and Wagner, 2005). (D) Starting model for the eIF4A/4G/4H interaction network. eIF4A-CTD is shown in surface representation and the eIF4G HEAT-1 contact surface (Oberer et al., 2005) is yellow. The interdomain orientation between eIF4A-CTD and eIF4A-NTD (navy ribbon), and the RNA (red wire) are modeled based on the structure of eIF4A3 (Andersen et al., 2006). The interdomain orientation of the HEAT domains of eIF4G (Marintchev and Wagner, 2005) is modeled based on the interdomain orientation in CBP80 (Mazza et al., 2002). The HEAT domains are color-coded as in panel C. Sites of mutations in eIF4G reported to affect eIF4A binding are shown as light blue (top cluster) and navy (bottom cluster) wires. Residues relevant for this work are labeled. The predicted binding sites of eIF4H, eIF4A-NTD and eIF4A-CTD are shown with arrows.
eIF4B and eIF4H (Figure 1B) are RNA-binding proteins that contain an RNA-recognition motif (RRM) domain and are homologous to each other over the entire sequence of eIF4H. eIF4B contains additional N- and C-terminal domains and has at least one additional RNA-binding region near its C-terminus. Both eIF4B and eIF4H stimulate the eIF4A helicase activity (reviewed in (Marintchev and Wagner, 2004)). eIF4H was reported to bind eIF4A, and its C-terminal region was found to be important for the interaction (Feng et al., 2005).
Human eIF4G contains three HEAT repeat domains (Bellsolell et al., 2006; Marcotrigiano et al., 2001) (Figure 1C). The first two of them, HEAT-1/MIF4G and HEAT-2/MA3 bind to eIF4A (Imataka and Sonenberg, 1997; Lamphear et al., 1995). HEAT-1 stimulates the helicase activity of eIF4A, whereas HEAT-2 has only a modulatory role (reviewed in (Marintchev and Wagner, 2004)).
The functions of the eIF4A/4G/4E complex (eIF4F), eIF4B and eIF4H in recruitment of the ribosome to the 5′-cap of mRNA and scanning have been known for over two decades. The structures of several individual components and isolated domains have been solved. However, despite continuing efforts, it has not been possible to solve the structure of the multiprotein Cap-binding complex; and the underlying regulatory mechanisms have remained elusive. The main challenge lies in the dynamic nature of the complex, which has to move along the 5′-UTR and unwind secondary structures in the mRNA (reviewed in (Jackson, 2000; Marintchev and Wagner, 2004, 2005)). Therefore, understanding the overall topology of the protein-protein interaction network centered around eIF4A and eIF4G requires alternative approaches.
We recently reported that the HEAT-1 domain of human eIF4G binds mainly to the C-terminal domain of eIF4A (eIF4A-CTD), but that the interaction with full-length eIF4A is stronger. We proposed that HEAT-1 binds weakly to eIF4A-NTD in the context of full-length eIF4A, promoting the formation of the closed, active ATP-bound conformation of eIF4A (Oberer et al., 2005). Furthermore, we established a distant evolutionary relationship of eIF4G and eIF4H with the CBP80 and CBP20 subunits, respectively, of the nuclear Cap-binding complex (CBC) and proposed that the domain organization of human eIF4G could be similar to that found in CBP80 (Marintchev and Wagner, 2005).
In this work, we used solution NMR, site-directed mutagenesis and biophysical assays to elucidate the topology of the human eIF4A/4G/4H helicase complex and the mechanisms of regulation of ATP binding to the helicase. We experimentally probed numerous interactions between individual domains of eIF4A, eIF4G and eIF4H, based on our previous studies on eIF4A and eIF4G (Marintchev and Wagner, 2005; Oberer et al., 2005). Our data show that the eIF4A/4G/4H complex is built by a dynamic network of multiple weak, but specific interactions. The results presented here provide insight into the mechanisms of regulation of the eIF4A helicase activity by eIF4G and eIF4H.
Results
Starting model for the organization of the eIF4A/4G/4H complex
We previously proposed that the domain organization of eIF4G may be similar to that of its distant relative CBP80 (Marintchev and Wagner, 2005). In the resulting model, shown in Figure 1D, right, the two eIF4A-binding domains of eIF4G (HEAT-1 and HEAT-2, painted yellow and orange, respectively) are positioned adjacent to each other. A number of point mutations in the eIF4G HEAT-1 and HEAT-2 domains have been reported to affect eIF4A binding. Remarkably, if our hypothesis about the inderdomain orientation in eIF4G is correct, these mutations form two clusters, each composed of residues from both eIF4G HEAT-1 and HEAT-2 (Figure 1D, right): circled in light blue (top) and in navy (bottom), respectively. eIF4A has two domains and each of them is too small to contact both clusters on the eIF4G surface simultaneously. Therefore, it is logical to propose that each of the two eIF4A domains contacts one of the clusters. Such arrangement predicts that both eIF4A-NTD and eIF4A-CTD interact with both eIF4G HEAT-1 and HEAT-2, forming four distinct contacts. Our NMR studies on the interaction between eIF4A-CTD and eIF4G HEAT-1 (Oberer et al., 2005) have shown that eIF4A-CTD binds near the N-terminus of HEAT-1, which is part of the top cluster in Figure 1D. Therefore, eIF4A-CTD should also bind to the N-terminal region of HEAT-2. To account for the remaining mutations, eIF4A-NTD would bind to the bottom cluster in Figure 1D. This arrangement also places eIF4A-CTD in proximity to eIF4H, if the latter is positioned similar to CBP20 in the CBP80/CBP20 complex. Finally, if the linker between the eIF4G HEAT-1 and HEAT-2 domains (not shown in Figure 1D) wraps around HEAT-1 as does the corresponding region of CBP80 (Mazza et al., 2002), it would also come in proximity to eIF4A-CTD, which would be a potential new eIF4A-binding region in eIF4G (see also Figure S3).
In summary, we hypothesize that the complex between the C-terminal segment of eIF4G, eIF4H and eIF4A adopts the topology outlined in Figure 1D. This model is based on homology to the nuclear cap-binding complex CBP80/CBP20 and mutational data. It predicts several interfaces between eIF4A and eIF4G, as well as binding of eIF4A-CTD to eIF4H. The domain orientations in the complex are expected to be dynamic and dependent on multiple protein-protein interactions. In the context of the multiprotein complex, even interactions that are weak in isolation can contribute to the stability and function, if the respective domains are brought in proximity by other interactions. We expect that all but the weakest interactions can be observed and studied in isolation, which would allow building up the topology of the complex from multiple pairs of interactions. In the following sections, we set out to experimentally test and further refine this model.
eIF4A-NTD binds to the C-terminal portion of the eIF4G HEAT-1/MIF4G domain
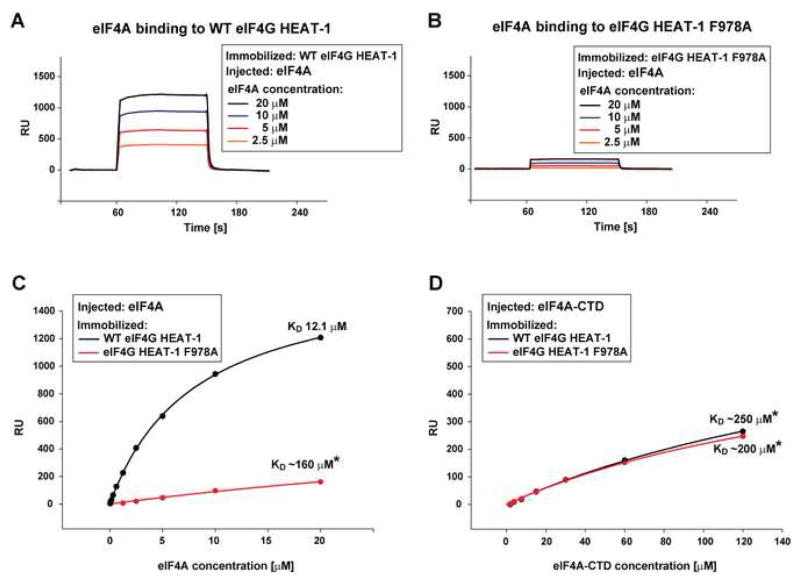
To test whether the C-terminal portion of eIF4G HEAT-1 contacts eIF4A-NTD, we studied eIF4A binding to WT eIF4G HEAT-1 and the F978A mutant using Surface Plasmon Resonance (SPR, see Supplemental Data for details). The F978A mutation near the C-terminus of eIF4G HEAT-1 has been reported to affect eIF4A binding (Imataka and Sonenberg, 1997; Morino et al., 2000), and according to our model (Figure 1D), should affect eIF4A-NTD binding.
Binding between HEAT-1 and the isolated eIF4A-NTD was too weak to study by SPR (data not shown). Therefore, we compared binding of WT eIF4G HEAT-1 and the F978A mutant to full-length eIF4A and to eIF4A-CTD: if our prediction is correct, the mutation would affect binding to full-length eIF4A, but not to eIF4A-CTD.
Consistent with previous work (Oberer et al., 2005), WT HEAT-1 bound with much higher affinity to full-length eIF4A (Figure 2C) than to eIF4A-CTD (Figure 2D). The F978A mutation strongly affected eIF4G HEAT-1 binding to full-length eIF4A, but had no effect on the interaction of eIF4G HEAT-1 with eIF4A-CTD. Based on these results, we concluded that the C-terminal region of eIF4G HEAT-1 interacts with eIF4A-NTD. Consistent with our results presented here, it was recently shown that in yeast, the eIF4G HEAT-1 domain contacts eIF4A-NTD and the residue corresponding to F978 in human eIF4G contacts eIF4A-NTD (Schutz et al., 2008).
Figure 2. The F978A mutation in eIF4G HEAT-1 affects binding to eIF4A, but not to eIF4A-CTD.
(A, B) Overlay of SPR sensorgrams showing binding of eIF4A to immobilized WT eIF4G HEAT-1 (A) and eIF4G HEAT-1 F978A
(B). (C, D) SPR graphs showing binding of full-length eIF4A (C) and eIF4A-CTD (D) to immobilized WT eIF4G HEAT-1 (black) and HEAT-1 F978A (red). KD values marked with a star should be considered only estimates, since concentrations higher than the KD could not be reached in the titration due to limited solubility.
Both domains of eIF4A bind to the eIF4G HEAT-2/MA3 domain
We used an NMR chemical shift perturbation assay to test whether eIF4A-NTD and eIF4A-CTD bind to eIF4G HEAT-2. This approach exploits the sensitivity of the peaks in NMR spectra to the changes in the environment around the respective nuclei. This method can detect specific interactions with KD values as high as 10 mM, limited by the solubility of the interacting partners (reviewed in (Marintchev et al., 2007), see Supplemental Data for details).
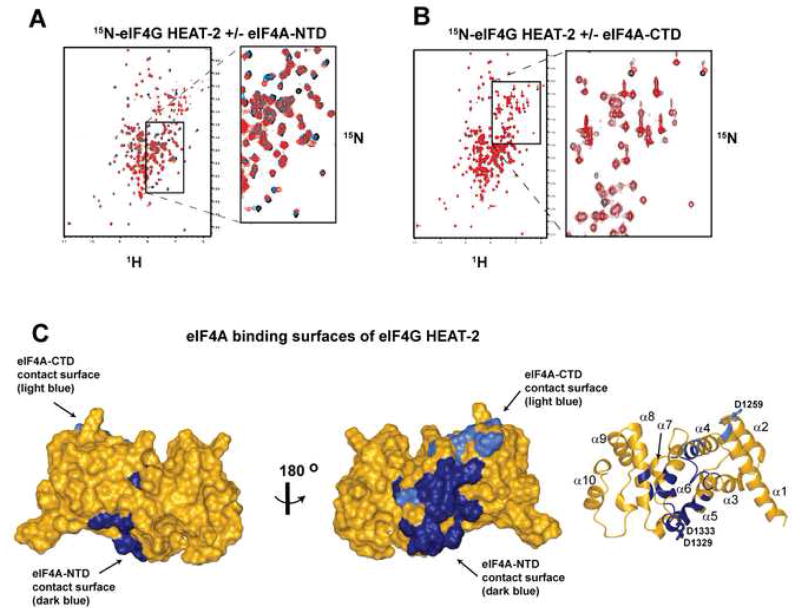
Using 15N-labeled HEAT-2 and unlabeled individual eIF4A domains, we found that HEAT-2 binds to both eIF4A-NTD (Figure 3A) and eIF4A-CTD (Figure 3B). The interaction with eIF4A-NTD appeared stronger than that with eIF4A-CTD, which was confirmed by SPR data (Figure S1 and data not shown).
Figure 3. eIF4G HEAT-2 binds to both domains of eIF4A.
(A) Overlay of 15N TROSY-HSQC spectra of 0.4 mM 15N-labeled eIF4G HEAT-2 alone (black) and in the presence of 0.15 mM (blue) or 0.4 mM (red) unlabeled eIF4A-NTD.
(B) Overlay of 15N HSQC spectra of 0.15 mM 15N-labeled eIF4G HEAT-2 alone (black) and in the presence of 0.15 mM unlabeled eIF4A-CTD (red).
(C) eIF4A binding surfaces of eIF4G HEAT-2. The orientation of HEAT-2 in the left panel is the same as that in Figure 1D, whereas in the middle panel it is rotated 180° along the Y-axis. The right panel shows the HEAT-2 domain in ribbon, in the same orientation as in the middle panel. Residues affected by eIF4A-NTD binding are painted in dark blue; residues affected by eIF4A-CTD binding are painted in light blue.
To map the eIF4A-interacting surfaces on the crystal structure of eIF4G HEAT-2 (Bellsolell et al., 2006), we assigned the backbone NMR resonances for eIF4G HEAT-2. NMR chemical shift mapping showed that eIF4A-NTD and eIF4A-CTD do indeed bind to HEAT-2 where predicted (Figure 3C, see Supplemental Data for details). As predicted from our model, residues in HEAT-2 where point mutations affected eIF4A binding (Yang et al., 2004) lie within the mapped eIF4A contact surfaces. Residues D1329 and D1333 (bottom cluster in Figure 1D) are part of the eIF4A-NTD contact surface, while residue D1259 (top cluster in Figure 1D) is part of the eIF4A-CTD contact surface.
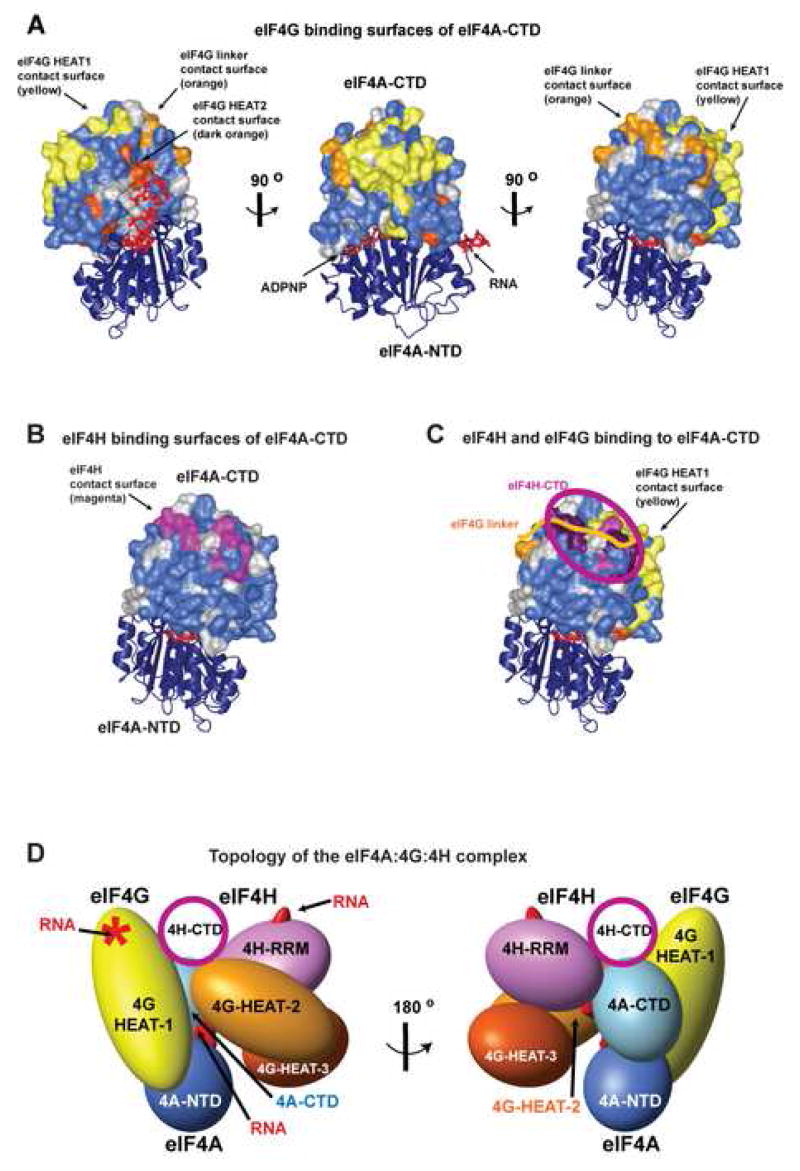
We used NMR chemical shift perturbation to map the residues of eIF4A-CTD that are affected by binding to the eIF4G HEAT-2 domain (Figure 4A, dark orange). The results indicated that the HEAT-2 binding site is near the previously mapped HEAT-1 binding site (Figure 4A, yellow) (Oberer et al., 2005). The HEAT-2 binding site appears to be adjacent to the RNA-binding surface. HEAT-2 binding also affected individual residues on the periphery of the interface with the eIF4A-NTD and ATP.
Figure 4. Topology of the eIF4A/4G/4H helicase complex.
(A) eIF4G-binding surfaces of eIF4A-CTD. The eIF4A structure is as in Figure 1D. Residues of eIF4A-CTD affected by binding of eIF4G are painted in yellow (for eIF4G HEAT-1), light orange (the linker between the eIF4G HEAT-1 and HEAT-2 domains) and dark orange (eIF4G HEAT-2). Residues in eIF4A-CTD not affected by any of the interactions are in blue; residues that could not be used for mapping (e.g., due to spectral overlap) are in grey.
(B) eIF4H-binding surface of eIF4A-CTD. eIF4A is shown in the same orientation as in the right panel in A, above. Residues in eIF4A-CTD affected by eIF4H binding are in magenta.
(C) Overlap between the eIF4H- and the eIF4 G linker-binding surfaces of eIF4A-CTD. Coloring of residues affected by eIF4H binding is as in panel B, and coloring of residues affected by eIF4G binding is as in panel A, except the residues affected by both eIF4H and eIF4G linker binding, which are painted in purple. The overlap is shown schematically by superimposing cartoons representing eIF4H-CTD (magenta oval) and the eIF4G interdomain linker (orange line).
(D)Topology of the eIF4A/4G/4H helicase complex. The mutual orientation of the eIF4G domains and eIF4H is modeled after the structure of the nuclear CBP80/CBP20 complex (Mazza et al., 2002). The orientation of eIF4A with respect to eIF4G is based on NMR chemical shift mapping (this work and (Oberer et al., 2005)). Domains, whose structures are known or could be modeled, are displayed as solid bodies with size and shape corresponding to their structures, providing the overall topology of the complex. eIF4H-CTD, whose structure is not known, is shown as a circle. The eIF4G interdomain linker (not shown) is expected to be wrapping around the eIF4G HEAT-1 domain (as does the corresponding linker in CBP80 (Mazza et al., 2002), see Figure S3A), and around eIF4A-CTD (as shown in this work).
The position of the RNA on eIF4A is modeled based on the structure of eIF4A3 (Andersen et al., 2006)). The RNA-binding site of the eIF4H RRM domain is based on that of its homolog CBP20 in the CBP80/CBP20 complex (Mazza et al., 2002). The RNA-binding site of the eIF4G HEAT-1 domain (marked with a star) is based on mutation data (Marcotrigiano et al., 2001).
eIF4A-CTD binds to the linker between eIF4G HEAT-1 and HEAT-2
We used NMR to test whether the N-terminal two-thirds of the linker between HEAT-1 and HEAT-2 interact with any of the eIF4A domains. This construct (called here eIF4G-CY, see Figure 1C) spans from the end of HEAT-1 (the H1-CT motif) to the region corresponding to the C-terminus of yeast eIF4G (the y4G-CT motif (Marintchev and Wagner, 2005)). The remainder of the interdomain linker shows little conservation among eIF4G proteins from different species. The linker region was soluble, but its 15N-HSQC spectrum showed little dispersion (data not shown), indicating that it is unfolded, at least in the absence of the rest of eIF4G. Titration of 15N-labeled eIF4A-CTD with unlabeled eIF4G-CY showed weak binding between the two proteins. Mapping of the residues affected by the interaction on the surface of eIF4A-CTD (Figure 4A, orange) indicated that the linker contacts a stretch of residues spanning from the vicinity of the HEAT-1 contact surface to near the eIF4A-NTD interface. No obvious binding was observed by NMR titration between eIF4A-NTD and eIF4G-CY (data not shown). The eIF4G linker appears to wrap on the outside of eIF4A, away from its interfaces with the HEAT domains (Figure 4A). The location of the linker-binding site on eIF4A-CTD is consistent with the proposed wrapping of the linker around eIF4G HEAT-1 (Marintchev and Wagner, 2005).
eIF4H binds to eIF4A-CTD
Using NMR chemical shift perturbation, we observed binding of eIF4H to eIF4A-CTD (Figures 4B and S2B), but not to eIF4A-NTD (data not shown). Interestingly, while the eIF4H-binding surface on eIF4A-CTD does not overlap with the binding sites for eIF4G HEAT-1 and HEAT-2, it does partially overlap with the binding site for the eIF4G interdomain linker (Figure 4C). Consistent with this observation, eIF4H can partially displace the eIF4G linker from eIF4A-CTD in NMR titration experiments (data not shown). No obvious binding was observed between eIF4H and any of the three eIF4G HEAT domains, or between the individual HEAT domains (data not shown), indicating that if these domains do interact in the context of full-length eIF4G, the binding of the isolated domains to each other is too weak to detect. The C-terminal 72-residue region of eIF4H was sufficient for the interaction with eIF4A-CTD, although eIF4A-CTD appeared to contact the N-terminal portion of eIF4H as well (Figure S2).
Topology of the eIF4A/4G/4H helicase complex
As seen in Figure 4A, the surfaces of eIF4A-CTD affected by binding of eIF4G HEAT-1, HEAT-2 and the linker are adjacent, but not overlapping, indicating that all three interactions can occur simultaneously. Thus eIF4A can act as a nucleation center bringing these three segments of eIF4G together. The mapped interaction interfaces of eIF4A with the eIF4G HEAT-1 and HEAT-2 domains (Figures 3 and 4) not only support the predicted overall topology of the complex, but more importantly provide sufficient information to orient the individual domains with respect to each other. We used our NMR data to build a rough model of the topology of the eIF4A/4G/4H helicase complex (Figure 4D). eIF4A is at the core of this assembly and holds the entire complex together through multiple interactions with eIF4H and eIF4G domains.
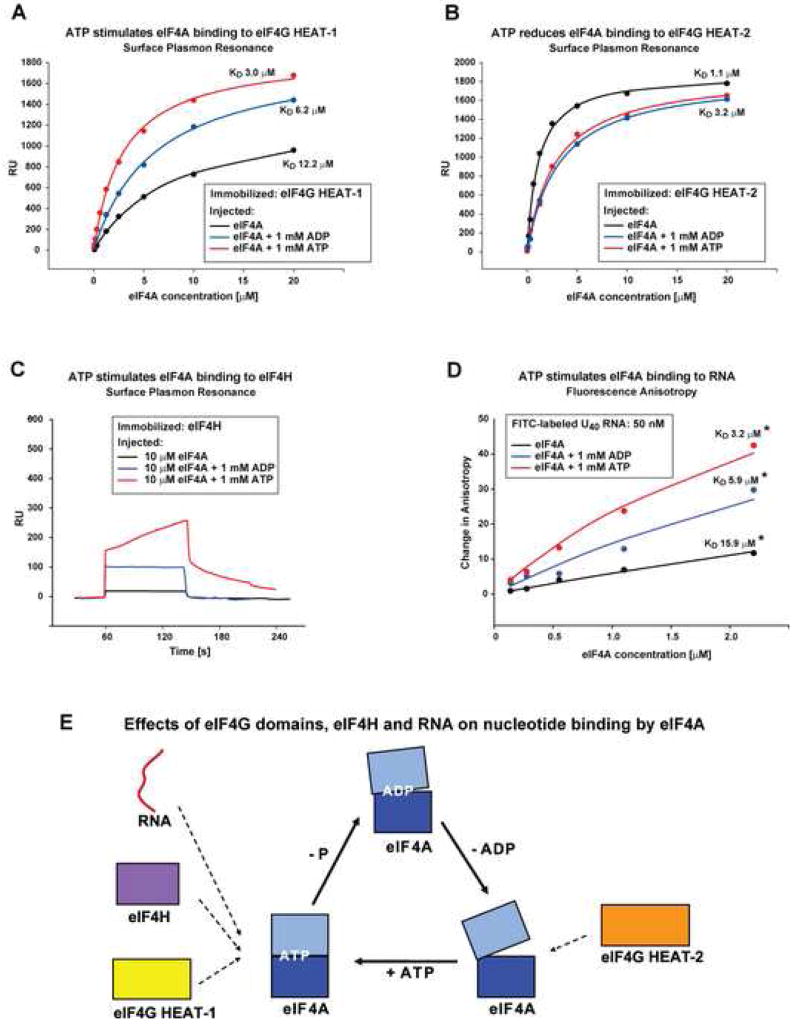
eIF4G HEAT-1, eIF4H and RNA all stimulate ATP binding to eIF4A
We used SPR to compare the binding of eIF4G HEAT-1 to eIF4A in the presence of ATP, ADP, and in the absence of nucleotide. ATP increased the affinity of eIF4A for eIF4G HEAT-1 fourfold. ADP also stimulated the interaction between eIF4A and HEAT-1, although to a lesser extent than ATP (Figure 5A). In contrast, ATP and ADP both decreased the affinity of eIF4A for eIF4G HEAT-2 threefold (Figure 5B). This implies that eIF4G HEAT-1 increases the affinity of eIF4A for ATP fourfold, compared to nucleotide-free eIF4A, whereas eIF4G HEAT-2 decreases it threefold (principle of detailed balance (Fersht, 1999)). Therefore, the overall affinity of eIF4A for ATP varies by an order of magnitude, depending on whether it is bound to the HEAT-1 or HEAT-2 domain of eIF4G, with free eIF4A having intermediate affinity for ATP.
Figure 5. Effects of ATP and ADP on the eIF4A/eIF4G and eIF4A/eIF4H interactions.
(A, B) SPR graphs of eIF4A binding to immobilized eIF4G HEAT-1 (A) or eIF4G HEAT-2 (B) in the absence of nucleotide (black) and in the presence of 1 mM ATP (red) or 1 mM ADP (blue).
(C) Overlay of SPR sensorgrams showing binding of 10 μM eIF4A to immobilized eIF4H in the absence of nucleotide (black), in the presence of 1 mM ATP (red) and 1 mM ADP (blue).
(D) Fluorescence anisotropy graphs of eIF4A binding to FITC-labeled U40 RNA oligonucleotide in the absence of nucleotide (black) and in the presence of 1 mM ATP (red) or 1 mM ADP (blue). The KD values in panel D, marked with a star, should be considered estimates because of the tendency of eIF4A to aggregate at higher concentrations. Binding curves at lower eIF4A concentrations are shown since those are least affected by eIF4A aggregation.
(E) Summary of the effects of eIF4G, eIF4H and RNA on the affinity of eIF4A for nucleotides. RNA, eIF4H, and eIF4G HEAT-1 (left) favor the closed ATP-bound conformation, whereas eIF4G HEAT-2 (right) prefers the open nucleotide-free conformation of eIF4A.
The binding of eIF4H to eIF4A-CTD or full-length eIF4A was too weak to study by SPR. However, eIF4H binding to full-length eIF4A could be observed in the presence of ATP (Figure 5C). Remarkably, the SPR curves are biphasic with a fast initial association followed by a slow process, and dissociation also exhibits a fast and a slow phase (Figures 5C and S2E, see Supplemental Data for more details). ADP also stimulated eIF4H binding to eIF4A, but unlike ATP, it did not cause formation of the slowly dissociating complex (Figure 5C). Stable binding of eIF4H to the eIF4A/eIF4G complex also requires the presence of ATP (see Figure S4).
We used Fluorescence anisotropy (Fluorescence polarization) to study the effect of ATP and ADP on eIF4A binding to an RNA oligonucleotide labeled with Fluorescein Isothiocyanate (FITC). Fluorescence anisotropy allows distinguishing the free FITC-labeled RNA that tumbles faster from the protein-bound FITC-labeled RNA, which is part of a larger complex and tumbles slower in solution. The effects of ATP and ADP on eIF4A binding to RNA (Figure 5D) were essentially the same as their effects on eIF4A binding to eIF4G HEAT-1 (Figure 5A): ATP stimulated RNA binding several-fold and ADP had a smaller effect. These results indicate that eIF4G HEAT-1, eIF4H and RNA all bind cooperatively with ATP, whereas eIF4G HEAT-2 does not (summarized in Figure 5E).
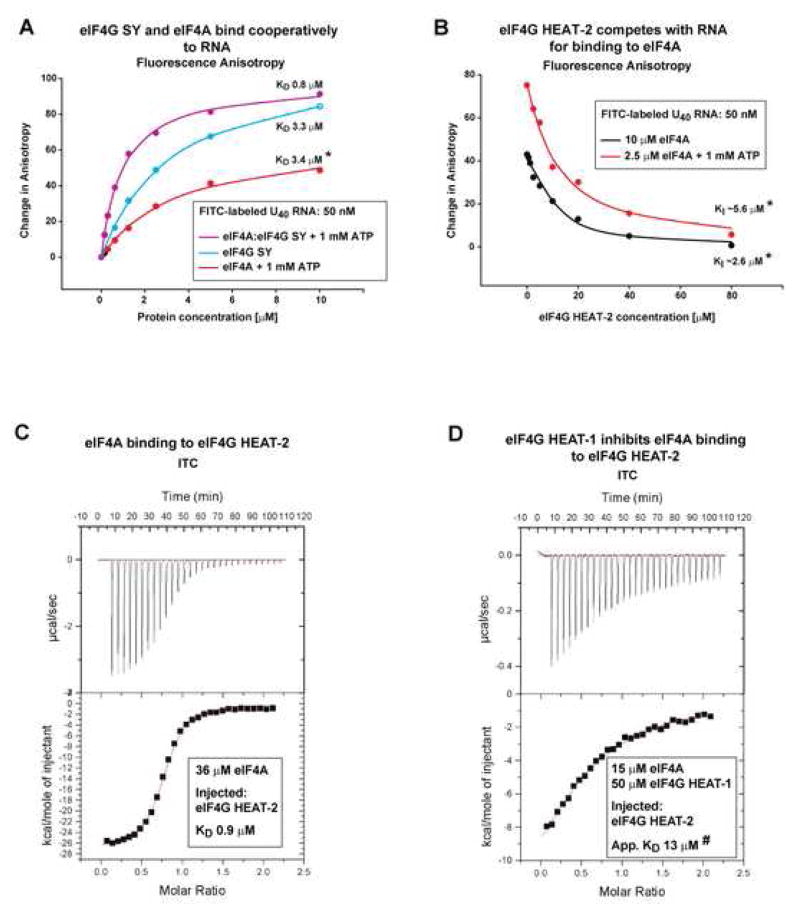
eIF4G HEAT-1 and eIF4G HEAT-2 have opposite effects on eIF4A binding to RNA
We used Fluorescence anisotropy to study the effects of eIF4G HEAT-1 and HEAT-2 on the affinity of eIF4A for RNA. Since eIF4G HEAT-1 itself binds RNA and its affinity for eIF4A is relatively weak, it was not possible to form a stable HEAT-1/eIF4A complex and ensure that eIF4A and eIF4G HEAT-1 bind RNA only as a complex and not also independently. Therefore, we used a larger eIF4G HEAT-1 containing construct, called here eIF4G SY (see Figure 1C). This construct corresponds to the minimal eIF4G fragment sufficient to support scanning; contains an additional RNA-binding region as well as two extra eIF4A-binding sites (see Figure 1C and Figure 4A); and binds to eIF4A with nanomolar affinity (Korneeva et al., 2001). The complex of eIF4G SY and eIF4A binds to RNA with higher affinity than the individual proteins (Figure 6A). In contrast, eIF4G HEAT-2 does not bind RNA at the concentrations used (data not shown) and competes with RNA for binding to eIF4A (Figure 6B). We also tested the effect of eIF4G HEAT-1 on the binding of eIF4A to eIF4G HEAT-2. The Isothermal Titration Calorimetry experiments in Figure 6CD show that eIF4A binding to eIF4G HEAT-2 is reduced by at least an order of magnitude in the presence of eIF4G HEAT-1.
Figure 6. Opposing effects of eIF4G SY and eIF4G HEAT-2 on RNA binding to eIF4A.
(A) Fluorescence anisotropy graphs showing binding of FITC-labeled U40 RNA oligonucleotide to eIF4A in the presence of 1 mM ATP (red); eIF4G SY (cyan); and the eIF4A/eIF4G SY complex (purple).
The eIF4G SY fragment (see Figure 1C) consists of the HEAT-1 domain and additional RNA- and eIF4A-binding segments.
(B) Fluorescence anisotropy graphs showing inhibition by eIF4G HEAT-2 of the binding of 10 μM eIF4A to 50 nM FITC-labeled U40 RNA in the absence of nucleotide (black) and 2.5 μM eIF4A to 50 nM FITC-labeled U40 RNA in the presence of 1 mM ATP (red).
(C, D) Isothermal Titration Calorimetry (ITC) graphs of eIF4G HEAT-2 binding to eIF4A in the absence (C) and presence (D) of eIF4G HEAT-1.
Note that in the experiment shown in panel D, the concentration of eIF4G HEAT-1 (50 μM) is not saturating: it is only ~4 times higher than the KD of the eIF4A/HEAT-1 interaction (12 μM, Figure 2C). Therefore, a fraction of eIF4A is not bound to HEAT-1 and the calculated apparent KD for the interaction between eIF4A and eIF4G HEAT-2 in the presence of HEAT-1 (marked with a star on panel D) should be considered a lower limit of the actual KD.
Discussion
Topology of the eIF4A/eIF4G/eIF4H helicase complex
eIF4A is the prototypic member of the DEAD box family of helicases, a subset of the Superfamily 2 (SF2) helicases. It exemplifies a minimal helicase, containing only the two RecA-like domains, where the conserved helicase motifs are located, and requires the accessory proteins eIF4G, eIF4H and eIF4B for function. Although the existence of a complex of eIF4A with other translation initiation factors was first reported almost 30 years ago, it has not so far been possible to solve its structure (reviewed in (Gingras et al., 1999; Marintchev and Wagner, 2004; Rogers et al., 2002)). Here, we combined modeling, NMR, site-directed mutagenesis, and quantitative binding assays to elucidate the topology of the eIF4A/4G/4H helicase complex (Figure 4D).
The eIF4H-binding surface of eIF4A-CTD does not overlap with the HEAT-1 or HEAT-2 binding surfaces (Figure 4) and eIF4H forms a complex with eIF4A and eIF4G (see Figure S4D). Stable association of eIF4H with eIF4A and eIF4G requires the presence of ATP (compare Figure S4 panels C and D), consistent with the stabilizing effect of ATP on the eIF4A/eIF4H interaction (Figure 5C). These results are in line with the ability of eIF4G and eIF4H to simultaneously stimulate the helicase activity of eIF4A (reviewed in (Rogers et al., 2002)). Although we could not detect binding between eIF4H and eIF4A-NTD, the ability of ATP and ADP to stimulate binding of eIF4H to full-length eIF4A (Figure 5C), but not eIF4A-CTD (data not shown) indicates that eIF4H binds to both domains of eIF4A, although its interaction with eIF4A-NTD appears to be dependent on the presence of nucleotide. This interpretation is consistent with a recent report that implicated eIF4A-NTD in eIF4H binding in the presence of RNA and AMPPNP (Rozovsky et al., 2008). While in this work we have focused our attention on eIF4H, our findings are likely also applicable to its homolog eIF4B (see Figure 1B), which appears to bind to the same site on eIF4A (Rozovsky et al., 2008).
eIF4G and eIF4H modulate the affinity of eIF4A for nucleotides
We were also able to get insights into the mechanisms of regulation of the eIF4A helicase activity by eIF4G and eIF4H. A common trend is observed among RNA, eIF4H and the two eIF4A-binding domains of eIF4G: they all bind to both eIF4A domains and bind most strongly to full-length eIF4A. In all cases, the interactions influence the affinity of eIF4A for ATP: eIF4H and eIF4G HEAT-1 stimulate the helicase activity of eIF4A in part by increasing its affinity for ATP, whereas eIF4G HEAT-2 lowers the affinity of eIF4A for nucleotides.
It is well established that eIF4A has different conformations in the ATP-bound state, the ADP-bound state and in the nucleotide-free state (Lorsch and Herschlag, 1998a, b). Presumably, the nucleotide-free form of eIF4A is open with no stable contacts between the two domains. The ATP- and ADP-bound states are both closed, but different from each other. The ADP-bound state could be “less closed”, which could in turn affect the formation of a contiguous RNA binding surface (Figure 5E). Accordingly, while RNA, eIF4G HEAT-1 and eIF4H favor the ATP-bound conformation of eIF4A over the nucleotide-free form, they have intermediate affinity for the ADP-bound conformation. It was recently suggested for yeast eIF4G that the domain corresponding to HEAT-1 in human may only contact eIF4A-NTD when eIF4A is nucleotide-free or bound to ADP, but would separate from the NTD when ATP is bound to eIF4A (Schutz et al., 2008). However, this idea clearly contradicts our results showing that HEAT-1 has even higher affinity for ATP-bound eIF4A than for ADP-bound or nucleotide-free eIF4A (Figure 5A). Therefore, eIF4G HEAT-1 appears to contact both domains of ATP-bound eIF4A, as well as ADP-bound and nucleotide-free eIF4A. It remains to be seen whether the conformation of eIF4A is identical or somewhat different when bound to ATP, eIF4G HEAT-1 or both.
We show that the interactions of eIF4A with eIF4G HEAT-1 and HEAT-2 are anti-cooperative (Figure 6CD), even though there is no overlap between the surfaces of eIF4A that bind HEAT-1 and HEAT-2 (Figure 3 and KE & GW, unpublished data). The opposite effects of ATP on HEAT-1 and HEAT-2 binding to eIF4A (Figure 5) indicate that this anticooperativity is mediated by the eIF4A conformation. HEAT-1 requires the closed, ATP-bound interdomain orientation in eIF4A for optimal binding, whereas HEAT-2 prefers the open, nucleotide-free conformation. Therefore, only one eIF4G HEAT domain at a time would be able to bind both eIF4A domains with high affinity, whereas the other HEAT domain would either be bound to only one of the eIF4A domains or its binding to the second domain would be suboptimal.
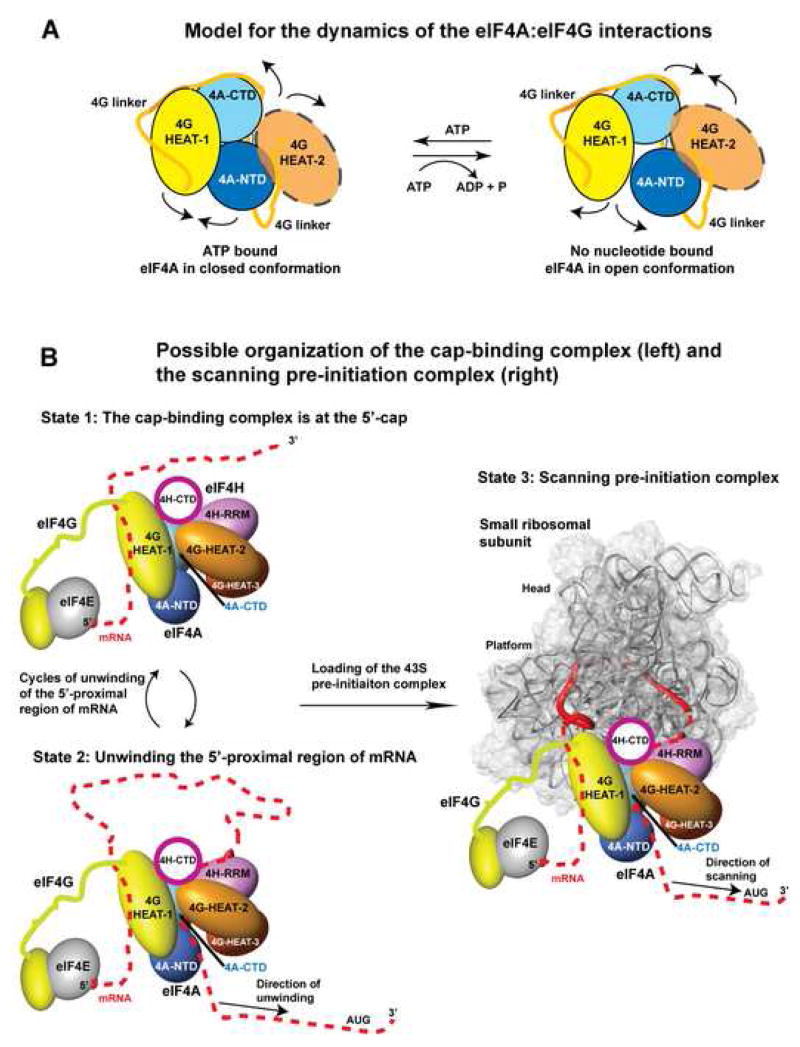
The interplay between the dynamics of the eIF4A/eIF4G interactions and the ATP binding/hydrolysis cycle of eIF4A is schematically represented by the model in Figure 7A (see also Movie S1). In the absence of nucleotide, eIF4A binds to eIF4G HEAT-2 about an order of magnitude tighter than to HEAT-1 (Figure 5AB), which would favor the open eIF4A state shown in the right panel of Figure 7A. ATP binds cooperatively with eIF4G HEAT-1 and anticooperatively with HEAT-2 (see Figure 5AB) and could partly shift the equilibrium toward the closed state shown on the left. Since RNA also binds cooperatively with ATP (Figure 5D) and competes with HEAT-2 (Figure 6B), RNA binding to eIF4A would displace HEAT-2 and drive the equilibrium toward the closed ATP-bound state of eIF4A (left panel of Figure 7A).
Figure 7. Models for the mechanisms of unwinding of mRNA and scanning.
(A) Model for the dynamics of the eIF4A/eIF4G interactions
eIF4G HEAT-1 (yellow) stimulates ATP binding and the helicase activity of eIF4A by simultaneous binding to both eIF4A domains in the closed ATP-bound conformation (left). eIF4G HEAT-2 (orange) favors the nucleotide-free state by simultaneous binding to both eIF4A domains in an open conformation (right). The interdomain linker of eIF4G (light orange) also binds eIF4A and stabilizes the complex. The arrows indicate directions of rearrangements during the ATP hydrolysis/nucleotide exchange cycle. ATP (not shown) binds at the interface between the two eIF4A domains. HEAT-2 is shown semi-transparent to emphasize that it is not required for the ATP-binding/hydrolysis cycle.
(B) Hypothetical model for the organization of the cap-binding complex and the scanning complex.
State 1. Model of the cap-binding complex bound at the 5′-cap. The orientation and coloring of eIF4A, eIF4G and eIF4H is as in Figure 4D. The linker between the eIF4G HEAT-1 and HEAT-2 domains is not shown. The mRNA is drawn as a dashed red line. The 5′-cap/eIF4E/4G complex structure shown is from yeast (Gross et al., 2003).
State 2. Model for the unwinding of the 5′-proximal region of mRNA.
The position and 5′-3′ polarity of the mRNA on eIF4A is modeled based on the structure of eIF4A3 (Andersen et al., 2006)). The direction of translocation/unwinding along the mRNA (5′ to 3′) is indicated by an arrow.
State 3. Model for the scanning complex.
The small ribosomal subunit (grey semi-transparent surface, with the rRNA backbone shown as ribbon) and the mRNA (red solid ribbon) are from 1JGP.pdb (Yusupova et al., 2001). The direction of scanning of the initiation complex along the mRNA (5′ to 3′) is indicated by an arrow. According to this model, eIF4A is on the front (3′-side) of the scanning initiation complex, in agreement with its helicase function.
ATP hydrolysis and ADP release would reduce the affinity of eIF4A for RNA and eIF4G HEAT-1, while increasing the affinity for HEAT-2 (see Figure 5). Therefore, ATP hydrolysis would shift the equilibrium toward the open state of eIF4A (Figure 7A, right panel). Binding of a new molecule ATP would again promote the closed eIF4A conformation (Figure 7A, left panel) and complete the cycle. This mechanism outlined in Figure 7A is enhanced by eIF4H. ATP bound to eIF4A causes formation of a tight complex with eIF4H while hydrolysis of ATP and displacement of nucleotide weakens the eIF4H/eIF4A interaction (Fig. 5C). While the mechanism described above fits well with the known data about the functions of eIF4A and eIF4G, the observed effects on nucleotide binding should only be considered qualitative, because the exact binding affinities could be significantly different in the context of full-length eIF4G.
The changes in the eIF4A domain orientation during the ATP binding/hydrolysis cycle of the helicase would cause movements in the domains of eIF4G and eIF4H that interact with the helicase. Thus, the entire eIF4A/4G/4H complex is a dynamic structure that cycles among three distinct states: ATP-bound, ADP-bound, and nucleotide-free (see also Figure 5E). eIF4G HEAT-2 prefers the open nucleotide-free conformation of eIF4A (Figure 5B), but the binding sites for eIF4A-NTD and eIF4A-CTD on the surface of HEAT-2 are adjacent to each other (Figure 3C). This indicates that while the eIF4A interdomain orientation in the eIF4A/HEAT-2 complex is open enough to disrupt the nucleotide-binding pocket, eIF4A-NTD and eIF4A-CTD remain in proximity to each other, instead of being far apart as in the crystal structure of yeast eIF4A (Caruthers et al., 2000). Therefore, the overall domain orientation in the eIF4A/4G/4H complex, shown on Figure 4D would remain roughly similar in all three states.
It must be pointed out that eIF4G HEAT-2 only serves a modulatory role in mammals, but is not essential for translation initiation (Morino et al., 2000) and has even been lost during the evolution of a number of fungal species, including budding yeast (reviewed in (Marintchev and Wagner, 2004)). Therefore, the ATP binding/hydrolysis cycle shown in Figure 7A can operate without HEAT-2 stabilizing the open nucleotide-free state of eIF4A. The effects of eIF4G HEAT-2 on nucleotide binding to eIF4A reported here suggest a possible modulatory role for HEAT-2. In the absence of RNA, the function of eIF4G HEAT-2 could be to lower the affinity of eIF4A for ATP and thus suppress idle ATPase activity in the eIF4A/eIF4G complex that would otherwise be partially stimulated by eIF4G HEAT-1.
The eIF4A/eIF4G/eIF4H helicase complex and the mechanism of mRNA unwinding
The RNA-binding surfaces of eIF4A are well-known from the structures of eIF4A homologs in complex with ATP and RNA (see e.g., (Andersen et al., 2006)). In the closed active conformation, the two RecA-like domains come in contact with each other, forming the ATP-binding pocket, and a contiguous RNA-binding surface. In all available structures of eIF4A homologs in complex with RNA, the NTD of the protein is oriented toward the 3′ end of the RNA, and the CTD is oriented toward the 5′ end. Therefore, when eIF4A is translocating from the 5′-cap of the mRNA in the 3′-direction, eIF4A-NTD would be on the front. eIF4G HEAT-1 also binds RNA and its N-terminal region is important for binding (Marcotrigiano et al., 2001). Since the HEAT-1 binding surface of eIF4A-CTD is opposite to its RNA-binding surface (Figure 4D), the eIF4G HEAT-1 and eIF4A likely do not bind to adjacent regions of the mRNA. Interestingly, the binding site of eIF4H on eIF4A (Figure 4D) indicates that its RRM domain can contact the mRNA immediately 5′ from eIF4A, which would place it behind the helicase with respect to the direction of translocation (Figure 7B, left panels). This suggests that while both eIF4G HEAT-1 and eIF4H stabilize ATP and RNA binding to eIF4A, they may employ different mechanisms for stimulation of eIF4A activity. The topology of the complex described here (Figure 4D) indicates that eIF4G HEAT-1 tethers eIF4A to mRNA, whereas binding of eIF4H to single-stranded mRNA behind eIF4A could also serve to prevent mRNA reannealing and promote processive unidirectional translocation of eIF4A. It should be noted here that eIF4G possesses at least one more RNA-binding region: located N-terminal from the HEAT-1 domain and important for scanning (labeled “S” on Figure 1C) (Prevot et al., 2003). It is not currently known where that region of eIF4G binds the mRNA with respect to eIF4A.
The cap-binding complex, composed of eIF4E, eIF4G, eIF4A, as well as eIF4B or eIF4H, helps recruit the small, 40S ribosomal subunit both through interactions with eIF3 and by unwinding any secondary structures in the mRNA in the vicinity of the 5′-cap (clearing the “ribosome landing pad”). In our model, the RNA-binding surface of eIF4G HEAT-1 in the complex with eIF4A appears to be almost on the opposite side with respect to the RNA-binding site of eIF4A. One can thus hypothesize that eIF4G HEAT-1 contacts a region of mRNA “behind” (5′ from) the helicase. The mRNA between eIF4G HEAT-1 and eIF4A could be looped out as the complex translocates in the 3′ direction while staying tethered to the 5′-cap. Although the cap-binding complex has greater helicase activity than free eIF4A, it still has limited processivity: it will eventually fall off the leading end of the mRNA and resume the cycle from the 5′-end, thus constantly keeping the 5′-proximal “landing pad” ready for 40S subunit binding (Figure 7B, left panels).
Model for the organization of the scanning pre-initiation complex
Since the Cap-binding complex is bound at the 5′-end of the mRNA, it is simple to assume that it remains behind (on the 5′ side) the 40S ribosomal subunit during scanning. However, mechanistically, it would be more logical for the helicase eIF4A to be in front (on the 3′ side) of the scanning complex. The organization of the eIF4A/4G/4H complex (Figure 4D), suggests that it could be simultaneously behind and in front of the 40S subunit during scanning. On the bacterial ribosome, the mRNA enters from the solvent side of the small subunit; wraps around the Neck coming to the interface with the large subunit; then exits again through the solvent side, ~50 Å away from the entry site (Yusupova et al., 2001). The path of the mRNA through the eukaryotic translation pre-initiation complex appears to be similar (Pisarev et al., 2008). The 40S subunit is recruited to the mRNA through the interaction between eIF4G and ribosome-bound eIF3, which is on the solvent side of the 40S subunit (Siridechadilok et al., 2005). Superimposing our model of the eIF4A/4G/4H complex on the crystal structure of the small subunit bound to mRNA (Yusupova et al., 2001) indicates that it is spatially possible for eIF4A to be in front of the scanning complex, while eIF4G HEAT-1 is near the mRNA on the 5′ side (Figure 7B, right panel). The polarity of mRNA on eIF4A is consistent with this hypothetical model. eIF4H could contact mRNA behind eIF4A, but in front of the 40S subunit. Wrapping of mRNA around the Neck of the 40S subunit would bring the two ends of the mRNA near each other and favor simultaneous interaction of the eIF4A/4G/4H complex with mRNA segments both on the front (3′) and back (5′) of the scanning complex. Once the scanning pre-initiation complex is assembled on mRNA, it can processively translocate in the 3′ direction in search of the start codon. eIF4G HEAT-1 could either retain its interaction with the 5′ portion of the mRNA or contact rRNA instead. eIF4E may remain associated with both the cap and eIF4G during scanning, at least transiently, as discussed previously (Jackson, 2000; Marintchev and Wagner, 2004). Since eIF4H and its homolog eIF4B appear to share a common binding site on eIF4A (Rozovsky et al., 2008) and similar function, in complexes containing eIF4B instead of eIF4H, eIF4B will likely take the place of eIF4H. Finally, the core components of the helicase complex (eIF4A, eIF4B and eIF4G HEAT-1) are conserved between human and budding yeast. Therefore, while budding yeast lack an eIF4H homolog and have lost the eIF4G HEAT-2 and HEAT-3 domains, the overall organization of the scanning complex is likely conserved with that in human.
Experimental Procedures
Vectors, protein expression and purification
The human eIF4A1 (406 residues), eIF4G1 (1600 residues), and eIF4H isoform 2 (228 residues) or subcloned fragments thereof were used. The constructs used in this study are shown in Figure 1. The expression constructs for human eIF4A1 and eIF4A-CTD are described in (Oberer et al., 2005). The clone expressing eIF4A-NTD has an N-terminal His-tag, followed by a TEV protease cleavage site. All other proteins and protein fragments used in this study have an N-terminal GB1 tag, followed by a His-tag and a TEV cleavage site. Expression and purification of unlabeled proteins, as well as of proteins uniformly labeled with stable isotopes for the NMR experiments, was carried out as previously described for eIF4A-CTD (Oberer et al., 2005).
NMR resonance assignments and chemical shift perturbation assay
NMR spectra were recorded at 298 K on a Bruker 750, 600, or 500 MHz, or a Varian Inova 600 or 500 MHz spectrometers, equipped with cryogenic probes. Unless stated otherwise, samples for NMR measurements contained 0.4–1 mM protein in buffer containing 20 mM Tris-HCl (pH 7.0), 150 mM KCl, 2 mM DTT, 1 mM EDTA, 0.01% NaN3, 0.2 mM AEBSF, and 10% D2O. The samples of eIF4A-CTD were at 150 μM or lower, due to the limited solubility of these proteins at physiological salt concentrations. Backbone resonance assignments were obtained for 100 % of the assignable residues in the eIF4G HEAT-2 domain, using standard triple-resonance experiments. Chemical shift mapping was done as described in (Marintchev et al., 2007).
Surface Plasmon Resonance (SPR)
SPR experiments were carried out using a BIAcore 3000 instrument (Biacore Inc., Piscataway, NJ). Wild-type or mutant eIF4G HEAT-1, eIF4G HEAT-2, or eIF4H was immobilized on a research grade CM5 sensor chip using amino coupling kit. The binding experiments were carried out in the buffer described above for the NMR experiments at 25°C, at flow rate of 20 μl/min. eIF4A, eIF4A-NTD, or eIF4A-CTD was injected at increasing concentrations. 1 mM ATP (or ADP) and 3 mM MgCl2 was added to the injected eIF4A samples, where indicated. Curve fitting was done with SigmaPlot (SPSS Inc.).
Fluorescence Anisotropy
Fluorescence anisotropy RNA binding experiments were carried out in 384-well plates on a PerkinElmer EnVision plate reader. The samples contained 50 nM FITC-labeled U40 RNA oligonucleotide (Dharmacon). The binding experiments were carried out in the buffer described above for the NMR experiments at 25°C. Curve fitting was done with SigmaPlot (SPSS Inc.), as described in (Roehrl et al., 2004).
Isothermal Titration Calorimetry (ITC)
ITC binding experiments were carried out on a VP-ITC machine (MicroCal) at 25°C by injecting aliquots of eIF4G HEAT-2 into a well containing eIF4A with and without HEAT-1 in the buffer described above for the NMR experiments, except that DTT was replaced with TCEP, and the buffer contained 5 mM MgCl2 and 5% glycerol. Data were processed using Origin software.
Structure analysis and modeling
The interdomain orientation between eIF4A-NTD, eIF4A-CTD, and the RNA were modeled based on the structure of eIF4A3 (2HYI.pdb, (Andersen et al., 2006)). For modeling, we used the crystal structure of human eIF4A-NTD (2G9N.pdb) and a homology model for human eIF4A-CTD (Oberer et al., 2005) based on the structure of yeast eIF4A. The interdomain orientation of the HEAT domains of eIF4G was as modeled previously (Marintchev and Wagner, 2005) based on the interdomain orientation in CBP80 in the nuclear Cap-binding complex (1HT2.pdb (Mazza et al., 2002)). The topology of the eIF4A/4G/4H complex was built interactively in MOLMOL, guided by the results from NMR chemical shift mapping and site-directed mutagenesis. Modeling of the position of the eIF4A/4G/4H complex on the ribosome was done in MOLMOL, which was also used to generate the resulting figures.
Supplementary Material
Supplemental Data include Supplemental Results, Discussion, Experimental Procedures, References, 4 figures, and one movie.
Acknowledgments
We thank Tatyana Pestova and Alan Hinnebusch for helpful discussions and comments. This work was supported by NIH grants CA68262 and GM47467 to GW. AM was supported by a Howard Temin K01 Career Award from the NCI (CA119107).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Everly DN, Jr, Read GS. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J Virol. 2005;79:9651–9664. doi: 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science. W. H. Freeman and Company; 1999. pp. 125–131. [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ. Comparative view of initiation site selection mechanisms. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control. Cold Spring harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 127–184. [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Merrick WC, Rhoads RE. Characterization of the two eIF4A-binding sites on human eIF4G-1. J Biol Chem. 2001;276:2872–2879. doi: 10.1074/jbc.M006345200. [DOI] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998a;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998b;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot D, Decimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- Roehrl MH, Wang JY, Wagner G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14:2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol. 2004;24:3894–3906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Supplemental Results, Discussion, Experimental Procedures, References, 4 figures, and one movie.