Abstract
Over the last 15 years, great improvements in genetic engineering and genetic manipulation strategies have led to significant advances in the understanding of the genetics governing embryological limb development. This field of science continues to develop, and the complex genetic interactions and signalling pathways are still not fully understood. In this review we will discuss the roles of the principle genes involved in the three-dimensional patterning of the developing limb and will discuss how errors in these signalling cascades correlate to congenital limb deformity in humans. This review is aimed at orthopaedic surgeons wishing to understand the principles of congenital limb deformity related to genetic signalling errors. It is by no means a comprehensive study of the molecular genetics governing the complex interactions involved in each step of limb development. There are however many syndromes involving limb deformity for which the molecular causes are unknown.
Keywords: Limb development, Limb patterning, Genetic regulation, Hox genes, Retinoic acid
Introduction
The genetic processes that control development of the limb, in both vertebrates and invertebrates, are complicated and are still not fully understood. The temporal and spatial expression of individual genes and gene families has been studied. This information supplemented with experimental evidence from gene manipulation studies has established the role of these genes in limb patterning in the developing embryo. Errors in the pathways controlling limb patterning are often associated with defects in the development of other organ systems within the embryo. The most commonly affected include: the gastro-intestinal tract, the heart, the central nervous system and the genito-urinary tract.
An understanding of the complex temporal and spatial co-ordination of genetically driven molecular pathways in the developing limb is crucial to understanding the pathology of congenital limb deformity. The orthopaedic surgeon must be aware of both the functional and psychosocial problems encountered by those affected. These conditions are often difficult to treat and can be associated with poor outcomes. In the future a complete understanding of the pathways and pathology involved in embryological human limb development may lead to the development of molecular genetic therapies that may prevent or improve these disabling abnormalities.
Our current understanding of the molecular genetics of human limb development is extrapolated from experiments manipulating genetic interactions of vertebrates and invertebrates. Vertebrate forelimbs and hindlimbs are morphologically similar across species, and there are broad similarities in the morphology of the upper and lower limbs. The developing limb can be divided into three principle zones; the proximal stylopod, the middle zeugopod and the distal autopod. In the developing human the stylopod region becomes the arm or thigh, the zeugopod becomes the forearm or leg and the autopod becomes the hand or foot. The “patterning” process controls the genetic signals governing limb development in three axes: proximal-distal, anterior–posterior, and dorsal–ventral. It also promotes differentiation of specific forelimb and hindlimb structures.
The human limb buds appear on day 26 for the upper limb and day 28 for the lower limb. Activation of the mesenchymal cells of the lateral mesodermal plate causes an outgrowth of the limb bud consisting of a mesenchymal core surrounded by ectoderm. The distal border of the ectoderm thickens to form the apical ectodermal ridge (AER). The progress zone lies beneath the apical ectodermal ridge, and signals from the AER promotes rapid undifferentiated growth of its mesenchyme. The AER’s influence can only extend to the adjacent cells of the progress zone and the more proximal mesenchymal cells, being further from the influence of the apical ectodermal ridge, start to differentiate into muscle and cartilage. By 6 weeks the hand and foot plates are apparent, separated from the rest of the limb bud by a circumferential constriction which becomes the wrist. Another, more proximal, constriction develops at the level of the elbow and knee. A joint inter-zone is induced, and after local chondrocyte proliferation the joint cavity is formed. Distally, apoptosis of the parts of the apical ectodermal ridge (AER) creates finger and toe buds.
Several gene families are involved in the spatially and temporally co-ordinated growth and differentiation of the developing limb bud. The products of these genes act as signals to turn on other “downstream” genetic pathways. Most genes influence the initiation and patterning of both the forelimb and the hindlimb, but some genes are differentially expressed in the developing forelimb and hindlimb.
The initiation of limb outgrowth and the role of Hox and T-box genes
There are 39 members of the HOX gene family, and they are arranged in four separate clusters. HOX genes are evolutionarily highly conserved and encode transcription factors of fundamental importance for body patterning. The HOXA cluster is located on chromosome 7, the HOXB cluster on chromosome 17, the HOXC cluster on chromosome 12 and the HOXD cluster is on chromosome 2. HOX genes contain a homeobox region of about 180 bp that encodes the homeodomain, a protein able to bind DNA. The homeobox genes encode protein transcription factors capable of switching on cascading genetic pathways. These homeodomain proteins act in synergy with other transcription factors in the promoter region of downstream genes. The promoter regions of genes are very specific and will only be activated if the correct combination of transcription factors is present. This gives a high degree of specificity to the promoter region and is fundamental to the control of the patterning process in the developing limb.
Hox genes are expressed along the rostro-caudal axis in the lateral mesoderm of all segmented animals and are fundamental to the positioning of the developing limbs or wings in the embryos of both vertebrates and invertebrates. In the limb fields, areas where the limb buds will develop, specific Hox genes become upregulated by retinoic acid. This initiates downstream genetic signalling, ensuring synchronised and progressive growth along the three axes of growth: anterio-posterior, dorso-ventral and proximo-distal. On the initiation of limb outgrowth other transcription factors are expressed to control specific areas of patterning, i.e. forelimb versus hindlimb.
Studies on a range of vertebrate limb models have identified three genes that appear to programme a limb to develop down either a forelimb or hindlimb pathway. Tbx5 and Tbx4 are T-box family transcription factors expressed in the forelimb and hindlimb, respectively. Pitx1, another transcription factor, is expressed in the developing hindlimb, but not the forelimb. The temporal and spatial expression patterns of Tbx5, Tbx4 and Pitx1 suggest they play an important role in programming the identity of the developing limb (Table 1).
Table 1.
Summary of site and mechanism of action and clinical syndromes associated with errors in expression of principle gene families involved in limb patterning
| Gene family | Example | Role | Area of action | Research | Clinical correlation |
|---|---|---|---|---|---|
| Fibroblast growth factors | FGF-4 and FGF-8 | Growth factors | AER | Induce accessory limbs in chick embryos | FGF-receptor3 mutations cause achondroplasia |
| Hedgehog | Sonic hedgehog (Shh) | Binds Ptc to release Smo and promotes expression of BMP, WNT, HOX, & Gli | ZPA | Ectopic activity causes duplications | |
| Patched (tumour suppressor gene) and smoothened | Ptc and Smo | Ptc is an Shh receptor. Smo is released when Shh binds Ptc. | Cell surface mesenchyme | ||
| Bone morphogenic proteins | BMP-2 BMP-7 |
Osteoblast differentiation Chondrocyte condensation and differentiation |
Mesenchyme and AER | Digit formation and apoptosis | Hunter-Thompson and Grebe type chondrodysplasias |
| WNT | Wnt-7a | Dorsalising gene. Turns on LMX-1. Repressed by engrailed-1. Promotes Shh expression | AER | ||
| Fringe | Radical fringe | Limits the AER to the tip of the limb bud | Dorsal AER | ||
| Homeobox (HOXA, B, C, D) | Hox-b8 | Controls limb positioning and patterning in developing zeugopod, autopod and stylopod | Overlapped pattern in lateral plate mesoderm and limb bud | Hand-foot-genital syndrome Synpolydactyly |
|
| T-box | Tbx4 Tbx5 |
Transcription factors, turned on by BMP2, BMP4 or Wnt, control limb identity | Early stages of limb outgrowth | Mis-expression changes limb morphology from forelimb to hindlimb or vice versa | Holt-Oram syndrome |
| SOX | SOX9 | Condensation and differentiation of chondroblasts | Early skeleton | Campomelic dysplasia |
Mis-expression experiments in chicks have provided further evidence for the role of these genes. When ectopically expressed in the hind limb, Tbx5 can alter the morphology of the developing limb to a more wing-like structure. Tbx4 and Pitx1 can change forelimb morphology into hindlimb morphology if mis-expresssed in the developing forelimb. T-box genes may alter the morphology of the limb by selectively inducing or repressing genes or markers that are specific to either the forelimb of hindlimb. For instance ectopically expressed Tbx5 can induce expression of the forelimb marker Hoxd9 and repress the hindlimb marker Hoxc9 [1, 2]. The important role these trancription factors plays is highlighted in experiments where Tbx5 is knocked out or inactivated. In these studies the embryos that develop do so with complete failure of formation of any elements of the forelimb. These studies also suggest that Tbx5 interacts with Fgf and Wnt to initiate outgrowth of the limb bud [3, 4].
T-box genes determine limb identity
Tbx5 and Tbx4 activate fibroblast growth factor-10 (Fgf10) in the forelimb and hindlimb, respectively, and it is noted that Tbx5 binding sites have been identified in the Fgf10 promoter sequence in mice and humans (Fig. 1). Fgf10 signals the ectoderm to induce Fgf8, which is instrumental in the formation of the apical ectodermal ridge (AER)—a thickened area of pseudo-stratified columnar epithelium at the tip of the developing limb bud. A positive feedback loop, regulated by the Wnt signalling pathway, is then established between Fgf8 and Fgf10, such that Fgf10 promotes Fgf8 expression and Fgf8 promotes Fgf10 expression [3]. If Fgf10 is knocked out in mice the resultant embryos develop without limbs, indicating the importance of this fibroblast growth factor [5].
Fig. 1.
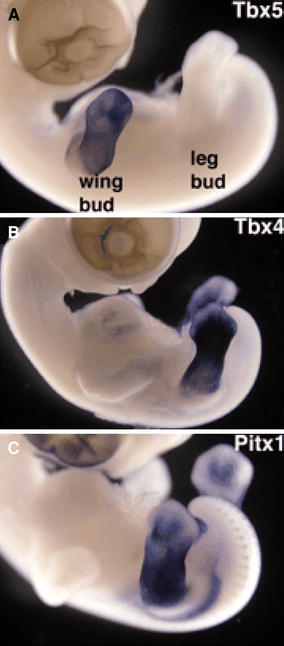
The role of T-box genes in limb identity: chick embryo study demonstrating the distribution of expression of Tbx5 (upper limb), Tbx4 (lower limb) and Pitx1 (lower limb) reproduced with permission of the Company of Biologists [34]
Holt-Oram syndrome, an autosomal-dominant condition affecting approximately 1 in 100,000 individuals characterised by forelimb abnormalities (absent radius, tri-phalangeal thumb, etc.) and cardiac defects (atrial-septal defect) is attributable to Tbx5 mutation in humans [6, 7] (Fig. 2). The majority of cases of Holt-Oram syndrome result from new mutations in the Tbx5 gene.
Fig. 2.

Tri-phalangeal “indicised” thumb typical of Holt–Oram syndrome (due to Tbx5 mutation)
The other gene that has an important role in establishing limb identity is Pitx1 (Fig. 1). It promotes the formation of hindlimb characteristics that can change forelimb characteristics to hindlimb characteristics if it is expressed ectopically in the developing forelimb. Pitx1 is not essential for hindlimb development, and if knocked out, the hindlimb will develop, but with a morphology less typical of a hindlimb and more like a forelimb. Pitx1 may contribute to expression of Tbx4 in the hindlimb, but it is not essential for Tbx4 induction [8].
The apical ectodermal ridge and control of proximo-distal patterning
The apical ectodermal ridge is the driving centre for limb bud development. If the AER in developing chick embryos is removed, the developing limb will be truncated, and the severity of the truncation is related to the gestational age at which the AER is removed [9].
Initially the AER is positioned and induced by Fgf8 and Fgf10; this is followed by expression of Fgf4 at the dorsal end of the limb bud AER. Eventually Fgf4 is expressed across the whole of the AER. This implies that the Fgf family has a role in temporo-spatial positioning within the developing limb [10]. The expression of Fgf4 and Fgf8 stimulates and maintains the rapid growth of the progress zone (PZ) and prevents the local mesenchymal cells from differentiating into chondrocytes [11]. Tissue proximal to the progress zone, being no longer influenced by the AER, becomes influenced by BMPs causing condensation and differentiation of the mesenchymal cells into groups of chondrocytes.
Radical fringe (R-fng), a homologue of drosphilia fringe, is expressed in the dorsal half of the limb and restricts the AER to the distal tip of the developing limb. Radical fringe causes expression of Ser2 (equivalent to drosphilia serrate), and Ser2 defines the border of the AER [12]. Engrailed-1 suppresses the expression of R-fng and therefore Ser2 and assists the formation of the AER.
As well as controlling the position of the limb fields as discussed above, the Hox gene family is instrumental in proximo-distal patterning and the formation of the skeletal precursors of the embryological skeleton. The temporo-spatial expression of different Hox genes in the limb buds of developing mice and gene targeting experiments have provided valuable information on the importance of Hox genes in regulating the types and shapes of the bones in the developing limb. Hox gene expression occurs in three phases in three places in the limb, corresponding with the development of the stylopod (humerus), the zeugopod (radius and ulna) and the autopod (carpus and hand) (Fig. 3a–c).
Fig. 3.
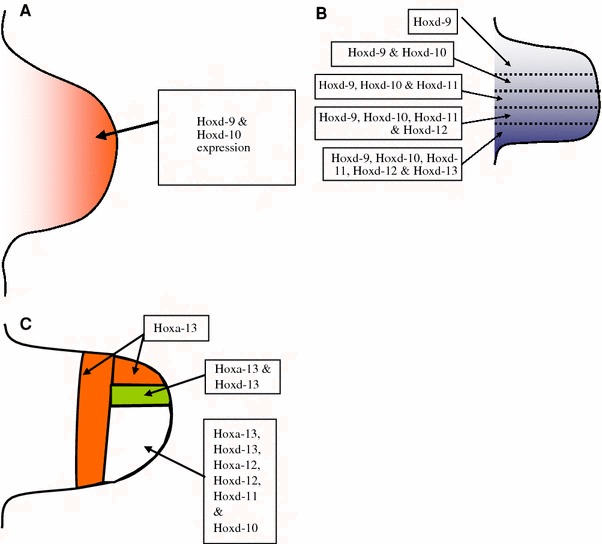
a Stylopod stage: expression patterns of Hox genes during formation of the humerus. b Zeugopod stage: overlapping expression of Hox genes pattern the developing radius and ulna. c Autopod stage: patterned Hox gene expression governs the structural identity of the developing hand
Genes from the Hox-A and Hox-D clusters control patterning and hence morphology of the developing limb in the human embryo. Individual Hox genes overlap to varying degrees, and this creates areas within the embryonic limb that are molecularly distinct from other areas. These areas will go on to develop different morphological profiles. Individual or groups of different Hox genes will turn on promoters in different genetic cascades, eventually instructing the developing mesenchymal cells to differentiate down specific developmental pathways. Hoxd9 is expressed across the whole of the limb during the zeugopod stage, but Hoxd13 is only expressed in the posterior part of the limb at that stage. Hoxd10, Hoxd 11 and Hoxd12 are expressed in an overlapping pattern between Hoxd9 and Hoxd13. This differential expression is responsible for the morphological differences between the radius and ulna in the forelimb and the tibia and fibula in the hindlimb [13].
Errors in human Hox gene expression can result in limb malformations. Synpolydactyly is a dominantly inherited condition with errors in expression of HOXD13. Affected individuals usually have middle/ring finger and 4th/5th toe syndactyly and duplication of a digit in the syndactylous web (Fig. 4). Hand-foot-genital syndrome results from mutations in HOXA13 and is characterised by genital abnormalities (hypospadias in males) and bilateral thumb and great toe hypoplasia. Other abnormalities of the digits are also seen including clinodactyly of the little fingers, hallux varus and small great toenails [14].
Fig. 4.
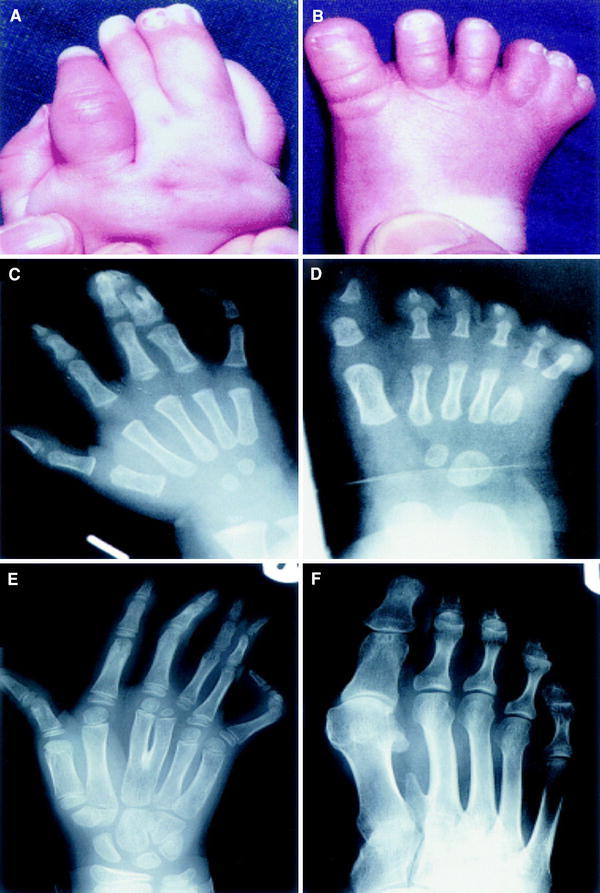
Synpolydactyly: examples of limb abnormalities caused by mutations in HOXD13
The zone of polarising activity and patterning of the antero-posterior axis
Patterning of the antero-posterior axis is controlled by the Zone of Polarising Activity (ZPA), a collection of cells at the posterior border of the limb close to the AER, adjacent to the body wall. The ZPA is initiated by the actions of Hoxb-8 and retinoic acid on the posterior mesoderm [15, 16]. Retinoic acid secreted by the cells of the ZPA upregulates Sonic Hedgehog (SHH) expression, and this controls the development of the antero-posterior axis [17]. As the AER grows distally, it pulls the ZPA with it (Fig. 5).
Fig. 5.
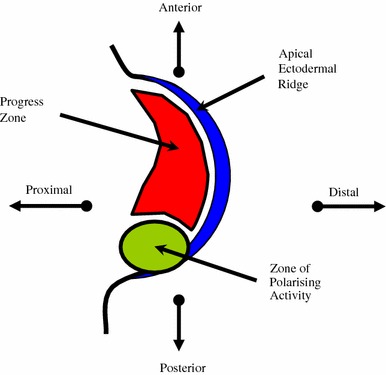
The developing limb bud showing relative positions of the zone of polarising activity, the apical ectodermal ridge and the progress zone
The influence of the ZPA on patterning in the antero-posterior axis has been demonstrated in elegant studies where an extra ZPA is transplanted to the anterior surface of chick limb buds, resulting in mirror-image duplication of the digits [18, 19].
A complicated set of positive feedback loops exists, in which Shh stimulates Fgfs in the AER, and Fgfs in AER activate Shh in the ZPA, thus influencing development in more than more than one axis. WNT7a maintains the Shh signal once it has been initiated [13, 20].
Development of the dorso-ventral axis
LMX1 is a transcription factor containing homeodomain, and when it is induced by WNT7a specifies cells to be dorsal. BMPs in the ventral ectoderm induce expression of Engrailed1, a transcription factor that represses Wnt7a expression [21]. It can therefore be deduced that the regulated expression or suppression of WNT7a controls patterning in the dorso-ventral axis. WNT7a also promotes SHH expression in the ZPA and therefore also influences antero-posterior patterning [22].
Progression of skeleto-genesis and the role of bone morphogenic proteins
As discussed above, HOX genes regulate the formation of the bones of the developing limb. SHH, FGF and WNT products all promote HOX gene expression. HOX-A and HOX-D clusters in particular control the formation of the stylopod, zeugopod and autopod. Alterations in the expression of their gene family members along with co-ordinated input from other transcription factors (e.g. T-Box) accounts for differences in the morphology of the upper and lower limbs.
Bone morphogenic proteins (BMPs) are part of the transforming growth factor beta super-family and act via cell surface bone morphogenic proteins receptors (BMPRs) to induce the formation of bone and cartilage. To date 16 BMPs have been discovered. BMP2, BMP4 and BMP7 are evident in the developing mesoderm and the AER and are the only BMPS that appear to have important roles in skeletal development. The bone morphogenic protein family also contains cartilage-derived morphogenetic proteins (CDMPs) and growth and differentiation factors (GDFs) reflecting specificities in their roles.
BMPs are expressed in response to the Shh signal pathway. These in turn trigger signal transduction via the cell surface BMPRs and induce the expression of the SMAD family of protein trancription factors. Noggin and Chordin are inhibitors of BMPs, and the fine balance between BMPs and Noggin/Chordin regulates the polarity of the developing embryo.
BMP2 (gene locus 20p 12) acts as a retinoid mediator, playing a key role in osteoblast differentiation and induction of bone formation. BMP4 (gene locus 14q22–q23) regulates the formation of limbs from the mesoderm and BMP7 (gene locus location: 20q13) is important in osteoblast differentiation and the induction of SMAD1 expression.
BMP2 and BMP7, under the influence of Shh, play a crucial role in digit identity and formation. Mice that have BMP-2 and BMP-7 chemically blocked or “knocked-out” develop extra digits, and BMPs are known to influence apoptosis of inter-digital tissue in chicks [23, 24]. It is also noted that mice lacking the BMP family member GDF-5 have severely shortened limbs and loss of phalangeal elements [25]. Mutations in BMPs and their inhibitors are associated with a number of human disorders that affect the skeleton, including Hunter-Thompson and Grebe types of chondrodysplasias [26].
Co-ordinated temporal and spatial control of progressive skeletal development is controlled by SOX9 and CBFA1 (runx-2), which is in turn regulated by cellular retinoid signalling [27]. SOX9 initiates the condensation and differentiation of chondroblasts in the embryonic limb. Campomelic dysplasia, a rare, often lethal, genetic condition characterized by short limbs, bowed legs, distinctive facial features, narrow chest and abnormal development of the sex organs in males, is caused by SOX9 mutations in humans. Cartilage fails to develop in limbs where SOX9 is inactivated [28, 29].
CBFA1, a member of the Runt transcription factor family, regulates chondrocyte maturation and osteoblast differentiation. Multiple bone-specific genes, including those for osteocalcin, osteopontin and collagen type 1, are induced by CBFA1 [27]. Cleidocranial dysostosis, a hereditary condition characterised by abnormal clavicles, delayed fusion of the bones in the skull, extra teeth, short stature and other skeletal changes, has been attributed to mutations in the CBFA1 gene in humans [30].
The role of retinoids and retinoic acid
Retinoic acid (vitamin A derivative) levels need to be carefully controlled during limb bud development to stimulate the correct growth of the limb along its three axes. Both high and low levels of retinoic acid have been implicated in a wide range of developmental abnormalities [31, 32], and control of concentrations of RA is finely controlled in vivo. Retinoic acid signalling is implicated in chrondroblast differentiation and chondrocyte hypertrophy [27].
It is known that retinoic acid upregulates the Hox genes in the limb fields and is instrumental in initiating limb bud development. Tadpoles having had their tails amputated and the stumps treated with retinoic acid develop legs instead of a tails at the amputation site [33]. Retinoic acid stimulates SHH up-regulation and influences the creation of the ZPA. It also controls the condensation and differentiation of chondroblasts and co-ordinates chondrocyte maturation, osteoblast differentiation and bone formation.
Principle control of RA levels is by metabolising enzymes; retinaldehyde dehydrogenase (RALDH) mediates the synthesis of retinoic acid, and cytochrome P450 (CYP26) breaks down RA. These enzymes are expressed in different amounts at different times and in different areas of the developing limb bud according to upstream genetic signals. The levels of available retinol or retinoic acid may be further regulated by cytoplasmic retinol/retinoic acid binding proteins.
A large family of retinoic acid receptors (RARs) control developmental gene transcription (i.e. homeobox) and can either enhance or repress the transcription process. The subfamilies and isoforms of the RARs are expressed in different parts of the developing limb, at differing times, and therefore dynamically control the wide range of nuclear effects of RA. This is discussed in greater detail in Weston’s review on the role of retinoid signalling in skeletal development [27].
Co-ordination of the three axes of development and conclusion
It can be seen that vast arrays of temporally and spatially co-ordinated genetic signals interact to control limb development in its three axes. Errors in the molecular control of this complicated pathway, seen both experimentally and clinically, have dramatic and devastating effects on limb development. Continuing research using mice, chicks and drosphilia will provide further information on the regulation of limb bud development in the human embryo, and this may lead to therapeutic interventions in the future.
Contributor Information
Guy Barham, Phone: +44-1202-295934, FAX: +44-1202-295934, Email: guybarham1@aol.com.
Nicholas M. P. Clarke, Phone: +44-2380-796140, FAX: +44-2380-796141, Email: ortho@soton.ac.uk
References
- 1.Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398:814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi JK, Koshiba-Takeuchi K, Matsumoto K, Vogel-Hopker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398:810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb filed in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- 4.Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- 5.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt–Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 7.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 8.Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 9.Summerbell D. A quantitative ananlysis of effect of excision of the AER from the chick limb bud. J Embryol Exp Morphol. 1974;32:651–660. [PubMed] [Google Scholar]
- 10.Tickle C. Patterning systems—from one end of the limb to the other. Dev Cell. 2003;4:449–458. doi: 10.1016/S1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 11.Vogel A, Rodriguez C, Izpisua-Belmonte JC. Involvement of FGF-8 in initiation, outgrowth and patterning of the vertebrate limb. Development. 1996;122(6):1737–1750. doi: 10.1242/dev.122.6.1737. [DOI] [PubMed] [Google Scholar]
- 12.Laufer E, Dahn R, Orozco OE, Yeo CY, Pisenti J, Henrique D, Abbott UK, Fallon JF, Tabin C. Expression of Radical fringe in limb bud ectoderm regulates apical ectodermal ridge formation. Nature. 1997;386:366–373. doi: 10.1038/386366a0. [DOI] [PubMed] [Google Scholar]
- 13.Sadler T (1998) Embryology, gene regulation of limb development. In: Herring JA, Birch JG (1998) The child with a limb deficiency, The American Academy of Orthopaedic Surgeons
- 14.Goodman FR. Limb malformations and the human HOX genes. Am J Med Genet. 2002;112:256–265. doi: 10.1002/ajmg.10776. [DOI] [PubMed] [Google Scholar]
- 15.Charite J, de Graaff W, Shen S, Deschamps J. Ectopic expression of Hoxb-8 causes duplication of the ZPA in the forelimb and homeotic transformation of axial structures. Cell. 1994;78:589–601. doi: 10.1016/0092-8674(94)90524-X. [DOI] [PubMed] [Google Scholar]
- 16.Scadding SR. Citral, an inhibitor of retinoic acid synthesis, modifies pattern formation during limb regeneration in the axolotl Ambystoma mexicanum. Can J Zool. 1999;77:1835–1837. doi: 10.1139/z99-147. [DOI] [Google Scholar]
- 17.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–16. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 18.Summerbell D. The zone of polarizing activity: evidence for a role in normal chick limb morphogenesis. J Embryol Exp Morphol. 1979;50:217–233. [PubMed] [Google Scholar]
- 19.Tickle C, Summerbell D, Wolpert L. Positional signalling and specification of digits in chick limb morphogenesis. Nature. 1975;254:199–202. doi: 10.1038/254199a0. [DOI] [PubMed] [Google Scholar]
- 20.Zuniga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 21.Dealy CN, Roth A, Ferrari D, Brown AM, Kosher RA. Wnt-5a and Wnt-7a are expressed in the developing chick limb bud in a manner suggesting roles in pattern formation along the proximodistal and dorsoventral axes. Mech Dev. 1993;43:175–186. doi: 10.1016/0925-4773(93)90034-U. [DOI] [PubMed] [Google Scholar]
- 22.Tickle C. Patterning systems—from one end of the limb to the other. Dev Cell. 2003;4:449–458. doi: 10.1016/S1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 23.Jena N, Martin-Seisdedos C, McCue P, Croce CM. BMP7 null mutation in mice: developmental defects in skeleton, kidney, and eye. Exp Cell Res. 1997;230:28–37. doi: 10.1006/excr.1996.3411. [DOI] [PubMed] [Google Scholar]
- 24.Zou H, Niswander L. Requirement for BMP signaling in interdigital apoptosis and scale formation. Science. 1996;272:738–741. doi: 10.1126/science.272.5262.738. [DOI] [PubMed] [Google Scholar]
- 25.Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 26.Faiyaz-Ul-Haque M, Ahmad W, Wahab A, Haque S, Azim AC, Zaidi SH, Teebi AS, Ahmad M, Cohn DH, Siddique T, Tsui LC. Frameshift mutation in the cartilagederived morphogenetic protein 1 (CDMP1) gene and severe acromesomelic chondrodysplasia resembling Grebe-type chondrodysplasia. Am J Med Genet. 2002;111:31–37. doi: 10.1002/ajmg.10501. [DOI] [PubMed] [Google Scholar]
- 27.Weston AD, Hoffman LM, Underhill TM. Revisiting the role of retinoid signaling in skeletal development. Birth Defects Res C Embryo Today Rev. 2003;69:156–173. doi: 10.1002/bdrc.10010. [DOI] [PubMed] [Google Scholar]
- 28.Foster JW. Mutations in SOX9 cause both autosomal sex reversal and campomelic dysplasia. Acta Paediatr Japn. 1996;38(4):405–411. doi: 10.1111/j.1442-200X.1996.tb03515.x. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundlos S. Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet. 1999;36(3):177–182. [PMC free article] [PubMed] [Google Scholar]
- 31.Kochhar DM. Limb development in mouse embryos. I. Analysis of teratogenic effects of retinoic acid. Teratology. 1973;7(3):289–298. doi: 10.1002/tera.1420070310. [DOI] [PubMed] [Google Scholar]
- 32.Rizzo R, Lammer EJ, Parano E, Pavone L, Argyle JC. Limb reduction defects in humans associated with prenatal isotretinoin exposure. Teratology. 1991;44(6):599–604. doi: 10.1002/tera.1420440602. [DOI] [PubMed] [Google Scholar]
- 33.Mohanty-Hejmadi P, Dutta SK, Mahapatra P. Limbs generated at site of tail amputation in marbled balloon frog after vitamin A treatment. Nature. 1992;355(6358):352–353. doi: 10.1038/355352a0. [DOI] [PubMed] [Google Scholar]
- 34.Logan M. Finger or toe: the molecular basis of limb identity. Development. 2003;130(26):6401–6410. doi: 10.1242/dev.00956. [DOI] [PubMed] [Google Scholar]


