Abstract
The antimicrobial peptide Attacin is an immune effector molecule that can inhibit the growth of gram-negative bacteria. In Glossina morsitans morsitans, which serves as the sole vectors of African trypanosomes, Attacins also play a role in trypanosome resistance, and in maintaining parasite numbers at homeostatic levels in infected individuals. We characterized the attacin encoding loci from a Bacterial Artificial Chromosome (BAC) library. The attacin genes are organized into three clusters. Cluster 1 contains two attacin (attA) genes located in head-to-head orientation, cluster 2 contains two closely related genes (attA and attB) located in a similar transcriptional orientation, and cluster 3 contains a single attacin gene (attD). Coding and transcription regulatory sequences of attA and attB are nearly identical, but differ significantly from attD. Putative AttA and AttB have signal peptide sequences, but lack the pro domain typically present in insect Attacins. Putative AttD lacks both domains. Analysis of attacin cDNA sequences shows polymorphisms that could arise either from allelic variations or from the presence of additional attacin genomic loci. Real time-PCR analysis reveals that attA and attB expression is induced in the fat body of flies per os challenged with Escherichia coli and parasitized with trypanosomes. In the midgut, expression of these attacins is similarly induced following microbial challenge, but reduced in response to parasite infections. Transcription of AttD is significantly less relative to the other two genes, and is preferentially induced in the fat body of parasitized flies. These results indicate that the different attacin genes may be differentially regulated.
Keywords: attacin loci, attacin expression, Glossina morsitans morsitans, trypanosome
Introduction
Attacins are glycine-rich immune proteins first reported in the moth Hyalophora cecropia in response to bacterial infection (Hultmark et al., 1983). They are thought to increase the permeability of the outer membrane of gram-negative bacteria by interacting with lipopolysaccharides (LPS) and inhibit the synthesis of outer membrane protein at the transcriptional level (Carlsson et al., 1998). The increased permeability facilitates other molecules such as lysozyme to access the inner membrane, setting up a synergistic action for the antibacterial peptides (Engstrom et al., 1984). Attacins have now been described from several lepidopteron and dipteran species. They are synthesized as pre/pro-mature peptides with the mature peptide typically being about 190 amino acids in length and forming a ‘random coil’ structure in solution (Gunne et al., 1990). This loose, flexible structure allows for ample amino acid substitutions, and has resulted in low amino acid identity among the Attacin homologs characterized from distant taxa. In the Drosophila genome, a family of four genes encodes Attacins, attA, B, C and D. AttC is similar to attA and attB, while attD is more divergent (Hedengren et al., 2000). The homolog of Drosophila attA has previously been described from the tsetse fly Glossina morsitans morsitans. Although the putative GmmAttA has a 5′-end located signal peptide domain (pre), similar to the Drosophila homolog, it lacks the pro-domain region typically associated with Attacins (Hao et al., 2001).
Previous studies in insects have shown that there are at least two major signal transduction pathways, Imd and Toll, which regulate the induced expression of the antimicrobial peptide (AMP) genes (Lemaitre et al., 1996; Georgel et al., 2001; Choe et al., 2002). Additional downstream pathways in Imd, such as JNK and IKK/Relish, can further modulate the induction kinetics of the effectors (Park et al., 2004; Kleino et al., 2005; Kim et al., 2005; Delaney et al., 2006). Several transcription factors have been implicated in AMP activation, specificity and termination, including the Dorsal, Dif and Relish and the Activator Protein 1 (AP1), which function in DNA binding, dimerization and nuclear localization (Senger et al., 2004; Kim et al., 2005). Correspondingly, the AMP promoter regions contain transcription factor binding motifs, such as AP1, NF-κB and GATA (Meister et al., 1994; Dushay et al., 2000; Zheng & Zheng, 2002; Cheng et al., 2006). The synergistic actions of the multiple transcription factors contribute to the specificity and the tight on/off regulation of insect immune responses.
In tsetse, vectors of African trypanosomes, Attacin has been implicated to play a role in immune defense against trypanosomes in addition to its antibacterial activity. Tsetse present multiple physical and biochemical barriers that mediate a high level of resistance to infection with trypanosomes. During the course of natural infections, the mammalian parasites (bloodstream form cells, BSF) acquired in an infected blood meal differentiate to procyclic form cells (PFC) in the midgut within 24-36 hours. A mechanism of resistance however eliminates infections in the majority of flies around 72 h after acquisition. In flies where parasites survive, parasites reach a high enough density to be microscopically detected with confidence in the midgut by days 6-10 post infection. After about a month, only a proportion of flies with gut infections give rise to mammalian infective metacyclic parasites in salivary glands. Hence, there are barriers to both midgut infections and then subsequently to parasite maturation in the salivary glands. It has been shown that AMP expression including attacins can contribute to tsetse’s resistance (Hao et al., 2001; Hu & Aksoy, 2006). While attacin expression is found to be induced within hours following provisioning Escherichia coli in an infected bloodmeal (per os challenge), it can only be detected after BSF parasites differentiate to PFC cells in the midgut several days post per os acquisition (Hao et al., 2001). Flies with gut parasite infections (infected), have high levels of attacin expression. In parasite resistant flies, the expression level of attacin was similar to that of the uninfected controls when analyzed 1 month post parasite acquisition (Hao et al., 2001). Tsetse attacins have been found to be regulated by the Imd pathway as expression can be abolished when the transcriptional activator relish is silenced by a double stranded RNA interference (dsRNAi) approach (Hu & Aksoy, 2006). Flies in which relish and attacin expression have been silenced exhibit significantly higher gut trypanosome infection prevalences and parasite intensities (Hu & Aksoy, 2006). Finally, the recombinant Attacin (recGmmAttA1) protein expressed in Drosophila S2 cells has been shown to have trypanocidal activity both in vitro and in vivo (Hu & Aksoy, 2005).
Here, we report on the genomic organization of the attacin gene family from G. m. morsitans. We describe the genomic encoding sequences for the multiple attacin transcripts, the transcriptional motifs associated with the upstream regulatory control regions and report on the tissue and pathogen specific nature of attacin expression profile.
Results
Tsetse BAC library characterization
To enable genomic studies, a large-insert BAC library was constructed from G. m. morsitans DNA (designated VMRC-29). Earlier attempts to construct the library from DNA extracted from tsetse that received normal bloodmeals led to high levels of contaminating clones, corresponding to tsetse’s symbiotic flora, in particular to Wolbachia. To reduce the density of the contaminating bacterial symbiont flora while maintaining host fertility, flies were given antibiotic tetracycline containing blood meals supplemented with a vitamin cocktail as described in Experimental procedures, below. The larva deposited by females were collected and DNA was prepared from newly emerged adults and used for the library construction. A number of different sources of fly material were tested for DNA preparation including whole flies, dissected tissues, larvae and pupae, and the best results were obtained from flash-frozen newly emerged whole adult flies. Fifty randomly selected BAC clones were end-sequenced and Blast analysis of the DNA sequences did not show any bacterial or other putative contaminating sequences (data not shown). DNA from 125 randomly selected BACs was digested with NotI restriction enzyme and analyzed by pulsed field gel electrophoresis. Results indicated that the median insert size was around 152 kb and the average insert size was 138 kb. This discrepancy may be due to a small fraction of smaller insertcontaining clones in the library, i.e. clones < 100 kb. The library containing 51 979 clones was picked into 384-well microtiter dishes and gridded onto nylon high-density colony filters for screening by hybridization. The library represents 13× coverage of the tsetse genome based on the approximately 613 MB haploid genome size of G. m. morsitans females (Aksoy et al., 2005).
Identification of attacin coding genomic loci
A PCR-amplified product corresponding to the previously described gmmattA1 cDNA was hybridized to the BAC library filters and one clone (39G22) was identified and confirmed to carry the attacin locus by PCR amplification with gene specific primers as well as by Southern hybridization analysis (data not shown). The sequence of the 154 071 kb BAC insert DNA was obtained and search of the known public databases indicated that it contained three clusters with identity to the previously characterized tsetse attacin cDNAs. Clusters 1 and 2 comprise bases 35 775 to 39 249 and 60 665 to 64 211, respectively, while cluster 3 corresponds to bases 71 918 to 72 559 (Fig. 1A). Cluster 1 carries two identical genes gmmatta11 and gmmatta12 (denoted attA11 and attA12) located in opposite transcriptional orientation with divergent promoters separated by a 2095 bp region. Cluster 2 also has two attacin genes, gmmatta21 and gmmattb (denoted attA21 and attB), which are similarly arranged in head-to-head orientation as in Cluster 1 but separated by a region of 2173 bp. Only one attacin gene, gmmattd (denoted attD) is present in the Cluster 3 segment.
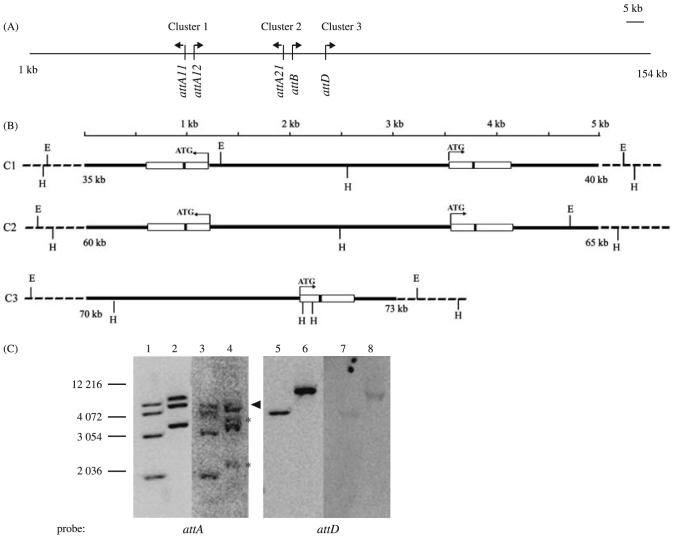
Figure 1.
Attacin gene organization on BAC 39G22. (A) The chromosomal organization of the three attacin clusters found on BAC39G22. Cluster 1 (C1) spans from 35 775 bp to 39 249 bp, cluster 2 (C2) from 60 665 to 64 211 bp. Cluster 3 (C3) spans coding sequence from 71 918 to 72 559 bp and includes upstream putative promoter region. The transcription units and direction of transcription of the three attacin clusters are indicated by arrows and named by attA11, attA12, attA21, attB and attD. (B) Detailed maps of restriction sites and transcription units on three clusters. The ORFs and the introns are depicted by boxes and black bars, respectively. The EcoRI recognition sites (E) are shown by vertical bars above and HindIII (H) sites are shown by vertical bars below. (C) Southern blot analysis of attacin gene organization in BAC39G22 and G. m morsitans genome. BAC39G22 DNA digested with HindIII (lane 1, 5) and EcoRI (lane 2, 6) and the genomic DNA by HindIII (lane 3, 7) and EcoRI (lane 4, 8), respectively. The blot was first hybridized to attA probe, stripped and rehybridized to attD. The stars denote the two additional bands detected with attA and the arrow indicates the band with less hybridization intensity (lane 4). The DNA size markers (bp) are shown.
Based on the known BAC39G22 sequence information, we expected four Hind III fragments of 1.7 kb, 2.8 kb, 3.9 kb and 4.8 kb and three EcoRI fragments of 3.1 kb, 4.6 kb and 5.6 kb that should hybridize to attA (Fig. 1B). Southern data confirmed the presence of these expected-sized DNA fragments in the BAC insert (Fig. 1C, lane 1 and 2, respectively). The BAC clone sequence also indicated the presence of one EcoRI fragment of 6.6 kb in size and three HindIII fragments of 3.8 kb, 1.9 kb and 122 bp in size with homology to attD (Fig. 1B). The presence of the 6.6 kb EcoRI fragment was confirmed (Fig. 1C, lane 6). Because there are only 47 base pairs homologous to attD within the 1.9 kb HindIII fragment and the 122 bp fragment is too small to be visualized on our gel system, only the 3.8 kb fragment could be detected by Southern hybridization (Fig. 1C, lane 5). We similarly analyzed the organization of attacin coding genes using G. m. morsitans genomic DNA that was digested with the same restriction enzymes, HindIII and EcoRI, respectively. With an attA cDNA probe, four fragments of about 1.7 kb, 2.8 kb, 3.9 kb and 4.8 kb could be detected with genomic DNA digested with HindIII enzyme, identical in size to those observed from the BAC insert (Fig. 1C, lane 3). With EcoRI digested genomic DNA, two fragments of approximately 4.6 kb and 3.1 kb in size were detected (Fig. 1C lane 4). The 5.6 kb fragment was detected as a fainter band, and two new fragments of around 3.5 kb and 2.2 kb were present in the EcoRI digested genomic DNA (Fig. 1C lane 4). While these new DNA fragments may arise from a different attacin locus in the genome, they may also represent restriction enzyme site polymorphisms present on one of the chromosomes of the diploid tsetse given the decreased intensity of the 5.6 kb fragment. When attD was used as the hybridization probe, the same 3.8 kb HindIII fragment and 6.6 kb EcoRI fragments were detected in genomic DNA (Fig. 1C lane 7 and 8, respectively). In summary, the three clusters present on BAC39G22 apparently represent the major attacin encoding loci in the tsetse genome.
Comparison of multiple attacin coding sequences
An identical intron sequence of 60 bp interrupts the coding sequence of both attA and attB genes at nucleotide 228. A divergent 77-bp-length intron is also present at the same site within the attD coding sequence. The complete open reading frames (ORFs) encode a putative preprotein of 208 amino acid residues for AttA and AttB, and 187 for AttD, respectively (Fig. 2A). The putative AttA peptide has a N-terminal located signal peptide of 20 amino acids, and G1 and G2 domains associated with the mature coding segment. The AttB peptide is identical to AttA with the exception of two amino acid substitutions, N187 and R195 instead of H187 and Q195, respectively. These amino acid differences result from three nucleotide substitutions from C559, A584 and A585 in AttA to A559, G584 and T585 in AttB, respectively.
Figure 2.
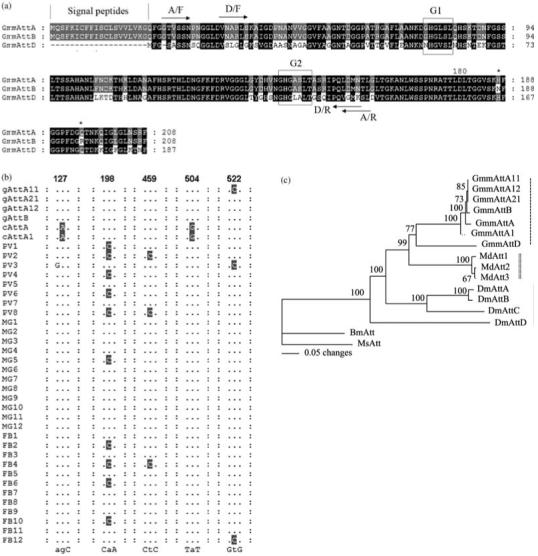
Sequence analysis of tsetse Attacin family. (A) Amino acid alignment. The shared residues between the three putative peptides are shaded in black, the conserved residues among two of them are shaded in grey. The amino acid residues differing between GmmAttA and GmmAttB are represented by star. The two primer pairs used in gene-specific RT-PCR amplification studies are indicated. A/F: GmmAttA forward primer, A/R: GmmAttA reverse primers; D/F: GmmAttD forward primer, D/R: GmmAttD reverse primer. Signal peptide is between two vertical lines. G1 and G2 domains are boxed. (B) The cDNA polymorphisms. Various attacin cDNAs and attacin genomic sequences are aligned over the regions where heterogeneities are observed. The variations are shaded, the consensus residues are indicated below, positions of variations in ORF are numbered above. Previously identified cDNAs: cAttA, AF368909 and cAttA1, AY607104; genomic sequences in this study: gAttA11, gAttA12, gAttA21, and gAttB; PV1-8: cDNAs from proventriculus; MG1-12: cDNAs from midgut; FB1-12: cDNAs from fatbody. (C) Phylogenetic tree showing different insect Attacin homologues. Accession number for the sequences used are: GmmAttA11; GmmAttA12; GmmAttA21; GmmAttB; GmmAttA (AF368909); GmmAttA1(AY607104); GmmAttD; Musca domestica (Md) Att1 (DQ062744), Att2 (AAY59540) and Att3 (AAR23786); Drosophila melanogaster (Dm) AttA (Z46893), AttB (AF220546), AttC (AF322249) and AttD (AF322250); Bombyx mori (Bm) Att (S78369); Manduca sexta (Ms) Att (DQ072728). Distance matrix analysis: bootstrap method with heuristic search. Bootstrap replicates = 1000. The clusters formed by tsetse, housefly and drosophila are marked by vertical dashes, double vertical dashed and vertical line, respectively.
Comparison of the previously reported attacin cDNA sequences (GmmAttA1: AY607104 and GmmAttA: AF368909) with the DNA sequence of attA and attB genes revealed two nucleotide substitutions at positions 127 and 504. To investigate whether allelic variations between the diploid chromosomes could account for the differences, we amplified attacin cDNAs from proventriculus, midgut and fatbody tissues of parasitized flies. A total of 32 clones isolated from five independent PCR reactions were characterized by sequence analysis. Alignment of cDNAs with those of the published cDNA sequences showed that none of them contained the polymorphisms associated with positions 127 and 504. We did however find two additional substitutions in multiple cDNAs that were different from the genomic attacin sequences; C198 instead of A198 was present in 19/32 clones and C459 instead of T459 was present in 3/32 clones (Fig. 2B). In addition, two cDNAs, one from proventriculus and the other from fat body, were similar to the genomic sequence attA11 in that they had a C residue instead of a T in position 522 (Fig. 2B). The polymorphisms observed in the cDNAs result in synonymous amino acid substitutions. It is difficult to rule out the presence of other attacin coding loci in the genome that may have given rise to the cDNA polymorphisms we observed. Given that our Southern mapping analysis indicates a strong conservation between the BAC and genomic DNA, the observed variations may also be attributed to allelic variations.
AttD has 71% and 70% identity to attA and attB nucleotide sequences and 68% and 67% identity to corresponding putative AttA and AttB proteins, respectively (Fig. 2A). To understand the evolutionary relationships among Attacins from tsetse and other related insects, we aligned and compared the putative Attacin protein sequences using parsimony analysis. The phylogenetic reconstruction analysis indicated that the Attacins analyzed from G. m. morsitans, Musca domestica, Drosophila melanogaster, Bombyx mori and Manduca sexta represent species-specific expansions of each gene family and their phylogenetic placement mirrors that of their host relationships (Fig. 2C). All tsetse protein sequences formed a single lineage most similar to those from M. domestica.
Characterization of attacin regulatory sequences
The 2095 bp DNA sequence between the head-to-head organized attacin genes in Cluster 1 is reverse complimentary to Cluster 2 with the exception of an additional 69 bp sequence present 323 bp upstream of attB in Cluster 2. It is likely that Clusters 1 and 2 represent a recent tandem gene duplication event followed by divergent evolution. The intragenic regions of Cluster 1 and 2 and the 2.5 kb sequence upstream of attD were analyzed for putative transcription regulatory sequences including AP1, NF-κB, and GATA, as well as TATA boxes (Fig. 3). Although the two attacin ORFs in each cluster are nearly identical, their upstream regulatory sequences vary appreciably. There are two NF-κB and GATA and one AP1 binding motif upstream of attA12 and attA21, while the upstream sequences associated with attA11 and attB have a single NF-κB and three GATA motifs. Both clusters have one GATA and one AP1 binding signatures located in the middle of the intragenic region as depicted in Fig. 3. The motifs we found in Cluster 3 were two GATAs located around 500 bp upstream of attD ORF.
Figure 3.
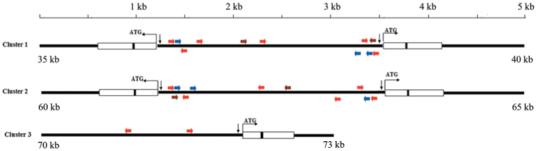
Promoter regions of clusters 1, 2 and 3. Putative transcriptional elements important for the immune activation are shown: NF-κB motifs marked by blue arrows, GATA by red arrows and AP1 by brown ones. TATA boxes are indicated by vertical arrows. The ORFs and the introns are depicted by boxes and black bars, respectively.
Expression profiles of Glossina attA, attB and attD
We used qRT-PCR method to analyze gene expression in midgut and fatbody tissues 24 hours after per os immune challenge with E. coli and trypanosomes as well as from microscopically confirmed parasite infected flies dissected 10 days post infection acquisition (Fig. 4). The expression data were compared to those obtained from the same aged control flies receiving normal blood meals. The primers we designed for amplification could not differentiate attA from attB due to their high identities, but they could distinguish transcripts corresponding to attD. Relative to the housekeeping gene β-tubulin level, expression of attA/B was found to be significantly higher than attD. The levels of attA/B and attD specific transcripts were more abundant in the fatbody than in the midgut. Following per os E. coli challenge, attA/B expression was induced in the fat body and midgut, while attD showed no significant immune activation (Fig. 4). When analyzed 24 hours after trypanosome feeding, there was no significant induction of attA/B or attD in either midgut or fatbody (Fig. 4). However, flies with microscopically confirmed parasite infections showed significant induction of both attA/B and attD expression in fatbody. Surprisingly, the same parasite infected flies showed a decreased level of attA/B expression in midgut tissue (Fig. 4). The expression data indicate that attA/B expression is responsive to both bacteria challenge and midgut parasite infections, while attD is expressed upon midgut parasite infection establishment.
Figure 4.
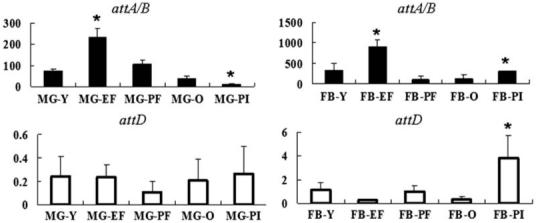
Regulation of Glossina attA/B and attD expression in midgut (MG) and fat body (FB) organs of normal, immune challenged and parasitized flies. Flies were challenged per os with Escherichia coli (EF) or trypanosomes (PF), respectively and compared to a control group of the same age flies (Y). In addition gene expression from flies microscopically confirmed to carry midgut infections (PI) was compared to a group of normal control flies of the same age (O). attA/B expression levels are indicated by black bars while attD is shown by white bars. Standard deviation values observed among the three samples analyzed are shown. Y axis indicates the relative expression values of attA/B or attD normalized by housekeeping gene tubulin (gmmtub) levels. Asterisk (*) indicates significant differences from the corresponding control group P < 0.05. P = 0.022 for attA/B for MG-EF; P = 0.023 for attA/B in MG-PI; P = 0.0063 for attA in FB-EF; P = 0.027 for attA/B in FB-PI and P = 0.039 for attD in FB-PI.
Discussion
The attacin gene family in tsetse is organized in two clusters 30 kb apart. The clusters encode two transcription units arranged in a head to head position within approximately a 5 kb DNA region. A third related locus is also present 5 kb upstream of Cluster 2. Clusters 1 and 2 are almost identical except for a 69 bp DNA insertion in the promoter region of cluster 2, indicative of a recent tandem gene duplication event. Interestingly, we found a mariner transposase gene approximately 200 bp downstream of cluster 2. It is possible that the two clusters originated through duplication of one ancestral cluster mediated by the mariner transposition.
The Drosophila genome data indicate that the AMPs are encoded by gene families; i.e. Drosomycins are encoded by seven genes, Attacins by four genes, Diptericins by two genes and Cecropins by three genes (Khush & Lemaitre, 2000). These findings are in agreement with the multiple transcription units we describe here for Attacins in tsetse. The multiple gene copies may be necessary for the insects to transcribe high levels of AMPs in response to a variety of immune stimuli. Alternatively, given the variations present in the transcription regulatory sequences, different members of the gene families may be activated in different tissues and/or in response to diverse stimuli.
The upstream transcription regulatory sequences and motifs associated with attA/B and attD as well as their protein coding sequences are significantly different. The promoter regions of attA/B have NF-κB, GATA and AP1 binding motifs similar to those identified in the regulatory regions of other AMPs in Drosophila (Senger et al., 2004; Kim et al., 2005; Senger et al., 2006). The NF-κB and GATA binding sites have been shown to be responsible for the intensity and tissue specific profile of expression of many AMPs upon immune challenge since its deletion or mutation results in significant reduction or even abolishment of gene expression (Meister et al., 1994; Senger et al., 2004). It has also been shown that AP1 binding sites near NF-κB can act as a repressor by recruiting the deacetylase complex to terminate the activation of a group of NF-κB target genes (Kim et al., 2005). A comparison between attA/B and Drosophila AMP control regions shows that overall there is conservation in both the orientation of the motifs and their approximate distances from the putative transcription start sites (Senger et al., 2004). In contrast, attD lacks the essential regulatory motives required for activation by the Rel protein except for GATA motifs. It is possible that in tsetse attD expression is controlled by a Rel-independent pathway. The amino acid sequences of tsetse AttA and AttB are nearly identical, while AttD is only 69% identical to the AttA/B form. In comparison to the Drosophila Attacins, all three tsetse peptides lack the propeptide domain that is involved in the secretion process in Drosophila (Gunne & Steiner, 1993). In addition AttD in tsetse lacks the 5′-end located signal peptide, similar to that reported for AttD in Drosophila. It is unknown how tsetse Attacins can undergo post translational modification during transport to the hemolymph.
The second difference between the attA/B and attD genes concerns their transcript levels with attA/B specific transcripts being hundreds of times more abundant than attD. Furthermore, expression of attA/B is induced in response to both gram negative bacteria and trypanosomes as reported before (Hao et al., 2001; Hu & Aksoy, 2006), while attD is only responsive in the fat body of flies with midgut parasite infections. It is interesting that attA/B expression in the parasitized midgut was significantly less than in the normal gut. Trypanosome infections in tsetse remain in the midgut without crossing into the hemoceol. It is possible that down regulation of host gut immune responses may be one mechanism parasites exploit to enhance their survival. It remains to be seen how parasite infections lead to the activation of attD in the fatbody given the absence of NF-κB motifs in its transcriptional control region. It has also been reported that Drosophila attD is controlled differently from its isoforms, attacin A, B and C (Hedengren et al., 2000; De Gregorio et al., 2002). Because the exclusive regulatory motif associated with attD is GATA, it is possible that GATA factors are responsible for the difference in the expression profile we observe. There are a total of five GATA factors in Drosophila: Pannier (dGATAa), Serpent (dGATAb), Grain (dGATAc), dGATAd and dGATAe. Serpent is the principal GATA factor expressed in the fat body, and it is evident that Rel-Serpent synergy is essential for a robust systemic immune response. dGATAe and Grain are expressed strongly in the gut (Senger et al., 2006). Mutating the three consensus GATA sites within Metchnikowin has abolished its expression in every tissue, while mutations in the three consensus Rel sites disrupted only its inducible expression in the anterior midgut, suggesting metchnikowin expression is regulated in a Rel factor dependent as well as independent way (Senger et al., 2006). We also found cDNAs corresponding to dGATAe and serpent factors in tsetse (Attardo et al., 2006). The phenomenon that attD can be activated by trypanosome in spite of lacking NF-κB motif or signal peptide also raises the possibility that serpent factor may stimulate attD expression in the fatbody via a different pathway.
At present, our results indicate that members of the Attacin family in tsetse are activated differently upon different immune stimuli. Given their trypanocidal activity and demonstrated role in the trypanosome infection establishment process, further research on the role of the different regulatory motifs associated with transcription control regions will be necessary to understand the mechanism of attacin expression and regulation in tsetse.
Experimental procedures
Insects and microorganisms
Newly emerged G. m. morsitans female flies received blood meals containing tetracycline (5 μg/ml) and ampicillin (each 20 μg/ml) in order to reduce the gut symbiotic microflora. Fertile flies were maintained on antibiotic containing blood meal supplemented with vitamins using a modification of the Nogge protocol (Nogge, 1976). The bloodmeal was supplemented with 1× MEM vitamin solution (Life Technologies catalog no. 11120-052, HyClone, Logan, UT), 100 μg/ml of thiazole, lipoic acid and biotin. We collected the first progeny deposited, allowed them to hatch and processed the adults for the BAC library DNA preparation. The reduction of symbiont densities in such progeny was documented by semiquantitative PCR analysis (data not shown). Trypanosoma brucei rhodesiense (strain YTat1.1) bloodstream form parasites were obtained from a rat infection and parasitemic blood was stored as frozen stabilates for fly feeding. Teneral flies received blood meals supplemented with 2 × 106/ml parasites and 1 × 106/ml E. coli DH5α, respectively. 24 hours post microbe challenge, midgut and fat body tissues were dissected from three flies per group. Flies carrying gut trypanosome infections were identified by microscopic dissections 10 days post infection acquisition and the midgut and fat body tissues from infected flies were collected for RNA isolation.
BAC library construction
Detailed information on DNA preparation, size selection, ligation and transformation, quality control and arraying conditions can be obtained on our laboratory web-page (aksoylab.yale.edu). Below we describe these protocols briefly.
DNA preparation
100 frozen flies were added to 20 ml of ice cold Homogenization Buffer (HB, 20 mM HEPES, 10 mM EDTA, 1 mM Spermidine, 0.5% NP40 detergent, pH 7.6) in a pre-chilled dounce homogenizer on ice. The homogenate was filtered through multiple successive cell strainers (100 μm and 70 μm BD Falcon), the cells were pelleted at 11 000 g at 4 °C for 10 min and resuspended in fresh ice cold HB. The quality of the cell suspension was microscopically observed and the solution was equilibrated at 37 °C for 10 min. One ml of the cell solution was added to 1 ml 2% Incert agarose and each well of a plug mold (BioRad, Hercules, CA) was filled with 80 μl suspension. The plugs were treated as described in Amemiya et al. (Amemiya et al., 1996).
Size selection
For size fragmentation, genomic DNA (1/2 plug per reaction) was partially digested at 37 °C for 30 min in 500 μl reaction volume containing five units of EcoRI (New England Biolabs, Ipswich, MA) and 30 units EcoRI methylase. The quantity of EcoRI and EcoRI methylase were optimally determined by titration experiments. DNA fragments were separated on a 1% agarose gel (Pulse-Field certified Bio-Rad) using pulse-field-gel (PFG)-electrophoresis (BioRad CHEF XA Mapper) and the following conditions: 30 hour run time, 10 s. initial time, 60 s final time, 120 reorientation angle, 5 V/cm field strength, 14 °C chamber temperature. Fractions ranging from 50-300 kb were excised from the agarose and DNA electroeluted according to Strong et al. (Strong et al., 1997; Osoegawa et al., 1998).
Ligation and transformation
A CopyControl BAC Cloning Kit (Epicentre Biotechnologies, Madison, WI) was used for cloning of the size-fractionated DNA fragments following methods outlined in Osoegawa et al. (Osoegawa et al., 1998). To remove salts, the ligation reactions were drop dialyzed on a 0.025 μm nitrocellulose filter (Millipore, Billerica, MA) for 1.5 h against ddH2O at 4 °C. The sample volume was then reduced to 20 μl by drop dialyzing against 30% PEG (PEG8000) in 0.5x TE at 4 °C. Using a BTX ECM 630 electroporator, 8 μl ligation mix was electrotransformed into 100 μl DH10B T1 resistant electrocompetent cells (Invitrogen) using 2 mm cuvettes and the following conditions: 2.5 kV field strength, 225 Ohms, 25uF. Transformed cells were allowed to recover in 10 ml SOC for 1 h at 37 °C.
Insert size determination and library arraying
Transformants that contained average insert size of 150 kb were picked into individual wells of 144 384-well micro-titer plates using a colony-picking robot (Norgren Systems, Fairlea, WV). A Total Array System (BioRobotics, Genomic Solutions, Ann Arbor, MI) was used for developing high-density nylon filter sets (22 cm × 22 cm) containing BAC DNA. In addition, the Total Array System was also used for the replication of the tsetse fly library. Each hybridization membrane represents over 18 000 distinct BAC clones stamped in duplicate (designated as VMRC-29, available at http://bacpac.chori.org/library.php?id = 354).
Selection of Gmmatt containing BAC clone
The full length cDNA for gmmattA was described previously (Hao et al., 2001). The gmmattA insert was PCR-amplified, gel-purified, labeled with [α-P32]dCTP using the Random Prime DNA Labeling Kit (Boehringer Mannheim, Indianapolis, IN, Cat no. 1 004 760) and was hybridized to one 22 × 22 cm nylon high-density BAC colony filter. A single hybridization positive BAC clone was obtained and DNA was prepared. The identity of the clone was followed by PCR and subsequent Southern hybridization analysis with attA. The 150 071 kb genomic DNA insert from BAC39G22 was sequenced at the Sanger Institute, UK as well as RIKEN Japan (Accession No. AM940018). For comparative analysis, genomic and cDNA attA sequences were aligned using ClustalX (5). The regulatory elements were searched for conserved motifs by using web-based TFSEARCH program (http://www.cbrc.jp/research/db/TFSEARCH.html), including NF-κB (GGGAA/TTCC/AC), GATA (A/TGATAA/G) and AP1 binding motif (A/CTGAGTT/CAG).
Southern analysis
High-molecular weight genomic and BAC39G22 DNAs were digested with HindIII and EcoRI, respectively and prepared for Southern analysis. For hybridization, PCR amplified 412 bp attA and 445 bp attD cDNA fragments were labeled using PCR DIG Synthesis Kit (Roche, Indianapolis, IN). The hybridized probe was immuno detected with anti-digoxigenin-alkaline phosphatase antibody and visualized using CSPD as substrate (Holtke et al., 1995).
Sequence of cDNA clones
Total RNA isolated from parasite infected midgut, fatbody and proventriculus tissues was transcribed with oligo-dT primer using SuperScriptTM (Invitrogen, SanDiego, CA), respectively. Two independent PCR reactions were carried out for each tissue for 25 cycles of 94 °C for 1 min, 50 °C for 1 min and 72 °C for 1 min in a volume of 25 μl with primer 5′ CGTCGTTCTAGTGAAAGGA 3′ and 5′ AACCCAAACCTATCTGT 3′. The PCR products were cloned into the pGEM-T vector (Promega, Madison, WI). 12 clones from each midgut and fatbody reactions and eight clones from each proventriculus reactions were sequenced.
Real time PCR expression analysis
Total RNA was prepared from midgut and fatbody of normal flies as well as E. coli and trypanosome 24 hours challenged as well as microscopic conformed parasitized flies. cDNAs were prepared by reverse transcription using oligo-dT and SuperScriptTM (Invitrogen). Aliquots of first strand cDNAs were used in PCR with gene specific primers: gmmattA (F/R): 5′ATGCCAACCTCTTCAACGAC 3′, 5′ CGTAACCTAAGCCTCCACCA 3′; gmmattD (F/R): 5′ ACGCCAATCTTTTGAAGACG 3′, 5′ ATCTGAGAACCAGTC 3′, and β-tubulin (F/R): 5′ CCATTCCCACGTCTTCACTT 3′, 5′ GACCATGACGTGGATCACAG 3′. Real time quantitative PCR was performed with an icycler iQ™ real time PCR detection system (BioRad). Normal flies same age as experimental groups were used as controls. All assays were carried out in duplicate with 3 flies for biological replicates for each experiment. T-tests were performed to determine gene expression significance from controls.
Acknowledgements
We would like to thank Mark Simmonds and Karren Mungall for sequencing the 39G22 BAC at the Sanger Institute. This research was supported by grants from the NIH AI51584 to SA. C.H. and J.W. were supported by the Li Foundation award to S.A. The BAC library construction was funded by NIH grant HG02526 (Chris T. Amemiya, BRI). MB is supported by the Wellcome Trust through their funding of the Pathogen Sequencing Unit.
References
- Aksoy S, Berriman M, Hall N, Hattori M, Hide W, Lehane MJ. A case for a Glossina genome project. Trends Parasitol. 2005;21:107–111. doi: 10.1016/j.pt.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Ota T, Litman GW. Construction P1 Artif chromosome (PAC) libraries from lower vertebrates. Academic Press; NY: 1996. pp. 223–256. [Google Scholar]
- Attardo GM, Strickler-dinglasan P, Perkin SA, Caler E, Bonaldo MF, Soares MB, et al. Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans. Insect Mol Biol. 2006;15:411–424. doi: 10.1111/j.1365-2583.2006.00649.x. [DOI] [PubMed] [Google Scholar]
- Carlsson A, Nystrom T, De cock H, Bennich H. Attacin - an insect immune protein - binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology. 1998;144(Pt 8):2179–2188. doi: 10.1099/00221287-144-8-2179. [DOI] [PubMed] [Google Scholar]
- Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, et al. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87:356–365. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- De gregoria E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Stoven S, Uvell H, Anderson KV, Engstrom Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. Embo J. 2006;25:3068–3077. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushay MS, Roethele JB, Chaverri JM, Dulek DE, Syed SK, Kitami T, et al. Two attacin antibacterial genes of Drosophila melanogaster. Gene. 2000;246:49–57. doi: 10.1016/s0378-1119(00)00041-x. [DOI] [PubMed] [Google Scholar]
- Engstrom P, Carlsson A, Engstrom A, Tao ZJ, Bennich H. The antibacterial effect of attacins from the silk moth Hyalophora cecropia is directed against the outer membrane of Escherichia coli. Embo J. 1984;3:3347–3351. doi: 10.1002/j.1460-2075.1984.tb02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Gunne H, Steiner H. Efficient secretion of attacin from insect fat-body cells requires proper processing of the prosequence. Eur J Biochem. 1993;214:287–293. doi: 10.1111/j.1432-1033.1993.tb17923.x. [DOI] [PubMed] [Google Scholar]
- Gunne H, Hellers M, Steiner H. Structure of pre-proattacin and its processing in insect cells infected with a recombinant baculovirus. Eur J Biochem. 1990;187:699–703. doi: 10.1111/j.1432-1033.1990.tb15356.x. [DOI] [PubMed] [Google Scholar]
- Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci USA. 2001;98:12648–12653. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedengren M, Borge K, Hultmark D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem Biophys Res Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- Holtke HJ, Ankenbauer W, Muhlegger K, Rein R, Sagner G, Seibl R, et al. The digoxigenin (DIG) system for non-radioactive labelling and detection of nucleic acids - an overview. Cell Mol Biol (Noisy-le-grand) 1995;41:883–905. [PubMed] [Google Scholar]
- Hu C, Aksoy S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol Microbiol. 2006;60:1194–1204. doi: 10.1111/j.1365-2958.2006.05180.x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Aksoy S. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem Mol Biol. 2005;35:105–115. doi: 10.1016/j.ibmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Hultmark D, Engstrom A, Andersson K, Steiner H, Bennich H, Boman HG. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. Embo J. 1983;2:571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush RS, Lemaitre B. Genes that fight infection: what the Drosophila genome says about animal immunity. Trends Genet. 2000;16:442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- Kim T, Yoon J, Cho H, Lee WB, Kim J, Song YH, et al. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol. 2005;6:211–218. doi: 10.1038/ni1159. [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. Embo J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Meister M, Braun A, Kappler C, Reichhart JM, Hoffmann JA. Insect immunity. A transgenic analysis in Drosophila defines several functional domains in the diptericin promoter. Embo J. 1994;13:5958–5966. doi: 10.1002/j.1460-2075.1994.tb06941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogge G. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia. 1976;32:995–996. doi: 10.1007/BF01933932. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, et al. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Park JM, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, et al. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004;18:584–594. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M. Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell. 2004;13:19–32. doi: 10.1016/s1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- Senger K, Harris K, Levine M. GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc Natl Acad Sci USA. 2006;103:15957–15962. doi: 10.1073/pnas.0607608103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong SJ, Ohta Y, Litman GW, Amemiya CT. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 1997;25:3959–3961. doi: 10.1093/nar/25.19.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XL, Zheng AL. Genomic organization and regulation of three cecropin genes in Anopheles gambiae. Insect Mol Biol. 2002;11:517–525. doi: 10.1046/j.1365-2583.2002.00360.x. [DOI] [PubMed] [Google Scholar]