Abstract
Estrogens are metabolized to active quinones that modify DNA and may lead to various cancers. To extend the analytical methodology for estrogen-modified purine bases, we report here a simple modification to existing synthetic procedures that use 2-iodoxybenzoic acid (IBX) as the oxidizing agent for the reference material and putative biomarker, 4-hydroxyestrone-1-N3Adenine (4-OH-E1-1-N3Ade). The reaction leads to two catechol estrogen quinones, CE1-2,3-Q and CE1-3,4-Q, both of which react via Michael additions to afford 4-OH-E1-1-N3Ade and other DNA adducts. LC separation permits the isolation of high-purity 4-OH-E1-1-N3Ade. With this method, we also prepared single-13C and uniformly-15N (U-15N) labeled 4-OH-E1-1-N3Ade with 8-13C-labeled Ade and U-15N-labeled adenosine 5′-monophosphate (AMP). The approach is also effective for the synthesis of 4-hydroxyestradiol-1-N3Adenine, 4-OH-E2-1-N3Ade, and 4-hydroxyestrone(estradiol)-1-N7Guanine, 4-OH-E1(E2)-1-N7Gua. The tandem mass spectra (MS2 and MS3) of 4-OH-E1-(unlabeled, 8-13C-, and U-15N-labeled)1-N3Ade and accurate mass measurements for MS2 product ions allow us to assign unambiguously the formula of fragments and delineate the fragmentation pathways. One important reaction is dehydration, which occurs at the ketone oxygen in C-17 position of estrone. Another is loss of NH3, a ubiquitous process for purines and modified purines, which is affected by the steroid modification. Evidence from MS/MS supports the migration of H-atom(s) from estrone in the loss of NH3. An interesting interaction occurs between the steroid and Ade in the modified base, promoting loss of CH2NH, a loss that distinguishes modified Ade from unmodified Ade. The synthesis of a stable-isotope-labeled 4-OH-E1-1-N3Ade and the understanding of the fragmentation processes will enable studies aimed at detection of naturally occurred 4-OH-E1-1-N3Ade in biological samples.
Introduction
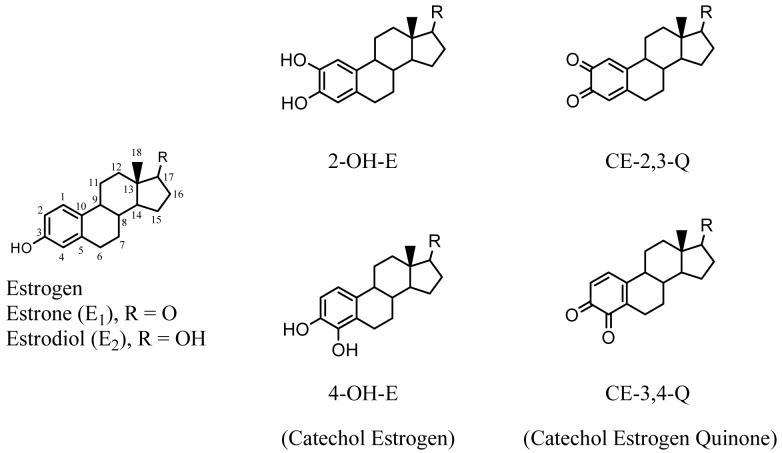
Exposure to estrogens contributes to an increased risk for breast cancer. Several mechanisms are proposed that include (1) estrogen-receptor-mediated increases in cell proliferation (1-7) (2) redox cycling of estrogen metabolites generating reactive oxygen species (2, 8-11) and (3) genotoxic processes leading to removal of nucleobases from DNA (10-21). According to the genotoxic hypothesis, two endogenous estrogens, estrone (E1) and 17β-estradiol (E2), are oxidized to catechol estrogens (CE) at the 2 or 4-position to form either 2-hydroxyestrogen (2-OH-E) or 4-hydroxyestrogen (4-OH-E) (see Figure 1 for structures). The two CEs are subsequently oxidized via semi-quinones to catechol estrogen-2,3-quinone (CE-2,3-Q) and catechol estrogen-3,4-quinone (CE-3,4-Q). These CEQs react both in vitro and in vivo with predominantly nucleophilic adenine (Ade) and guanine (Gua) bases by a 1,4-Michael addition, often leading to rapid loss of the purine by glycosyl-bond cleavage to leave abasic sites in the DNA (13, 17, 22). Although these sites can be repaired enzymatically, an overabundance may lead to DNA mutations and ultimately cancer. If the genotoxic mechanism is correct, products of the depurination (i.e., purine bases modified by catechol estrogens) may be a measure of the DNA damage extent and serve as biomarkers for early diagnosis of breast cancer, long before there are any overt signs of the disease.
Figure 1.
Structures of estrogen, catechol estrogens and catechol estrogen quinones.
To facilitate future studies of the mechanism of tumor initiation by endogenous estrogens and of the prospect for developing a biomarker for breast cancer, we report a simple synthesis of these modified nucleobases for use as reference materials and a characterization of their mass spectral fragmentations. The published chemical methods for preparing catechol-estrogen modified nucleobases utilize multi-step reactions (23-25). In light of the recently reported one-step reaction to convert estrogen to CEQ by using 2-iodoxybenzoic acid (IBX) as oxidant (26), we implemented a simpler method to synthesize and isolate purine bases modified by catechol estrogens by using only one purification step to obtain the putative biomarker, 4-hydroxyestrone-1-N3Adenine (4-OH-E1-1-N3Ade).
The successful synthesis offers a route to isotopically labeled reference materials, and we take the opportunity to investigate the LC/MS and particularly the LC/MS/MS and MS3 of unlabeled, 8-13C- and U-15N-labeled 4-OH-E1-1-N3Ade. Previous tandem mass spectrometric studies of 4-OH-E1(E2)-1-N3Ade showed formation of fragments via ring cleavage of estrogen (17). The new results provide a foundation for understanding the fragmentation processes that will accompany the highly specific detection of the modified bases in biological samples.
Experimental
Materials
Oxone, 2-iodobenzoic acid (IBA), dimethyl formamide (DMF), double-distilled acetic acid (Teflon grade, 99.5%), adenine (Ade) and 2′-deoxynucleoguanosine (dG) were purchased from Sigma-Aldrich (St. Louis, MO). Estrone (E1) and estradiol (E2) were purchased from Steraloids (Newport, Rhode Island). Stable-isotope-labeled adenine, (8-13C) adenine and (U-15N) adenine 5′-monophosphate (AMP), were purchased from Cambridge Isotope Laboratory (Andover, MA). Milli-Q water was used for the synthesis, semi-prep HPLC separation, and the LC/MS analysis. Omnisolv acetonitrile (ACN), methanol (MeOH) and LC/MS grade formic acid (FA) were purchased from Sigma-Aldrich and used for the LC/MS analysis and HPLC separation/purification. The oxidant, 2-iodoxybenzoic acid (IBX), was synthesized from IBA and oxone, as previously described (27).
(U-15N)Ade
To convert (U-15N) AMP to the free (U-15N) adenine base, (U-15N)-AMP (60 mg) was hydrolyzed in 12 M HCl for 3 h at room temperature. The solvent was then removed under reduced pressure with a SpeedVac (Savant 110, Thermo Fisher). The hydrolyte was reconstituted in 1 mL of water and purified by HPLC by using a semi-prep C18 column (10 × 150 mm, 3 Dm) from P. J. Cobert (St. Louis, MO) on a SpectraSystem 2000 HPLC (Thermo Fisher, San Diego, CA).
Synthesis of 4-OH-E1-1-N3Ade Adducts
IBX (5 mg) was added to a stirred solution of estrone (5 mg) in DMF (500 μL), and a red color began to develop in ∼ 10 s; the solution became dark red after 25-30 mins. An excess of Ade (5 mg) in 1 mL of acetic acid and water (1/1, v/v) was added and allowed to react for ∼ 3 h with stirring at room temperature (Scheme 1). The solvent was then removed at reduced pressure, and the residue was reconstituted in 1 mL of methanol and centrifuged at 14,000 rpm for 5 min. The yellow aqueous phase was divided into 10 aliquots and submitted to semi-preparative HPLC separation (10 aliquots were taken for analysis by MS to insure that the chromatogram was well defined). The same HPLC system (SpectraSystem, Thermo) and column (C18, P. J. Cobert) that were used for purification of (U-15N)Ade was used for the separation of 4-OH-E1-1-N3Ade. The gradient was started with 100% phase A (H2O, 0.1% FA) to 5% B (ACN, 0.1%FA) in 5 min and was ramped to 100% B in 25 min and held in B for 5 min. The eluent was monitored by UV spectrophotometic detection at wavelengths of 270 and 291 nm. Unlabeled, 8-13C- and U-15N-labeled 4-OH-E1-1-N3Ade adducts were isolated by this method, and approximately 1 mg amounts of adducts were obtained for each. To obtain high-purity 4-OH-E1-1-N3(U-15N)Ade, the separation was refined by using an Agilent Zorbax phenyl-modified C18 column (5 μm, 9.4 × 250 mm) from P. J. Cobert. The gradient was started with 100% A for 5 min and was ramped to 100% B in 30 min and held in B for 10 min.
Scheme 1.
Synthesis of catechol estrone adenine adduct, 4-OH-E1-1-N3Ade, from estrone (E1). E1 is oxidized by IBX to form two catechol quinones: CE1-2,3-Q and CE1-3,4-Q at 2,3- and 3,4-positions, respectively. Addition of Ade results in major DNA adduct, 4-OH-E1-1-N3Ade, formed between CE1-3,4-Q and Ade. Stable isotope-labeled adducts: 4-OH-E1-(8-13C)1-N3Ade and 4-OH-E1-1-N3(U-15N)Ade are synthesized according to the same scheme.
Mass Spectrometry Analysis
Both nano ESI-MSn and LC-ESI/MSn (n = 1 - 3) were conducted on a LCQ Deca quadrupole ion-trap mass spectrometer (Thermo-Fisher, San Jose, CA) coupled to a capillary LC (Waters Corp.). For direct nano ESI, an uncoated 360/50 μm OD/ID fused-silica capillary column with 15 μm PicoFrit™ nano tip (New Objective Inc., Woburn, MA) was used. Isocratic mobile phases with 70% solvent A (97% H2O, 3% ACN, 0.1% FA) and 30% B (3% H2O, 97% ACN, 0.1% FA) were used to deliver the analytes. Analyte solutions were loaded in 10 μL, and the column flow was 250 nL/min. For LC-ESI, an uncoated 360/75 μm OD/ID fused-silica capillary column with 15 μm PicoFrit™ nano tip (New Objective Inc., Woburn, MA) was custom-packed with C18(2) particles (Luna 3 μm 100 Å, Phenomenex, Torrance, CA) to afford a column that was ∼ 12 cm in length. The sample solution (5 μL) was loaded and analyzed with solvent A and B starting with 97% A for 8 min and then a 37 min linear gradient to 100% B. The eluent with a total flow rate of 8 μL/min was split before the column to achieve a flow rate of 270 nL/min at the tip.
The mass spectrometer was operated in the positive-ion mode. The spray voltage was set to 1.8 - 2.5 kV. No nitrogen sheath gas was used. The maximum time for injection of ions into the trap was 150 ms, and the number of microscans was one. The mass range for the scan was m/z 100 to 1000. For MS2 experiments, the collision energy was optimized to be 40% of the maximum energy available (∼ 5 eV, laboratory frame). For MS3 experiments, MS2 ions generated with 40% of the maximum collision energy were further fragmented with collision energies of 35, 40 and 45%, respectively.
Accurate mass measurements were carried out with an ion-trap/FT mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA). Samples were dissolved in H2O/ACN/FA (30/70/0.1), directly injected and sprayed with nano-ESI. The 4-OH-E1-1-N3Ade modified base ion was selected and collisionally activated at 22% energy in the linear ion trap. The maximum number of total ions stored in the trap was 3 × 108 in the MS mode. The elevated threshold for ion storage in the MS mode would permit more targeted ions to be accumulated in MS/MS mode. With the increased ion storage, the maxima time for injection of ions into the trap can reach 0.3 - 6 s. The transient time in the FT-ICR trap was set to match to the time of accumulating ions in the linear trap. A resolving power of FT was 1,000,000 (at m/z 400), and the mass was scanned from m/z 100 - 450.
Hydrogen/Deuterium (H/D) Exchange
After removing the analyte solvent under reduced pressure, the residue was reconstituted in D2O/ACN (70/30, v/v) and incubated over night at room temperature. The solution was used for MS analysis without any acid modifier.
Results and Discussion
Synthesis, Purification, and LC/MS of Reference Materials
The reaction of catechol estrogen quinones (CEQs), CE -3,4-Q, with AMP does not afford catechol estrogen adenine adducts, 4-OH-E-1-N3Ade. This is probably due to steric hindrance caused by the ribose to the approach of a CEQ to the N-3 of Ade in AMP (17, 24). The reactions of CEQs with the Ade free base or with Ade in a DNA double helix, however, do lead to 4-OH-E-1-N3Ade by 1,4-Michael additions. To synthesize the U-15N-labeled Ade base modified with a catechol estrone, we hydrolyzed U-15N-labeled AMP in 12 M HCl, converting the AMP to the free U-15N-labeled Ade base without affecting the Ade moiety. HPLC and MS analyses showed that the conversion of (U-15N) AMP to (U-15N) Ade and Ade·HCl was > 98%. Approximately 15 mg of purified (U-15N) Ade was obtained (70% yield), calculated on the basis of the starting (U-15N)-AMP amount.
A limitation to the synthesis via the quinones is that IBX oxidizes estrone to both CE-2,3-Q and CE-3,4-Q (Scheme 1), which are unstable in aqueous solution, preventing separation prior to reaction with Ade. Previous approaches reduce the quinones to catechols, separate them and reoxidize the pure catechols to quinones. The approach we have taken is to carry out the reaction in “one pot”, and purify the final mixture of 4-OH-E1-1-N3Ade and other catechol estrone adenine adducts by preparatory HPLC to afford high-purity 4-OH-E1-1-N3Ade. The entire reaction can be finished in 3.5 h at room temperature. The modified nucleobases are purified by preparative HPLC.
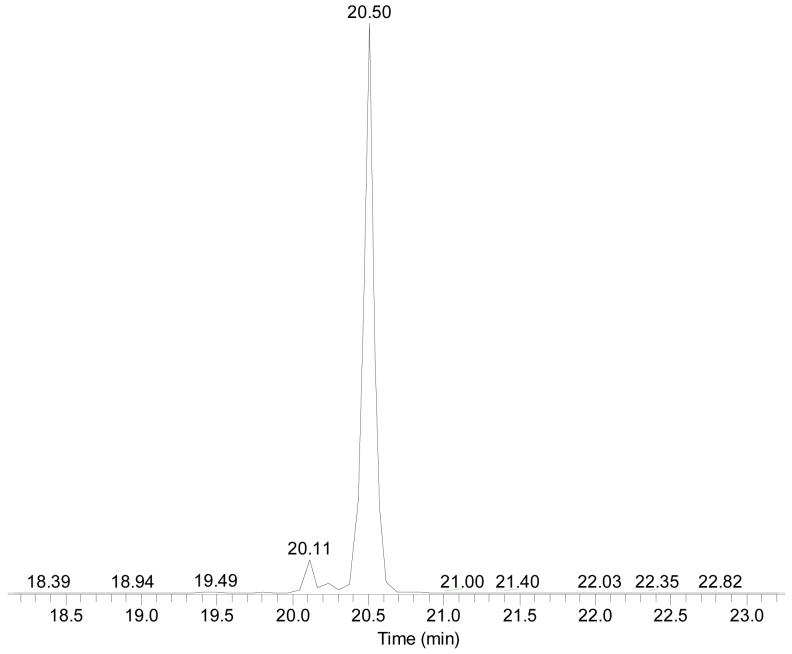
The keys to the success of this procedure are the LC separation and the MS detection of catechol estrogen DNA adducts; the chromatography affords an effective separation for both analytical (preparatory) and capillary (with microspray) LC combined with MS. The use of a mobile phase of H2O and 0.1% TFA, as one component, and MeOH and 0.1%TFA, as the other, afforded sharp chromatographic peaks. The use of this mobile phase, however, gave inadequate separation of 4-OH-E1-1-N3Ade from the other adducts. In addition, the TFA suppressed the signals for mass spectrometry analysis. Alternatively, a mobile phase of MeOH/H2O with 0.1% FA allowed the MS response to be increased by approximately eight fold. The chromatographic peaks, however, were twice as broad when FA was used instead of TFA. We then tested a mobile phase of A (H2O with 0.1% FA) and B (ACN with 0.1% FA); the MS response was nearly identical to that of MeOH/H2O/0.1% FA. Moreover, the ACN/H2O/0.1%FA system, allowed the chromatographic resolution to be nearly as good as achieved with MeOH/H2O/0.1% TFA and provided adequate separation of isomeric DNA adducts. To achieve both good LC separation and sensitive MS detection, we chose the mobile phase of ACN/H2O with 0.1% FA for both the nano-LC/MS/MS analysis and for the semi-prep HPLC separation and purification. Given that other relatively abundant DNA adducts elute later than 4-OH-E1-1-N3Ade, the products can be cleanly separated by reverse-phase preparatory HPLC (see Figure 2 for the LC of purified 4-OH-E1-1-N3(U-15N)Ade). The limit of detection for 4-OH-E1-1-N3Ade was 250 attomole (signal-to-noise ratio greater or equal to 3) for the LC/MS analysis with LCQ/Deca.
Figure 2.
LC/MS analysis of 4-OH-E1-1-N3(U-15N-labeled)Ade (tR = 20.50 min) shows the purity of the U-15N-labeled material. The minor component eluting at tR = 20.11 min is also an adduct formed between catechol estrone quinone and Ade.
We also isolated other estrogen purine adducts, 4-OH-E2-1-N3Ade, 4-OH-E1-1-N7Gua and 4-OH-E2-1-N7Gua, by this method. Unlike 4-OH-E-1-N3Ade, which forms between CE-3,4-Q and Ade free base, the adduct of 4-OH-E-1-N7Gua does arise in a reaction between CE-3,4-Q and dG. CE-3,4-Q binds preferentially to the N-7 of Gua, minimizing the steric hindrance caused by ribose for the binding to N-3 of Ade. Upon solvent removal, the glycosyl bond cleaved readily, resulting in 4-OH-E-1-N7Gua without the ribose.
Tandem Mass-spectrometric studies of 4-OH-E1-1-N3Ade and its isotopomers
Accurate mass measurements and evidence from the MS/MS of the isotopomers allow us to gain insight into the fragmentation pathways of estrogen-modified nucleobases, specifically of the 4-OH-E1-1-N3Ade. The LC traces of unlabeled, 8-13C-labeled (12C replaced by 13C at C-8 of Ade), and U-15N-labeled (all five 14N in Ade replaced by 15N) show that the three isomers co-eluted with peak widths at half height of approximately 5 s, underscoring that the isotopomers are appropriate internal standards.
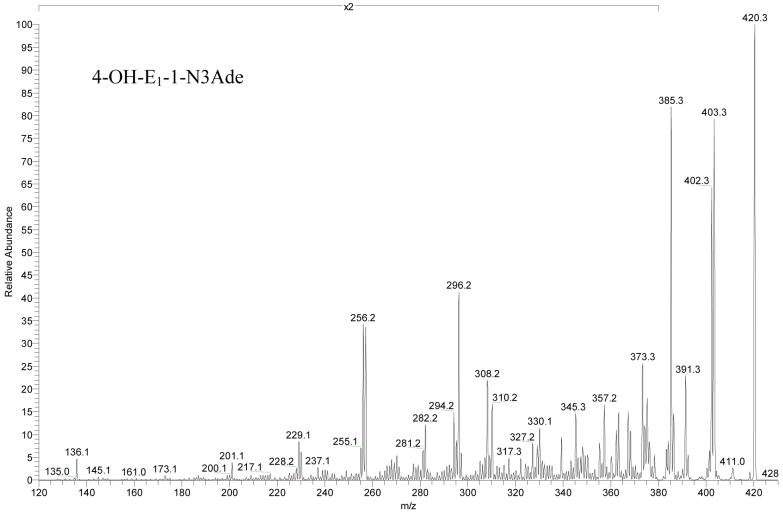
The product-ion spectrum of the unlabeled species (Figure 3) shows major fragments from deamination (loss of 17 u), dehydration (loss of 18 u), dehydration + deamination (loss of 35 u), and loss of methylimine (29 u) from the protonated molecules. H2O and 15NH3 have the same nominal mass of 18; thus, the doublet representing deamination and dehydration merged into a singlet for the 15N-labeled precursor of m/z 425.
Figure 3.
Product ion spectra (MS2) of [4-OH-E1-1-N3Ade + H]+ at a collision energy of 40% of the maximum for unlabeled adducts. The spectra are amplified by two times from m/z 120 to 380.
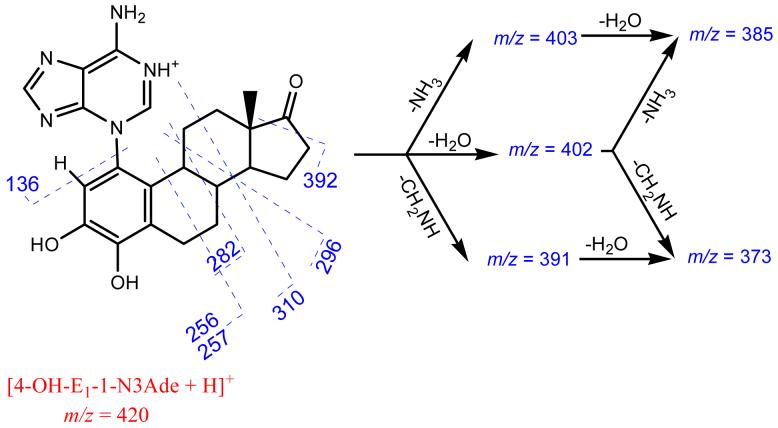
Competing weakly with the losses of small molecules (Scheme 2) are through-ring cleavages of the steroid moiety. Given that both fragment ions of m/z 256 and 257 contains five N-atoms, that of m/z 257 is a radical cation, in violation of the even-electron ion rule. A likely explanation is that the unpaired spin is localized as a benzyl radical, stabilized by spin delocalization into the aromatic ring A. The fragment of m/z 136 is the only species clearly identified in the low mass range, and it is likely protonated adenine, consistent with the labeling and accurate mass evidence. The postulated formulae of all fragments produced in MS2 are consistent with accurate mass measurements (Table 1); the mass accuracies for all product ions of unlabeled [4-OH-E1-1-N3Ade + H]+ are within 1 ppm of the theoretical masses (Table S1 in supporting information).
Scheme 2.
Proposed mechanism of major fragmentations for [4-OH-E1-1-N3Ade + H]+ parent ion. Identities of the product ions were confirmed with both U-15N labeling and accurate mass measurements.
Table 1.
Product ions and representative losses for [4-OH-E1-(un-, 8-13C- and U-15N-labeled)-1-N3Ade + H]+. Number of 15N loss is shown in parenthesis for [4-OH-E1-1-N3(U-15N)Ade + H]+. Relative abundances of dual species with same nominal mass are shown in brackets
| un-(m/z = 420) | 8-13C (m/z = 421) | U-15N (m/z = 425) | Neutral loss from parent |
|---|---|---|---|
| 418 | 419 | 423 (0) | H2 |
| 403 | 404 | 407 (1) | NH3 |
| 402 | 403 | 407 (0) | H2O |
| 401 | 402 | 405 (1) | NH3 + H2 |
| 400 | 401 | 405 (0) | H2O + H2 |
| 392 | 393 | 397 (0) | CO [60%] |
| 391 | 392 | 395 (1) | CH2NH |
| 386 | 387 | 389 (2) | (NH3)2 |
| 385 | 386 | 389 (1) | NH3 + H2O |
| 383 | 384 | 387 (1) | NH3 + H2O + H2 |
| 375 | 376 | 379 (1) | NH3 + CO |
| 373 | 374 | 377 (1) | H2O + CH2NH |
| 368 | 369 | 371 (2) | H2O + (NH3)2 |
| 367 | 368 | 371 (1) | (H2O)2 + NH3 |
| 363 | 364 | 367 (1) | CH2NH + CO |
| 362 | 363 | 365 (2) | (CH2NH)2 [45%] |
| 367 (0) | C3H6O [55%] | ||
| 357 | 358 | 361 (1) | NH3 + H2O + CO [90%] |
| NH3 + H2O + C2H4 [10%] | |||
| 355 | 356 | 359 (1) | CH2NH + (H2O)2 |
| 345 | 346 | 349 (1) | CH2NH + H2O + CO [70%] |
| CH2NH + H2O + C2H4 [30%] | |||
| 339 | 340 | 343 (1) | NH3 + (H2O)2 + CO |
| 327 | 328 | 331 (1) | (H2O)2 + CH2NH + CO [90%] |
| (H2O)2 + CH2NH + C2H4 [10%] | |||
| 310 | 311 | 315 (0) | C7H10O |
| 308 | 309 | 313 (0) | C7H10O + H2 |
| 296 | 297 | 301 (0) | C8H12O |
| 294 | 295 | 299 (0) | C8H14O |
| 282 | 283 | 287 (0) | C9H14O |
| 281 | 282 | 285 (1) | CH2NH + C7H10O |
| 257 | 258 | 262 (0) | C11H15O |
| 256 | 257 | 261 (0) | C11H16O |
| 255 | 256 | 259 (1) | CH2NH + C9H12O |
| 230 | 231 | 234 (1) | CH2NH + C11H13O |
| 229 | 230 | 233 (1) | CH2NH + C11H14O |
| 201 | 202 | 205 (1) | C13H17O2Na |
| 136 | 137 | 141 (0) | catechol estrone quinone a |
We carried out both direct-infusion and liquid-chromatographic ESI tandem mass-spectrometric (MS3) studies in which the major fragments produced by MS2 were activated for further decomposition at three collisional energies (35, 40 and 45% of maximum) to reveal novel fragmentations that are characteristic of the steroid-modified DNA base. We propose that the most low-mass fragment ions produced by MS2 of 4-OH-E1-1-N3Ade arise following the losses of water, ammonia, water plus ammonia, and methylimine. Therefore, we chose to focus on these important fragmentations, as discussed in the following sections.
(1) Loss of H2O
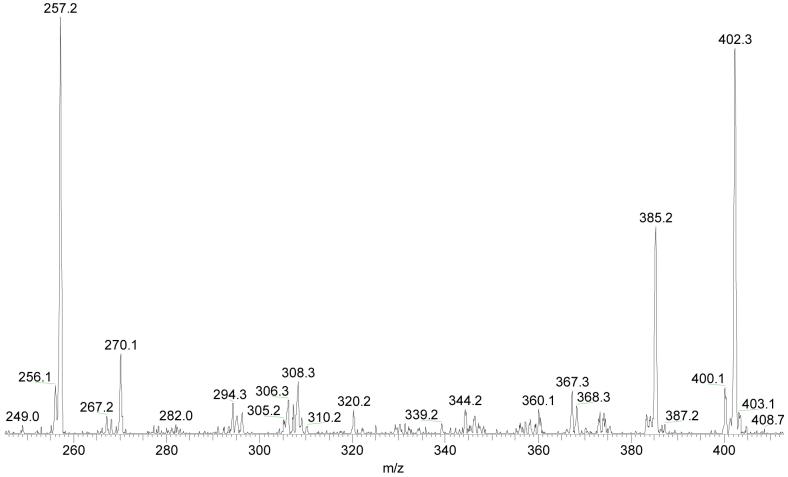
Loss of H2O from [4-OH-E1-1-N3Ade + H]+ leads to the product ion of m/z 402. There are three potential sites for H2O loss, two from the catechol oxygens and one involving the ketone oxygen at C-17. An MS3 study allows identification of the primary dehydration site for the steroid-modified nucleobase. The product-ion (MS3) spectrum (Figure 4) of the dehydrated species (of m/z 402) shows an abundant loss of NH3 to give an m/z 385 ion and, at lower m/z, product ions of m/z 256 and 257. The spectra of U-15N labeled material show that both these fragments contain five nitrogen atoms. Similar to the fragmentation of 4-OH-E1-1-N3Ade, the ions of m/z 256 and 257 arise from B-ring cleavage of an estrone containing the original two OH groups in the catechol ring A. In contrast, the ions would be of m/z 238 and 239 if the original dehydration had involved the catechol OH groups. This outcome shows that the primary H2O loss involves the ketone oxygen at C-17.
Figure 4.
Tandem MS3 of dehydrated product ion for unlabeled 4-OH-E1-1-N3Ade (m/z = 402) at a collision energy that is 40% of the maximum.
(2) Loss of NH3
Loss of NH3 occurs for both unmodified [Ade + H]+ and catechol-estrone-modified Ade, [4-OH-E1-1-N3Ade + H]+. Previous studies of unmodified [Ade + H]+ reveal that deamination of [Ade + H]+ involves the endocyclic N-1 and exocyclic N-10 with nearly equal probability (28). The steroid modification of Ade at the N-3 position, however, prohibits NH3 loss from that position. Second-generation loss of NH3 also occurs, as shown by MS3, from the product ions of m/z 310, 308 and 296 that are generated from cleavages of the steroid C-ring, but barely from the product ions of m/z 257 and not at all from the m/z 256 product, also generated from through B-ring cleavages of the steroid portion. These observations support that the H at C-9 of the steroid is necessary for the loss of NH3.
(3) Loss of CH2NH
Besides deamination and dehydration, the loss of methylimine (CH2NH), confirmed by accurate mass measurement, is a major fragmentation to give ions of m/z 391, 392, and 395 for un-, 8-13C- and U-15N-labeled adducts, respectively. Interestingly, this loss does not occur from [Ade + H]+ (28, 29). The loss from the 8-13C-labeled modified base produces exclusively an m/z 392 ion, indicating no involvement of C-8 of Ade. This is in accord with theoretical calculations that indicate that the most favorable position for protonation is N-1 of Ade (30), and that it is this nitrogen that is contained in expelled methylimine. To expel CH2NH, migration of two hydrogen atoms is needed. A plausible mechanism for the hydrogen migration involves the transfer of an H-atom from the steroid moiety to reduce the Ade moiety and oxidize the steroid.
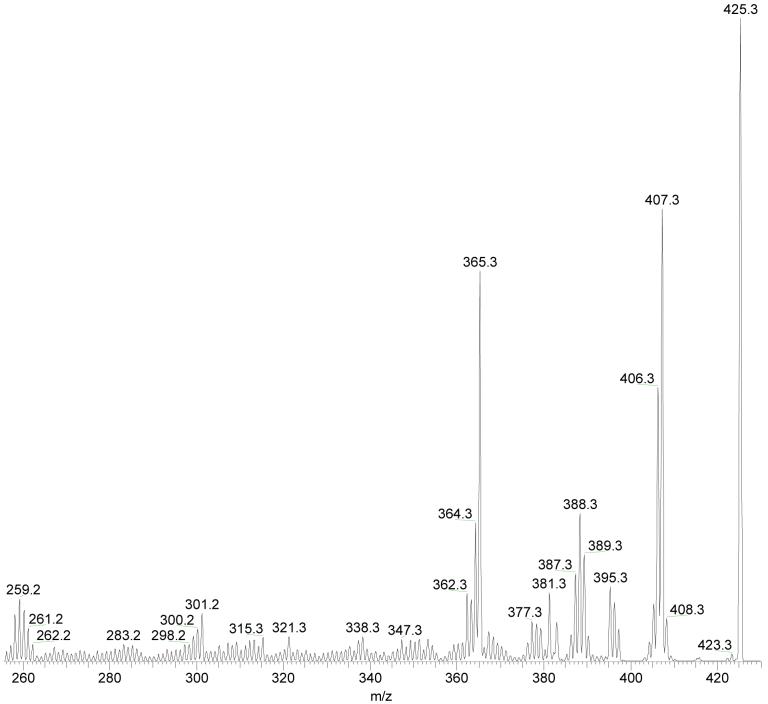
H/D exchange was carried out to determine the number of non-exchangeable H-atoms in CH2NH. For the precursor [4-OH-E1-1-N3Ade + H]+, there are five exchangeable H atoms: two exocyclic amine H-atoms at N-10 of Ade, two catechol H-atoms at C-3 and C-4 of E1, and the added proton that provides the +1 charge. The m/z 391 ion produced from undeuterated [4-OH-E1-1-N3Ade + H]+ shifts to m/z 395 and 396 in the MS2 of fully deuterated [4-OH-E1-1-N3Ade + H]+: d5-[4-OH-E1-1-N3Ade + H]+ (Figure 5). Accurate mass measurement proves that the losses are of singly deuterated and non-deutetrated CH2NH, respectively, from the d5 precursor. Interestingly, no losses of CHD2N or CD3N occur, indicating that two non-exchangeable H-atoms included in the CH2NH fragment from modified Ade (presumably the original H at position 2 of Ade and one H from the steroid).
Figure 5.
Product-ion spectrum (MS2) of fully exchanged d5-[4-OH-E1-1-N3Ade + H]+ at a collision energy of 40% of the maximum.
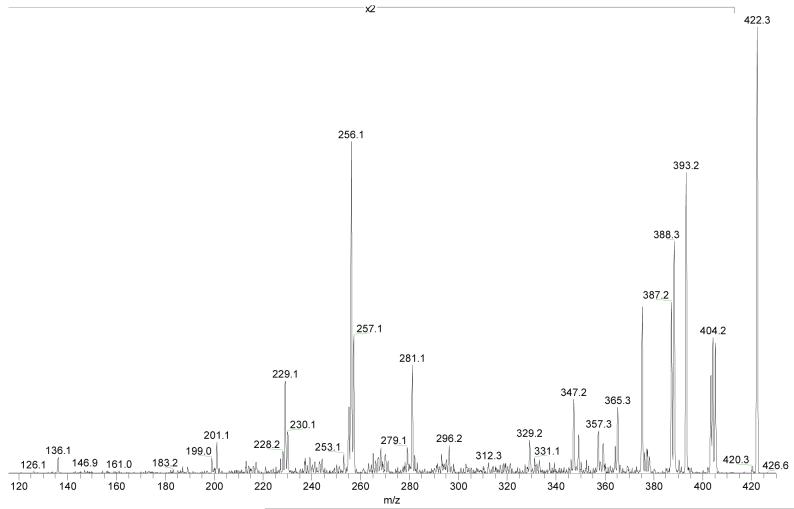
The reduction (i.e., formation of double bond) of the steroid is further supported by MS3 of the species formed by loss of CH2NH (Figure 6). Two fragmentations give closed-shell and radical-cation product ions of m/z 229 and 230 (see Scheme 3), respectively, in accord with the proposal that the steroid moiety has transferred one hydrogen atom to make the methylimine and another to N-3 of Ade. The corresponding through-B-ring cleavages of the unlabeled protonated molecule, [4-OH-E1-1-N3Ade + H]+, produce ions of m/z 256 and 257. Two related cleavages also leading to a closed-shell and a radical-cation product are those that involve ring C and give ions of m/z 295 and 294. The formation of a radical cation may be driven by the incipient B-ring conjugated double bonds between C-9 and C-11 of estrone that stabilize the radical site formed in the C-ring.
Figure 6.
Product-ion spectrum (MS3) of the [M + H - CH2NH]+ of m/z 391 for unlabeled 4-OH-E1-1-N3Ade at a collision energy of 35% maximum.
Scheme 3.
Proposed pathways for formation of [4-OH-E1-1-N3Ade + H - CH2NH]+ (m/z 391) and its subsequent MS3 fragmentation. Structures of product ions are postulated. The number of nitrogen atoms is confirmed with U-15N labeled species.
Of all the high-mass fragment ions, only that of m/z 391 fragments to give the m/z 255 product ion, which is also seen in the MS2 of [4-OH-E1-1-N3Ade + H]+. That the m/z 255 ion contains four nitrogen atoms (formula affirmed with 15N labeling and accurate mass measurement) and that it arises from the m/z 391 ion affirm that it is formed via B,C-through-ring cleavage of the 4-OH-E1-1-N3Ade following CH2NH loss.
The m/z 391 ion, formed by loss of methylimine, is an important precursor for subsequent fragmentation of the modified Ade. It gives rise, as shown by MS3, to product ions of m/z 373, 363, 355, 345, 294, 281, 255, 230, 229 and 201, which are also seen in the MS2 product ion spectrum of [4-OH-E1-1-N3Ade + H]+. A scheme for the formation and subsequent fragmentation of m/z 391 ion (Scheme 3) shows that the abstraction of the proximate hydride (H-) from steroid C-9 to Ade leads to the delocalization of charge at C-9 and can be stabilized by the phenol ring A of the steroid. Migration of a second non-exchangeable proton from steroid C-11 to Ade N-1 relays the charge back to Ade, leading to the formation of reduced Ade. The cleavage of the N1-C6 bond in the reduced Ade leads to expulsion of CH2NH.
It is noteworthy that loss of HCN does not occur from the modified Ade whereas it is a major loss for protonated adenine. This is another indication that introduction of the steroid moiety alters the fragmentation of adenine.
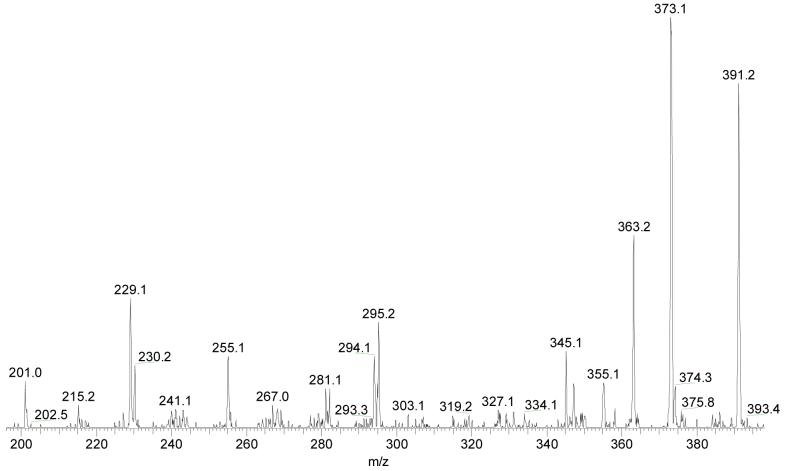
Fragmentation of [4-OH-E2-1-N3Ade + H]+
Delineation of the major product ions of [4-OH-E1-1-N3Ade + H]+ brings by analogy understanding of the fragmentation of 4-OH-E2-1-N3Ade without the need to synthesize stable-isotope-labeled analogs (see Figure 7 for the product-ion spectrum). Not surprisingly, the fragmentation of protonated 4-OH-E2-1-N3Ade resembles that of 4-OH-E1-1-N3Ade (a comparison of losses of neutral species from 4-OH-E1-1-N3Ade and 4-OH-E2-1-N3Ade is shown in the supplemental information). The triplet of ions at m/z 405-403 is attributed to losses of NH3, H2O and NH3 + H2, respectively. The fragment ion of m/z 393 arises from the distinctive loss of CH2NH from the protonated molecule, 4-OH-E2-1-N3Ade, suggesting that this CH2NH loss may play an important role in the fragmentation of similarly modified nucleobases. Given that the only difference between nucleobases modified with E1 vs. E2 is the substituent on the D ring, through-ring cleavages of the B and C steroid rings of both leads to same product ions of m/z 256, 257 and 296. The loss of CH2NH combined with through-ring cleavages leads to product ions of m/z 201, 229, 230, 255 and 281. The ions formed by losses of small molecules are relatively less abundant than those from the E1 adduct for reasons we do not understand.
Figure 7.
Product-ion spectrum (MS2) of unlabeled [4-OH-E2-1-N3Ade + H]+. The spectrum is amplified by 2 times in the range m/z 115 to 410.
Conclusions
Purine bases modified by catechol estrogen (CE1 and CE2) can be readily synthesized via a one-pot synthesis employing IBX as the oxidant and purified in a final HPLC step. The procedure is appropriate for preparation of valuable modified bases: 4-OH-E1(E2)-1-N3Ade, 4-OH-E1(E2)-1-N7Gua and for stable-isotope-labeled adenines that are modified by catechol estrone: 4-OH-E1(E2)-1-N3Ade, with 8-13C and U-15N labeled Ade.
Comparison of MS2 and MS3 of un-, 8-13C- and U-15N-labeled 4-OH-E1(E2)-1-N3Ade reveals that the primary H2O loss occurs at the C-17 ketone oxygen of estrone moiety. Competitive with the losses of water and ammonia are through-ring cleavages of steroid rings B, C and D. The hydrogens involved in the loss of ammonia do not originate solely from the Ade moiety but rather require migration of H-atom(s) from the steroid moiety. An interesting loss of methylimine, CH2NH, apparently involves transfer a pair of H-atoms from the steroid moiety of both E1 and E2 to the nucleobase, causing reduction of the base and oxidation of the steroid. This process may be characteristic of modified purines. The 4-OH-E2-1-N3Ade undergoes similar fragmentations, suggesting that many of the processes reported here are general.
Supplementary Material
Acknowledgement
The work was supported by the National Centers for Research Resources of the NIH (Grant No. 2P41RR000954).
References
- (1).Nandi S, Guzman RC, Yang J. Hormones and Mammary Carcinogenesis in Mice, Rats, and Humans - a Unifying Hypothesis. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3650–3657. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yager JD, Liehr JG. Molecular mechanisms of estrogen carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- (3).Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- (4).Flototto T, Djahansouzi S, Glaser M, Hanstein B, Niederacher D, Brumm C, Beckmann MW. Hormones and hormone antagonists: Mechanisms of action in carcinogenesis of endometrial and breast cancer. Horm. Metab. Res. 2001;33:451–457. doi: 10.1055/s-2001-16936. [DOI] [PubMed] [Google Scholar]
- (5).Clemons M, Goss P. Mechanisms of disease - Estrogen and the risk of breast cancer. N. Engl. J. Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- (6).Santen RJ. To block estrogen’s synthesis or action: That is the question. J. Clin. Endocrinol. Metab. 2002;87:3007–3012. doi: 10.1210/jcem.87.7.8589. [DOI] [PubMed] [Google Scholar]
- (7).Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit. Rev. Oncol./Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- (8).Liehr JG, Roy D. Free-Radical Generation by Redox Cycling of Estrogens. Free Radic. Biol. Med. 1990;8:415–423. doi: 10.1016/0891-5849(90)90108-u. [DOI] [PubMed] [Google Scholar]
- (9).Han XL, Liehr JG. 8-Hydroxylation of Guanine Bases in Kidney and Liver DNA of Hamsters Treated with Estradiol - Role of Free-Radicals in Estrogen-Induced Carcinogenesis. Cancer Research. 1994;54:5515–5517. [PubMed] [Google Scholar]
- (10).Liehr JG. Is estradiol a genotoxic mutagenic carcinogen? Endocr. Rev. 2000;21:40–54. doi: 10.1210/edrv.21.1.0386. [DOI] [PubMed] [Google Scholar]
- (11).Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem. Res. Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- (12).Liehr JG, Avitts TA, Randerath E, Randerath K. Estrogen-induced endogenous DNA adduction: possible mechanism of hormonal cancer. Proc. Natl. Acad. Sci. U. S. A. 1986;83:5301–5305. doi: 10.1073/pnas.83.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, Johansson SL, Patil KD, Gross ML, Gooden JK, Ramanathan R, Cerny RL, Rogan EG. Molecular origin of cancer: Catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Akanni A, AbulHajj YJ. Estrogen nucleic acid adducts: Reaction of 3,4-estrone-o-quinone radical anion with deoxyribonucleosides. Chem. Res. Toxicol. 1997;10:760–766. doi: 10.1021/tx970026c. [DOI] [PubMed] [Google Scholar]
- (15).Liehr JG. Role of DNA adducts in hormonal carcinogenesis. Regul. Toxicol. Pharmacol. 2000;32:276–282. doi: 10.1006/rtph.2000.1432. [DOI] [PubMed] [Google Scholar]
- (16).Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WR, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL. Genotoxic metabolites of estradiol in breast: potential mechanism of estradiol induced carcinogenesis. J. Steroid Biochem. Mol. Biol. 2003;86:477–486. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]
- (17).Li KM, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- (18).Zhao ZL, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks PG, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem. Res. Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- (19).Yager JD, Davidson NE. Mechanisms of disease: Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- (20).Bolton JL, Thatcher GRJ. Potential mechanisms of estrogen quinone carcinogenesis. Chem. Res. Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Belous AR, Hachey DL, Dawling S, Roodi N, Parl FF. Cytochrome P4501B1-mediated estrogen metabolism results in estrogen-deoxyribonucleoside adduct formation. Cancer Research. 2007;67:812–817. doi: 10.1158/0008-5472.CAN-06-2133. [DOI] [PubMed] [Google Scholar]
- (22).Todorovic R, Devanesan P, Higginbotham S, Zhao J, Gross ML, Rogan EG, Cavalieri EL. Analysis of potential biomarkers of estrogen-initiated cancer in the urine of Syrian golden hamsters treated with 4-hydroxyestradiol. Carcinogenesis. 2001;22:905–911. doi: 10.1093/carcin/22.6.905. [DOI] [PubMed] [Google Scholar]
- (23).Dwivedy I, Devanesan P, Cremonesi P, Rogan E, Cavalieri E. Synthesis And Characterization Of Estrogen 2,3-Quinones And 3,4-Quinones - Comparison Of Dna Adducts Formed By The Quinones Versus Horseradish Peroxidase-Activated Catechol Estrogens. Chem. Res. Toxicol. 1992;5:828–833. doi: 10.1021/tx00030a016. [DOI] [PubMed] [Google Scholar]
- (24).Stack DE, Byun J, Gross ML, Rogan EG, Cavalieri EL. Molecular characteristics of catechol estrogen quinones in reactions with deoxyribonucleosides. Chem. Res. Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- (25).Borges C, Lemiere F, Embrechts J, Van Dongen W, Esmans EL. Characterisation of estrone-nucleic acid adducts formed by reaction of 3,4-estrone-o-quinone with 2 ′-deoxynucleosides/deoxynucleotides using capillary liquid chromatography/electro spray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:2191–2200. doi: 10.1002/rcm.1610. [DOI] [PubMed] [Google Scholar]
- (26).Saeed M, Zahid M, Rogan E, Cavalieri E. Synthesis of the catechols of natural and synthetic estrogens by using 2-iodoxybenzoic acid (IBX) as the oxidizing agent. Steroids. 2005;70:173–178. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- (27).Frigerio M, Santagostino M, Sputore S. A user-friendly entry to 2-iodoxybenzoic acid (IBX) Journal Of Organic Chemistry. 1999;64:4537–4538. [Google Scholar]
- (28).Nelson CC, McCloskey JA. Collision-Induced Dissociation Of Adenine. J. Am. Chem. Soc. 1992;114:3661–3668. [Google Scholar]
- (29).Alexander AJ, Kebarle P, Fuciarelli AF, Raleigh JA. Characterization of radiation-induced damage to polyadenylic acid using high-performance liquid chromatography/tandem mass spectrometry. Anal. Chem. 1987;59:2484–2491. doi: 10.1021/ac00147a010. [DOI] [PubMed] [Google Scholar]
- (30).Turecek F, Chen XH. Protonated adenine: Tautomers, solvated clusters, and dissociation mechanisms. J. Am. Soc. Mass Spectrom. 2005;16:1713–1726. doi: 10.1016/j.jasms.2005.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.