Abstract
Using an expression protein library of a hyperthermophilic archaeon, Pyrococcus furiosus, we identified a gene (PF0027) that encodes a protein with heat-stable cyclic nucleotide phosphodiesterase (CPDase) activity. The PF0027 gene encoded a 21-kDa protein and an amino acid sequence that showed ∼27% identity to that of the 2′-5′ tRNA ligase protein, ligT (20 kDa), from Escherichia coli. We found that the purified PF0027 protein possessed GTP-dependent RNA ligase activity and that synthetic tRNA halves bearing 2′,3′-cyclic phosphate and 5′-OH termini were substrates for the ligation reaction in vitro. GTP hydrolysis was not required for the reaction, and GTPγS enhanced the tRNA ligation activity of PF0027 protein, suggesting that the ligation step is regulated by a novel mechanism. In comparison to the strong CPDase activity of the PF0027 protein, the RNA ligase activity itself was quite weak, and the ligation product was unstable during in vitro reaction. Finally, we used NMR to determine the solution structure of the PF0027 protein and discuss the implications of our results in understanding the role of the PF0027 protein.
Keywords: cyclic-nucleotide-phosphodiesterase, RNA ligase, archaea, expression cloning, solution structure
INTRODUCTION
It has been proposed that the last universal common ancestor (LUCA) was a thermophile or a hyperthermophile (Achenbach-Richter et al. 1987; Brown et al. 2001; Di Giulio 2003). The hyperthermophilic archaeon Pyrococcus species thrives at temperatures above 95°C and is thought to be an evolutionarily ancient organism. Thus, analyzing genome-wide gene functions in Pyrococcus could lead to new insights into the fundamental knowledge of life. For example, the results of DNA microarray analysis of Pyrococcus furiosus (Schut et al. 2001; Schut et al. 2003) and of a genome-wide protein–protein interaction study in Pyrococcus horikoshii OT3 (Usui et al. 2005) have contributed to our general understanding of archaeal biology, in addition to pointing to features unique to these species. We have been studying RNA metabolism in P. furiosus (Kanai et al. 2003, 2006; Sato et al. 2003; Fujishima et al. 2007). It has been hypothesized that RNA played a central role in the very beginning of life (Forterre 2005; Gesteland et al. 2006); thus, analyzing gene regulation at the RNA level of the organism may contribute to a better understanding of the basis of gene function. Our strategy has been to discover novel RNA-binding proteins and RNA-related enzymes of P. furiosus. In a previous study (Kanai et al. 2003, 2006), we screened a recombinant protein library for P. furiosus proteins that can bind RNA. Using gel-shift assays to detect RNA binding, we found a gene product, which we named FAU-1 (P. furiosus AU-binding protein), that demonstrated the strongest binding activity of all the genomic library-derived proteins tested in our study against an AU-rich RNA sequence, r(A-U)10 (Kanai et al. 2003). We also demonstrated that a flavin-dependent thymidylate synthase, Pf-Thy1, possessed RNA-binding activity at the stem–loop structure located near the translational start codon AUG of its own mRNA (Kanai et al. 2006).
In the present study, we found that the PF0027 gene in P. furiosus encodes a highly heat-stable cyclic nucleotide phosphodiesterase (CPDase). The amino acid sequence of the PF0027 protein showed ∼27% identity to that of the Escherichia coli 2′-5′ RNA ligase, ligT protein, which is known to be capable of joining yeast Saccharomyces cerevisiae half-tRNAs (Arn and Abelson 1996). Thus far, all known archaea express two or more intron-containing pre-tRNA species whose introns are excised by the well-characterized tRNA splicing endonuclease (Tocchini-Valentini et al. 2005a,b). In the P. furiosus genome, two pre-tRNA genes (Met, Trp) have introns in their anticodon loops (Marck and Grosjean 2003). However, the enzyme involved in the ligation of tRNA halves is as yet unknown in the archaea kingdom. Therefore, we used an in vitro reconstitution system to analyze Pyrococcus tRNA ligation. Using this system, we found that the PF0027 protein possesses GTP-dependent tRNA ligation activity and that synthetic tRNA halves with 2′,3′-cyclic phosphate and 5′-OH end structures were a suitable substrate. These are well-known terminal structures of tRNA exons after the removal of tRNA introns by tRNA splicing endonuclease. We also found that GTP hydrolysis is not required for the ligation reaction, suggesting that the ligation step is regulated by a novel mechanism, although the ligation activity itself is quite low. Moreover, in order to help determine the structure–function relationship between the protein and its substrate, we analyzed the solution structure and interaction of PF0027 protein by NMR. Recently, the structures of PF0027-related proteins from Thermus thermophilus and Pyrococcus horikoshii have been determined by X-ray crystallography (Kato et al. 2003; Rehse and Tahirov 2005). A possible mechanism of PF0027 protein activity is discussed based on comparison of the protein to other RNA ligase and CPDase proteins.
RESULTS
Identification of P. furiosus gene with cyclic nucleotide phosphodiesterase activity
Previously, we have systematically identified novel RNA-binding proteins in a protein library overexpressing the P. furiosus genome (Kanai et al. 2003, 2006). In the present research, we found a protein, PF0027, in the same library with heat-stable cyclic nucleotide phosphodiesterase (CPDase) activity (Fig. 1A,B; also see Supplemental Fig. 1; Supplemental Table 1). The PF0027 protein consists of a 184-amino-acid protein that has a calculated molecular mass of 21.0 kDa. After searching the NCBI protein database using the PSI-BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/), we found that the PF0027 protein has a high degree of amino acid similarity to 2′-5′ RNA ligases from many kinds of archaea (Supplemental Fig. 2). For example, the PF0027 protein shows 89% identity and 96% similarity to the 184-amino acid-long 2′-5′ RNA ligase-like protein from P. horikoshii (NCBI protein database accession number NP_142114) and 76% identity and 88% similarity to the 184-amino acid-long 2′-5′ RNA ligase-like protein from Thermococcus kodakarensis KOD1 (NCBI protein database accession number YP_184151). Because the data for all these predicted related proteins came from the archaeal genome or structural projects, we next compared the amino acid sequence of the PF0027 protein with that of an experimentally characterized protein from E. coli, 2′-5′ RNA ligase protein, or ligT (formerly known as yadP) (Arn and Abelson 1996). A lower level of sequence identity (27%) and similarity (66%) was observed in the N-terminal 131-amino acid-long region between proteins PF0027 and ligT as compared with the predicted proteins from archaea. However, it is notable that the amino acid sequences and positions for two HXTX motifs (X: hydrophobic) were conserved between the two enzymes. This motif has been reported to be strictly conserved among members of the 2H phosphoesterase superfamily (Kato et al. 2003; Sakamoto et al. 2005). Biophysical analysis of the three phosphodiesterases (from Arabidopsis thaliana, yeast, and E. coli) has shown that CPDase activity depends on the presence of HXTX motifs (Hofmann et al. 2002).
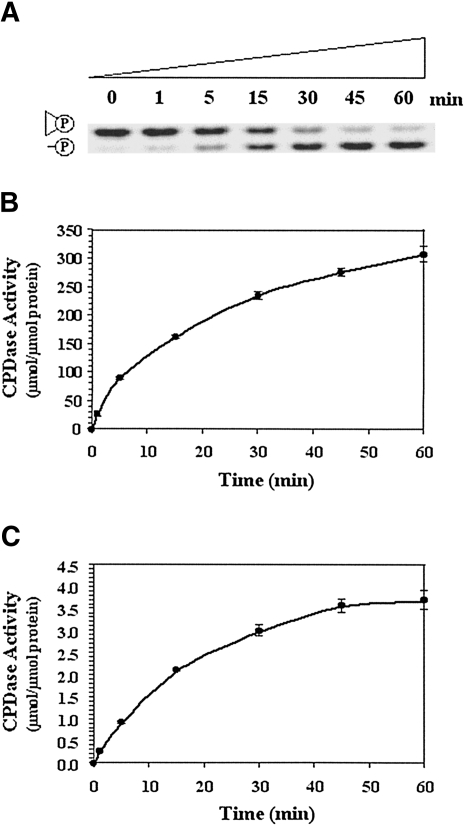
FIGURE 1.
Time-course analyses of cyclic nucleotide phosphodiesterase (CPDase) activities of PF0027 and ligT proteins. (A) Representative time-course analysis of CPDase activity on 8 M urea–20% PAGE gel. The reaction was carried out with 125 ng/mL purified PF0027 protein at 60°C using the FAM-R8 probe digested by RNase T1. Quantitative plots of the time-course analysis for (B) PF0027 protein and (C) ligT protein are shown. Data shown are mean ± SEM of three experiments.
To obtain large amounts of PF0027 and ligT proteins for biochemical and biophysical study, we overexpressed recombinant C-terminal His-tagged PF0027 and ligT proteins in E. coli and purified the protein to near-homogeneity. The purified His-tagged recombinant PF0027 and ligT proteins had molecular masses of ∼22 and 20 kDa, respectively, as determined by SDS-polyacrylamide gel electrophoresis (Supplemental Fig. 3). We used a ribonuclease T1-digested tetraribonucleotide (see Supplemental Fig. 1b,c) in order to further characterize the CPDase activity of the PF0027 protein. We conducted a CE-TOFMS study of the substrate oligoribonucleotides before and after incubation with the PF0027 protein. In the elution profile, we detected two close peaks that appeared to correspond to the two bands (positions II and III) on the 20% polyacrylamide gel containing 8 M urea shown in Supplemental Figure 1c. The molecular masses of the substrates corresponding to these peaks were 1285.1666 and 1303.1771, respectively, and the relative amounts of these peaks had traded places (Supplemental Table 1). These results show that the difference between the molecular masses of the two substrates was ∼18 and suggest that the former substrate was r(CAUG>p), where >p refers to the 2′,3′-cyclic phosphate group, and that the latter substrate was the hydrolyzed form, r(CAUG-2′-p).
To determine whether a 2′ or 3′ phosphate group at the 3′ terminus of an RNA oligomer was produced by the PF0027 protein, we next investigated the structure of the reaction products by NMR. Most of the non-exchangeable 1H and 31P resonances were assigned successfully. For the reference sample (Supplemental Fig. 4a), the resonances at 5.6 and 5.4 ppm were derived from H2′ and H3′ of G4 of r(CAUG>p), respectively. The HP-COSY experiment showed the connectivity between a low-field-shifted phosphorus resonance (17.5 ppm) and both the H2′ and H3′ resonances, confirming the formation of a 2′,3′-cyclic phosphate group at G4. In the case of the PF0027-treated sample (Supplemental Fig. 4b), although the H3′ resonance overlapped with the water signal (4.8 ppm), the HP-COSY experiment showed that the H2′, H3′, and P resonances were up-field shifted and a 2′-phosphate group had formed. Thus, the PF0027 protein was found to cleave the 2′,3′-cyclic phosphate group to produce a 2′-phosphate group at the 3′ terminus of an RNA oligomer.
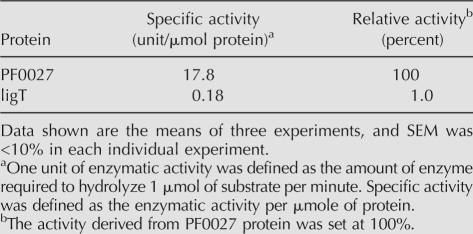
We also found that purified ligT protein possessed the same activity (Fig. 1C). However, the specific activity at the optimal temperature of the PF0027 protein (60°C) was 100 times that of the ligT protein (22°C) (Table 1). As the CPDase activity of the PF0027 protein was strong over a wide range of temperatures (37°–70°C) (data not shown), the 100-fold difference between the specific activities of the two enzymes is not likely to be due to using a suboptimal temperature for PF0027 (Supplemental Fig. 5). Moreover, the PF0027 protein was still active at 95°C, whereas activity of the ligT protein was lost at high temperatures, indicating that the PF0027 protein possesses highly heat-stable CPDase activity.
TABLE 1.
Comparison of cyclic nucleotide phosphodiesterase (CPDase) activities of PF0027 and ligT proteins
PF0027 protein possesses GTP-dependent tRNA ligase activity
Since the PF0027 protein has a degree of amino acid similarity to an experimentally characterized 2′-5′ RNA ligase protein, E. coli ligT, we next tested the PF0027 protein for RNA ligase activity (Fig. 2). Previous studies have shown that the ligT protein requires tRNA substrates for its ligation activity (Arn and Abelson 1996), and yeast tRNA ligase preferentially binds pre-tRNA substrates (Apostol and Greer 1991). Thus, we chose synthetic tRNA half oligoribonucleotides that corresponded to the 5′ and 3′ halves of P. furiosus tRNAtrp. We also checked the effect of terminal structures of the tRNA substrate (3′ end of 5′ half-tRNA, 2′ or 3′ phosphate or 2′,3′-cyclic phosphate; and 5′ end of the 3′ half-tRNA, 5′-OH or 5′-phosphate) on tRNA ligase activity.
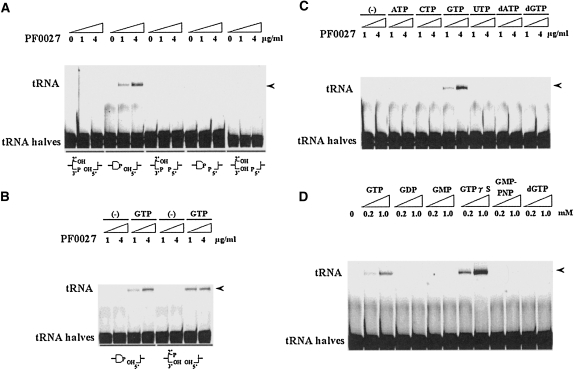
FIGURE 2.
GTP-dependent tRNA ligase activity of PF0027 protein. (A) tRNA ligase activity using synthetic tRNA halves with different terminal structures. (Bottom) A combination of the 5′- and 3′-end structures. The reaction was carried out with 1 or 4 μg/mL purified PF0027 protein for 15 min at 37°C in the presence of 1 mM GTP. (B) GTP (1 mM)-dependent tRNA ligase activity. (C) Effect of various ribonucleotides and deoxyribonucleotides (1 mM each) on tRNA ligase activity. (D) Effect of GTP-analogs on tRNA ligase (4 μg/mL) activity. Synthetic tRNA halves bearing 2′,3′-cyclic phosphate and 5′-OH termini were used for Experiments C and D. (Arrowheads) Indicate the positions of ligated product (tRNA).
As shown in Figure 2, A and B, tRNA substrates containing tRNA halves with 2′,3′-cyclic phosphate or 2′ phosphate at the 3′ end and -OH at the 5′ end were appropriate substrates for ligation by PF0027. The ligation reaction was completely dependent on the addition of GTP to the reaction mixture. Although we could not determine the chemical linkage at the ligation junction directly (discussed below), the results suggested that the PF0027 protein possesses 2′-5′ tRNA ligase activity. This is supported by the observation that the PF0027 protein is similar to the known 2′-5′ RNA ligase protein from E. coli, ligT, and by the data presented here showing that the PF0027 protein can cleave the 2′,3′-cyclic phosphate group to produce a 2′-phosphate group at the 3′-terminus of an RNA oligomer. Since ligT did not require any ribonucleotide for the RNA ligation reaction, we further checked the effect of several ribonucleotides and deoxyribonucleotides on the tRNA ligation activity of the PF0027 protein in vitro and found that GTP is specific and indispensable for the tRNA ligation reaction (Fig. 2C): ATP, CTP, UTP, dATP, and dGTP cannot substitute for GTP. Interestingly, GTPγS, a GTP analog, enhanced the RNA ligation activity, while GMPPNP, another GTP analog, did not. Neither GDP nor GMP enhanced the ligation activity (Fig. 2D). Since it is reported that γ-S NTPs are capable of serving as a substrate for nucleotide hydrolysis (Shuman et al. 1980; Peck and Herschlag 2003), we further conducted a trace experiment using fluorescence-labeled GTP (Mant-GTP) and confirmed that GTP hydrolysis did not occur during the ligation step (although we detected almost the same ligation activity as GTP). The observation was also confirmed in our NMR experiment (data not shown). Thus, these results suggested that the GTP structure is important but GTP hydrolysis is not required for the RNA ligation reaction by the PF0027 protein. Using our in vitro system, we confirmed that purified ligT protein possessed tRNA ligase activity (Arn and Abelson 1996); however, the presence of an auxiliary ribonucleotide was not required for tRNA ligase activity. Next, we checked if GMPPNP or GDP has any inhibitory effect on the ligation activity of PF0027 protein in the presence of GTP or GTPγS (Fig. 3). For this purpose, we first titrated the amount of GTP or GTPγS and calculated their apparent K m values for the ligation reactions as ∼0.5 mM each, respectively. Competition experiments with GMPPNP or GDP showed that >60% of the ligation product was still detected even when the same amounts of competitors were used. These results suggested that the ligation step is not regulated by the GTP–GDP switching mechanism.
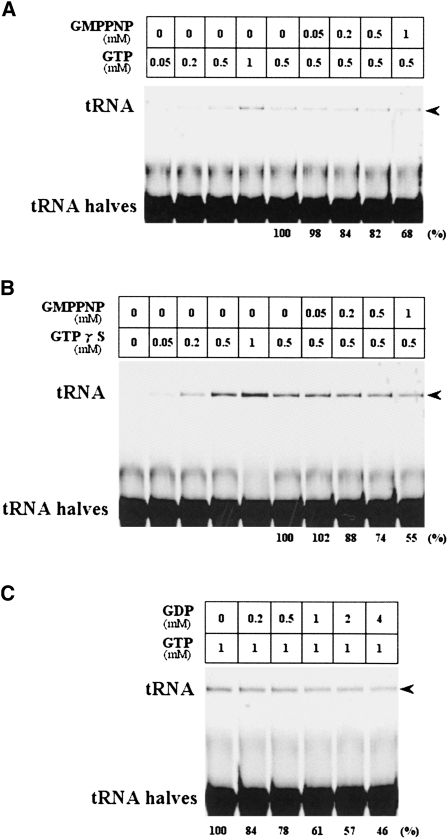
FIGURE 3.
Effect of GMPPNP and GDP on tRNA ligation activity of PF0027 protein. The effect of GMPPNP on (A) GTP-dependent and (B) GTPγS-dependent tRNA ligation activities and the effect of (C) GDP on GTP-dependent tRNA ligation activity were examined. The reaction was carried out with 4 μg/mL purified PF0027 protein for 15 min at 37°C with the indicated concentration of GMPPNP, GTP, or GDP. Synthetic tRNA halves bearing 2′,3′-cyclic phosphate and 5′-OH termini were used for all the experiments. (Arrowheads) Indicate the positions of ligated product (tRNA). (Bottom) For competition experiments, the relative amounts (%) of ligation products are indicated. Similar results were obtained from at least two independent experiments.
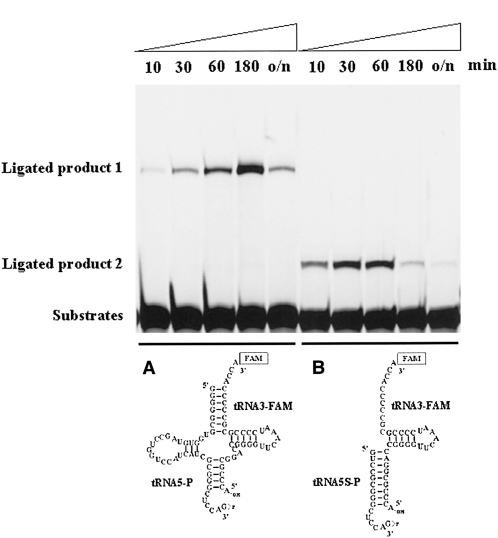
Because the amount of ligation product produced by the PF0027 protein was quite low, we characterized the equilibrium of the RNA ligation reaction (Fig. 4). Time-course analysis of the ligation reaction indicated that the PF0027 protein possessed both forward and reverse (and/or nuclease) activities. In fact, we detected degradation products of the substrate oligoribonucleotide after incubation with PF0027 protein (data not shown). This observation was consistent with a previous report of the characteristics of the ligT protein in E. coli (Arn and Abelson 1996). Therefore, the PF0027 protein may perform both ligation and cleavage in vitro, just like the ligT protein did. Interestingly, the peak of the ligation product in time course analysis was dependent on the substrate structures. For the wild-type tRNA substrate, the peak was 180 min after the start of incubation (Fig. 4A), while for the 5′ deletion mutant of the tRNA substrate, the peak was between 30 and 60 min after the start of incubation (Fig. 4B). Since the PF0027 protein could ligate the 5′ deletion mutant substrate, we further restricted the ligation substrate to determine the specificity. The results shown in Figure 5A indicated that a small stem–loop structure substrate mimicking the anticodon loop of the tRNA was sufficient for a detectable amount of the ligation product. It is of note that the ribonucleotide sequences in this substrate followed the consensus sequences found in the tRNAs (Marck and Grosjean 2002). For example, in Figure 5A, the fifth ribonucleotide from the 3′ end of the 5′ half-substrate should be a pyrimidine base (C or U), and the last ribonucleotide of the 5′ half-substrate should not be a C base. When mutations were introduced into the ribonucleotides in the substrates, the ligation efficiency was decreased (Fig. 5B,C). Especially, the two ribonucleotide replacements shown in Figure 5C markedly reduced the RNA ligation activity. These results suggest that the consensus sequences in the tRNA anticodon loop region are important for the efficiency of ligation by the PF0027 protein. Based on all these results, it is suggested that the PF0027 protein may be involved in tRNA-related role(s), but the protein alone is not an exact candidate for the tRNA ligase in archaea (discussed below).
FIGURE 4.
Time course of forward and reverse RNA ligation reaction of PF0027 protein. (A,B) Two different substrates bearing 2′,3′-cyclic phosphate and 5′-OH termini were used for the ligation reaction. The reaction was carried out with 1 μg/mL purified PF0027 protein at 37°C in the presence of 1 mM GTP.
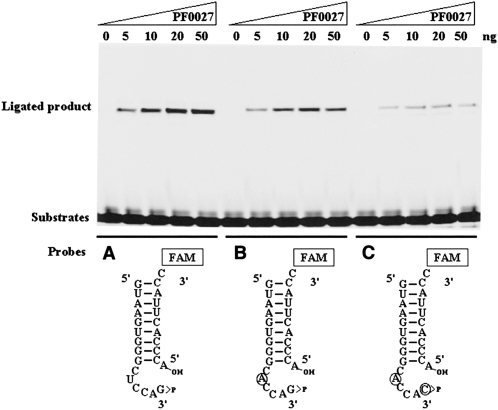
FIGURE 5.
Consensus ribonucleotide sequences in tRNA anticodon loop structure are required for ligation reaction by PF0027 protein. Three different substrates bearing 2′,3′-cyclic phosphate and 5′-OH termini were used for the ligation reaction. The reaction was carried out with 0–50 ng of purified PF0027 protein for 15 min at 37°C in the presence of 1 mM GTPγS. (Circles) Mutated ribonucleotides. The substrates used were (A) tRNA5S-P and tRNA3S-FAM, (B) tRNA5SM1-P and tRNA3S-FAM, and (C) tRNA5SM2-P and tRNA3S-FAM.
Solution structure and interaction of PF0027 protein
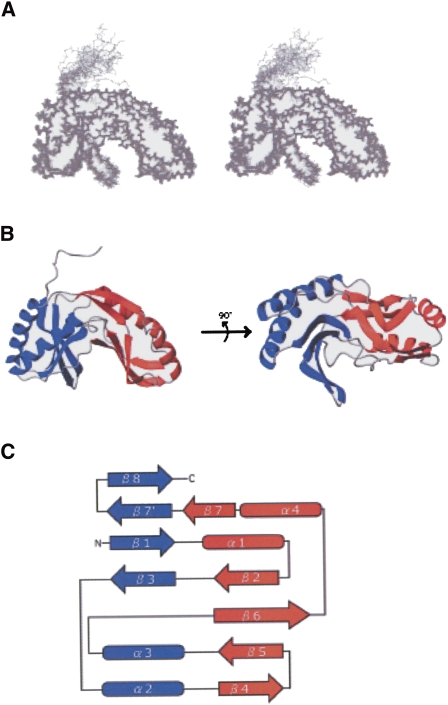
In order to examine the structure–function relationship, the solution structure and interaction with oligonucleotide as well as with GTP were analyzed by NMR methods. Using standard triple-resonance assignment methodology, backbone assignments could be made for almost all residues (except for 15N of Ile100; 15N and 13Cα of Ser10; and 13CO of Arg2, Gly79, and Ile100), as described previously (Okada et al. 2006). A total of 3615 NOE, 220 dihedral angles, and 10 hydrogen bond restraints were obtained from the NMR data, and 20 structures converged to low total energy (Fig. 6A). These 20 structures were well defined, with a heavy atom and backbone root-mean-square deviation (RMSD) of 1.32 ± 0.17 Å and 0.68 ± 0.16 Å, respectively (Supplemental Table 2). The converged structures did not contain experimental distance violation of >0.3 Å or dihedral violation of >5°. The convergence was further assessed by the (α, α) spacing for all selected structures. In a Ramachandran plot, the backbone dihedral angles for all the residues of the final converged structures fell either in the most favored region or in the additional allowed region.
FIGURE 6.
Tertiary structure of PF0027 protein. (A) Stereo view of superposition of final 20 structures obtained by NMR structure determination. (B) Ribbon diagram. (Blue) The terminal lobe; (red) the transit lobe. (C) Schematic diagram of secondary structure topology. (Arrows) Secondary structure elements (β-strands) and cylinders (α-helices).
The solution structure of the PF0027 protein included four α-helices and nine β-strands, and was characterized by two lobes with similar architecture (Fig. 6B,C). Following the previously established nomenclature, the lobe containing the N- and C-terminal regions was referred to as the terminal lobe (blue in Fig. 6), whereas the other lobe was referred to as the transit lobe (red in Fig. 6). The terminal lobe consisted of an antiparallel β-sheet (β 1, 3, 8, and 9) and two α-helices (α 2 and 3), and the transit lobe also consisted of an antiparallel β-sheet (β 2, 4, 5, 6, and 7) and two α-helices (α 1 and 4). The antiparallel β-sheets came together to form a β-barrel. A putative active site that included the two HXTX motifs existed in a cavity at the center of the β-barrel. Moreover, there were four helices on the outside of each lobe. The relationship between the secondary structure elements and the sequence is shown in Supplemental Figure 2.
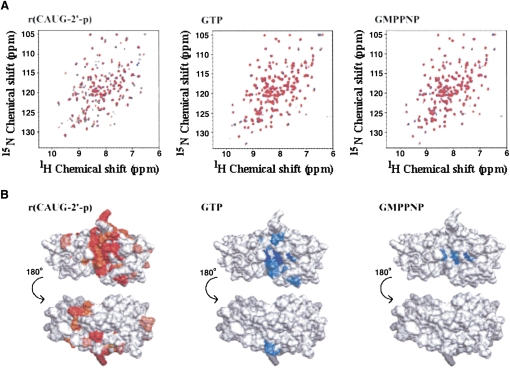
In order to analyze the interaction between the PF0027 protein and its substrate RNA, uniformly 13C- and 15N-labeled PF0027 was titrated against the r(CAUG-2′-p) oligoribonucleotide. Upon addition of the oligoribonucleotide, some of the amide 1H and 15N resonances showed a chemical shift change (Fig. 7A, left panel), and, thus, the amino acid residues of PF0027 involved in binding of the oligoribonucleotide could be assigned. Residues of the PF0027 protein could be classified into four categories: 1, disappearance of resonances; 2, large (>0.05); 3, moderate (0.02–0.05 ppm) changes in average amide chemical shifts; and 4, small or no change (<0.02 ppm) in chemical shift. Unassigned residues, mainly due to signal overlap, were included in the no change category. The locations of perturbed residues in the structure of PF0027 are shown in red in Figure 7B (left panel). Furthermore, uniformly 15N-labeled PF0027 protein was titrated against GTP or GMPPNP, and the results are also shown in Figure 7 (middle and right panels, respectively). Most of the perturbed residues were located at the periphery of the two HXTX motifs. In the case of GTP as well as GMPPNP, perturbed residues were located only in the vicinity of the HXTX motifs. The magnitude of perturbation for GMPPNP was significantly smaller than that for GTP.
FIGURE 7.
Analysis of interaction between PF0027 protein and substrates. (A) HSQC spectra of PF0027 protein. For r(CAUG-2′-p), uniformly 13C-labeled and 15N-labeled PF0027 protein was used at a concentration of 0.1 mM. For GTP or GMPPNP, uniformly 15N-labeled PF0027 protein was used at a concentration of 0.3 mM. The molar ratios of PF0027 protein and substrates were 0, 0.5, and 1 for r(CAUG-2′-p) and 0, 1, and 3 for GTP and GMPPNP as indicated by black, blue, and red, respectively. (B) Chemical shift changes of NH signals mapped on PF0027 protein structures. For r(CAUG-2′-p) or GTP/GMPPNP, the residues whose signals disappeared are shown in dark red or blue, respectively. Large (>0.05 ppm) and moderate (0.02–0.05 ppm) average amide chemical shifts (Δδav) are indicated by the brightness of the red or blue color of each residue. (White) Residues showing small (<0.02 ppm) or no changes. Changes in the average amide chemical shifts were calculated using Δδav = { 0.5 [(0.2ΔδN)2 + ΔδH2]}½, where ΔδN and ΔδH are the amide nitrogen and amide proton chemical shift differences between the free and bound states of the protein.
DISCUSSION
In this study, we present the biochemical and structural characterization of the PF0027 protein. As the PF0027 protein could ligate synthetic tRNA halves with a cyclic phosphate at the 3′ end and –OH at the 5′ end, tRNA ligation appears to be markedly dependent on the substrate terminal structures. However, the RNA ligation activity itself was low as compared to the activity of the ligT protein (data not shown). Since we used synthetic tRNA substrates, there were no base modifications. E. coli 2′-5′ RNA ligase required tRNA half-molecules containing nucleotide base modifications (Arn and Abelson 1996), suggesting that base modifications in pre-tRNA molecules are also important for the ligation step in archaea. In addition to this, we detected a reversible reaction (and/or nuclease activity) mediated by the PF0027 protein. The combination of these factors may produce low amounts of ligation product. Because of this, we could not determine the chemical linkage at the junction. A huge amount of background derived from unligated substrates obscured the junction determination in several experiments (data not shown).
In eukaryotes, the yeast tRNA splicing endonuclease cleaves pre-tRNA, resulting in an intron and two tRNA half-molecules bearing 2′-3′ cyclic phosphate and 5′-OH termini (Trotta et al. 1997). The tRNA ligase is reported to be a multifunctional enzyme with cyclic phosphodiesterase activity (CPDase activity), NTP-dependent polynucleotide kinase activity, and ATP-dependent RNA-ligase activity (Sawaya et al. 2003; Wang and Shuman 2005). Comparable characteristics were reported for a plant tRNA ligase (Englert and Beier 2005). Similarly, the CPDase activity of the PF0027 protein may open the 2′-3′ cyclic phosphate of tRNA after digestion by archaeal tRNA endonuclease. Looking at the amino acid sequence of archaeal and bacterial F0027 protein family proteins, however, the presence of an NTP-dependent polynucleotide kinase domain similar to that found in eukaryotes is not obvious. Moreover, eukaryotic tRNA ligase produced a standard 3′,5′-phosphodiester linkage in mature tRNA. However, our present study suggested that P. furiosus possesses more bacterial type 2′-5′ tRNA ligase. The GTP dependency of the ligation reaction is another novel feature of the PF0027 protein. Eukaryotic tRNA ligase also requires either ATP or GTP for the ligation reaction. In this case, the eukaryotic tRNA ligase transfers the γ-phosphate of an NTP cofactor to the donor 5′ end of the tRNA exon (Wang and Shuman 2005), whereas E. coli 2′-5′ tRNA ligase is not required by the NTP cofactor. Our present study indicated that the archaeal PF0027 protein shows intermediate features between bacteria and eukaryotes, in that the enzyme requires the cofactor GTP, but not the transfer of γ-phosphate.
We considered that the 2′-5′ linkage in the splicing junction site is not appropriate for providing functional tRNAs in the cell. In fact, the ligation junction was quite unstable during in vitro reaction mediated by the PF0027 protein. Since we have characterized the tRNA ligation reaction using purified PF0027 protein, it is also possible that another factor(s) may be required for the stable formation of the ligation junction, and possibly forms the 3′-5′ linkage. Recently, Weitzer and Martinez (2007a,b) reported that the human RNA kinase hClp1 phosphorylated not only the 5′ end of short interfering RNAs (siRNAs) but also the 5′ end of the 3′ exon during human tRNA splicing. They showed that silencing by the siRNA against this enzyme impaired mature tRNA formation. We have screened the proteome in P. furiosus and found that the PF0112 gene encodes a putative RNA kinase that has a degree of similarity to hClp1 (data not shown). Furthermore, in the P. furiosus genome, one of the genes that neighbors PF0027, PF0026, encodes a putative tRNA nucleotidyltransferase or CCA-adding enzyme (NCBI protein database accession number NP_577755). An E. coli tRNA nucleotidyltransferase has been reported to possess three different activities: 2′,3′-cyclic phosphodiesterase activity, 2′-nucleotidase activity, and phosphatase activity (Yakunin et al. 2004). Based on the report and our present observations, the genomic DNA regions of PF0026 and PF0027 may correspond to a tRNA-processing pathway, including tRNA splicing and repair of tRNA breakage, in this organism. Because a genetic engineering system for P. furiosus is not yet available, it is also true that analysis of PF0027 protein function in vivo remains very difficult now.
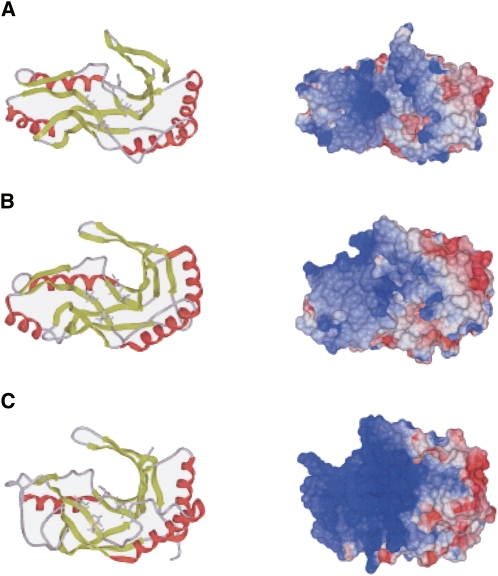
Alignment of PF0027 protein family member sequences revealed that the proteins are approximately 200 amino acid residues long and are well conserved between bacteria and archaea (Supplemental Fig. 2). Comparison of the 3D structures of PF0027-related proteins from T. thermophilus (PDB entry 1IUH) (Kato et al. 2003), P. horikoshii (PDB entries 1VDX and 1VGJ) (Rehse and Tahirov 2005), and P. furiosus (this study) showed that the global folds in these proteins are similar to one another, although α-helix 4, which makes up the C-terminus lobe in PF0027 and the PF0027-related protein from P. horikoshii, is absent in the PF0027-related protein from T. thermophilus (Fig. 8). Furthermore, the local conformations of the HXTX motifs are similar to one another, and the electrostatic surface potentials are also similar (Fig. 8), indicating that the catalytic properties of these PF0027-related proteins are also similar. On the other hand, there are local structural differences in the C-terminal β 7′ and β 8 regions. In fact, the backbone RMSD between PF0027 and the PF0027-related protein from P. horikoshii (PDB entry 1VDX) is 2.23 Å for residues 1 to 184 (the backbone RMSD for residues 1–160 is 1.55 Å). The structure of the β 7′ and β 8 regions did not converge well (data not shown). Furthermore, the B-factor of this region is large in the crystal structure. These observations suggested that the β 7′ and β 8 regions are highly flexible. Moreover, chemical shift perturbation analysis of the PF0027 protein using the r(CAUG-2′-p) oligoribonucleotide suggested that the tRNA binding site is located at the 9 β-strand cleft (Fig. 7), at the base of the HXTX motifs (His40-Leu43 and His125-Leu128). In the case of GTP, most of the perturbed residues were located near the HXTX motifs, suggesting direct involvement of GTP in the reaction. The magnitude of perturbation for GMPPNP is significantly smaller than that for GTP, indicating that GMPPNP binds the PF0027 protein with low affinity, probably due to the difference in conformation and/or charge distribution between GTP and GMPPNP. These data are consistent with the result that GMPPNP did not enhance the ligation activity. Although Kato et al. suggested that the large positive cleft of the PF0027-related proteins from T. thermophilus is essential for recognition of the large negatively charged tRNA molecule, the active site cleft of the PF0027 protein is both deeper and more narrow than that of the PF0027-related proteins from T. thermophilus (Kato et al. 2003; Rehse and Tahirov 2005). Thus, conformational change in the β 7′ and β 8 regions is likely to be required for effective recognition of the tRNA halves. In the crystal structure of a complex of CPDase from Arabidopsis thaliana and 2′,3′-cyclic uridine vanadate, there are two nucleoside-binding sites: the abovementioned active site and the second binding site, which is located outside of the cleft and close to the β 7′ and β 8 regions (Hofmann et al. 2002). Figure 7 shows that the region corresponding to the second binding site is affected by the addition of r(CAUG-2′-p). Together with the fact that the β 7′ and β 8 regions are flexible, the cleft including the second binding site forms a large platform for the oligonucleotide ligands. NMR titration of CPDase from goldfish with 3′-AMP showed that only the active site was affected (Kozlov et al. 2007), probably because the affinity of GTP for PF0027 protein is higher than that of 3′-AMP for CPDase from goldfish.
FIGURE 8.
Comparison of tertiary structure, (left) HXTX motif, and (right) electrostatic surface potential of PF0027 and two structurally related proteins. (A) PF0027 (PDB entry 2FYH), (B) PF0027-related protein from Pyrococcus horikoshii (PDB entry 1VDX), and (C) PF0027-related protein from Thermus thermophilus (PDB entry 1IUH). (Red) The α-helices and (light green) β-strands. The histidine and threonine residues in the HXTX motif are shown in stick model format. Images were produced by ICM Pro (Molsoft Inc.). Another structure for the PF0027-related protein from P. horikoshii (PDB entry 1VGJ) is essentially the same as the 1VDX structure shown.
MATERIALS AND METHODS
Screen for oligoribonucleotide migration activity using P. furiosus protein library, construction of expression vectors, and expression of recombinant protein
A protein library of P. furiosus DSM3638 was constructed using the expression vector pRSET-A (Invitrogen) as reported previously (Kanai et al. 2003). We made 40 protein pools (P1–P40), with each pool consisting of 20 independent lysates from E. coli bacteria expressing the P. furiosus clones. A total of 800 independent clones were screened, a number expected to cover the 2-Mb genome of P. furiosus. The protein pools were screened using an oligoribonucleotide probe (see Supplemental Fig. 1). A protein was considered positive in the assay if it changed the migration of the oligoribonucleotide probe in polyacrylamide gel containing 8 M urea.
To construct deletion mutants based on the positive clone pAL485, a Sau3AI–PstI 4.4-kb fragment and a PstI–Sau3AI 6.2-kb fragment from the original clone were ligated separately into the pRSET-A plasmid vector. To purify proteins corresponding to each of these, we constructed expression plasmids (pET-PF0027 through pET-PF0030) as previously described (Sato et al. 2003). Each of the resulting vectors encodes a full-length protein with a six-histidine tag at its C-terminal end. Similarly, in order to obtain an expression vector that encodes the ligT protein (formerly, yadP; NCBI protein database accession number NP_414689), genomic DNA from E. coli W3110 was prepared and then used as a template to clone ligT by PCR. Recombinant proteins were prepared as described previously (Kanai et al. 2006).
The primers used for PCR cloning were as follows:
27S, 5′-d(ACACATATGAGGGCGTTTATAGCTATAGACG)-3′;
27A, 5′-d(ATTCTCGAGTTCGCTTAATTCGAATCTTGC)-3′;
28S, 5′-d(ACCCATATGGAATACACAATAGTTGAC)-3′;
28A, 5′-d(ATCCTCGAGGATAGGCTTTTCCATTATCAGTG)-3′;
29S, 5′-d(CCGCATATGCTTCTCAAGTGCTTTGAC)-3′;
29A, 5′-d(TCACTCGAGACTATTGTGTATTCCATGGAGG)-3′;
30S, 5′-d(TTGCATATGGAAAAGGCCCTAATCCTAG)-3′;
30A, 5′-d(ATGCTCGAGAAGCACTTGAGAAGCATTTACG)-3′;
ligTS, 5′-d(CGACATATGTCTGAACCGCAACGTC)-3′; and
ligTA, 5′-d(CGACTCGAGTTGCGTTAGCGCCCAGCG)-3′.
Protein ID (NCBI protein accession number): PF0027 (NP_577756), PF0028 (NP_577757), PF0029 (NP_577758), and PF0030 (NP_577759).
Cyclic nucleotide phosphodiesterase (CPDase) assay
RNA band-shift activity or cyclic nucleotide phosphodiesterase (CPDase) activity was assayed by analyzing the FAM-labeled oligoribonucleotide probe on a 20% polyacrylamide gel containing 8 M urea. Basically, the 5′-end FAM-labeled oligoribonucleotide, FAM-R8 [5′-FAM-r(CAUGUCAC)-3′], was chemically synthesized by Hokkaido System Science and digested with ribonuclease T1 for 15 min at 22°C (2 units of enzyme was used per 700 pmol of FAM-R8 probe) to make a FAM-labeled tetramer 5′-FAM-r(CAUG)-3′ with cyclic guanosine at the 3′-end (Zegers et al. 1994). Reactions were performed in 20 μL of reaction buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 50 mM KCl, 1 mM MgCl2, 35 pmol of substrate, and purified recombinant enzymes (0.05–2.5 μg/mL for PF0027 protein and 5–25 μg/mL for ligT protein), or E. coli lysate expressing P. furiosus genomic DNA. Samples were incubated for 15 min at 60°C (PF0027 protein) or at 22°C (ligT protein). Next, an equal volume of stop solution was added (8 M urea, 1 M Tris-HCl at pH 7.2) along with a small amount of blue dextran (Sigma Chemical). The reaction mixtures were heated for 3 min to 70°C and cooled on ice, and then loaded onto 20% polyacrylamide gel containing 8 M urea. So that kinetic parameters could be determined, the amount of enzyme was controlled such that the cyclic phosphate to liberalized phosphate exchange rate at the 3′ end of the substrate did not exceed 30%.
RNA ligase assay
The tRNA half oligoribonucleotides were chemically synthesized by Hokkaido System Science. The oligoribonucleotide sequence was based on tRNAtrp in P. furiosus, which possesses a single intron in the anticodon loop region. The intronic region was identified and removed in silico using the SPLITS program (Sugahara et al. 2006). Typically, preparation of substrates and ligation reactions were conducted as follows: We treated the tRNA5-P oligoribonucleotide with recombinant PF1549 protein, a P. furiosus RNA-3′-phosphate cyclase, to generate a 2′,3′-cyclic phosphate in the 3′ terminus (data not shown). To generate the 2′-phosphate in the 3′ terminus, we further treated the tRNA5 oligo having a 2′,3′-cyclic phosphate with the PF0027 protein. In some cases, the 5′ terminus of the tRNA3-FITC oligoribonucleotide was phosphorylated by T4 polynucleotide kinase (Takara Bio. Inc.). These oligoribonucleotides were purified by CHROMA SPIN+TE-10 Column (Clontech) and used for the ligation assay. After annealing the two half oligoribonucleotides, a ligation reaction was performed in 20 μL of reaction buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 50 mM KCl, 1 mM MgCl2, 33 pmol of substrate, and 1 or 4 μg/mL purified recombinant enzyme in the presence of 1 mM GTP. Samples were incubated for 15 min at 37°C (PF0027 protein) or at 22°C (ligT protein). The ligation products were analyzed as described for the CPDase assay.
The oligoribonucleotides used were as follows (mutated ribonucleotides are underlined):
tRNA5-P, 5′-r(GGGGGCGUGGUGUAGCCUGGUCCAUCAUCGCGGGCUCCAG)-phosphate-3′;
tRNA5S-P, 5′-r(GUCCGCGGGCUCCAG)-phosphate-3′;
tRNA5SM1-P, 5′-r(GUCCGCGGGCACCAG)-phosphate-3′;
tRNA5SM2-P, 5′-r(GUCCGCGGGCACCAC)-phosphate-3′;
tRNA3-FAM, 5′-r(ACCCGCGGACCGGGGUUCAAAUCCCCGCGCCCCCACCA)-FAM-3′; and
tRNA3S-FAM, 5′-r(ACCCACUUACC)-FAM-3′.
CE-TOFMS analysis
Capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) was performed using an Agilent CE capillary electrophoresis apparatus (Agilent Technologies) with an Agilent G3250AA LC/MSD TOF system (Agilent Technologies). A cationic polymer-coated SMILE (+) capillary was used as the separation capillary. The electrolyte for CE separation was 50 mM ammonium acetate (pH 8.5) (Soga et al. 2002). After digestion of the R9 oligoribonucleotide [5′-r(CAUGUCACU)-3′] with ribonuclease T1, the resulting substrate (2.5 nmol) was incubated in 100 mL of reaction buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 50 mM KCl, and 1 mM MgCl2, with or without purified recombinant PF0027 protein (10 μg) for 15 min at 50°C. The reaction mixtures were passed through an Ultrafree-MC 5000 NMWL filter (Millipore) and injected at an injection pressure of 50 mbar for 30 sec.
Solution structure determination and NMR titration with oligonucleotide and nucleotides
13C-labeled and 15N-labeled PF0027 protein was generated in an E. coli expression system and purified by Ni2+ affinity chromatography. The NMR spectrum of the PF0027 protein is characterized by fine dispersion at higher temperature. Thus, all NMR experiments were preformed at 313 K. NMR for the PF0027 protein was performed on AVANCE 700 and 800 MHz spectrometers (BRUKER Biospin). NMR spectra were analyzed using several programs. The experimental details were described previously (Okada et al. 2006). The 1H, 13C, and 15N chemical shift assignments for the PF0027 protein have been deposited in the BioMagnResBank (Accession number 10004).
For structure determination, automated NOE cross-peak assignments (Herrmann et al. 2002) and structure calculations with torsion angle dynamics were performed using the software package CYANA 2.017 (Guntert et al. 1997). Peak lists for the two NOESY spectra were generated as input using the program NMRview (Johnson and Blevins 1994). The input further contained a chemical shift list corresponding to sequence-specific assignments. Dihedral angle restraints were derived using the program TALOS (Cornilescu et al. 1999). Ten hydrogen bond restraints were used. A total of 100 structures were independently calculated, and the 20 final structures were obtained from the CYANA cycle 7 with the lowest final CYANA target function values. The structures were validated using PROCHECK-NMR (Laskowski et al. 1996).
For PF0027 protein and substrate (oligoribonucleotide, GTP, and GMPPNP) interaction analysis, 13C-labeled and 15N-labeled PF0027 protein and 15N-labeled PF0027 protein concentrations were set at 0.1 and 0.3 mM, respectively, in 20 mM sodium phosphate buffer (pH 6.5) with 200 mM sodium chloride. The r(CAUG-2′-p) oligoribonucleotide was prepared from r(CAUG>p) oligoribonucleotide via PF0027 protein activity, and was then purified by polyacrylamide gel electrophoresis. The r(CAUG-2′-p) oligoribonucleotide was added in six steps, at molar ratios {[r(CAUG-2′-p) oligoribonucleotide]/[PF0027 protein]} of 0, 0.1, 0.2, 0.5, 0.7, and 1.0. GTP or GMPPNP was added in five steps, at molar ratios ([GTP or GMPPNP]/[PF0027 protein]) of 0, 0.1, 0.5, 1.0, and 3.0. 15N–1H HSQC spectra were measured at a probe temperature of 313 K using DRX-500 spectrometers (BRUKER Biospin). The backbone assignments at pH 4.7 have been determined previously (Kato et al. 2003), and we performed pH titration experiments from pH 4.7 to pH 6.5 to determine the backbone assignments at pH 6.5 (data not shown). Changes in average amide chemical shifts were calculated using Δδav = { 0.5 [(0.2ΔδN)2 + ΔδH2]}½, where ΔδN and ΔδH are the amide nitrogen and amide proton chemical shift differences between the free and bound states of the protein.
Accession number
The Protein Data bank (http://www.rscb.org/pdb) accession number for the coordinates of PF0027 protein is 2FYH.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Takamasa Ishikawa and Tomoyoshi Soga (Keio University) for their technical assistance with CE-TOFMS analyses and N. Kobayashi (RIKEN GSC) for allowing us to use the Kujira program for NMR data analysis. We also thank Ramesh Gupta (Southern Illinois University); Yoichi Watanabe and Shigeo Yoshinari (University of Tokyo); and Kosuke Fujishima, Yasushi Ishihama, and Yuji Kakazu (Keio University) for their helpful discussions. This research was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas, a Grant-in-Aid from the 21st Century Center of Excellence (COE) Program entitled, “Understanding and Control of Life's Function via Systems Biology (Keio University)”; research funds from the Yamagata Prefectural Government and Tsuruoka City; the RIKEN Structural Genomics/Proteomics Initiative (RSGI); and the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
Footnotes
Abbreviations: CPDase, cyclic nucleotide phosphodiesterase; NCBI, National Center for Biotechnology Information; PDB, protein data bank; FAM, 6-carboxyfluorescein; CE-TOFMS, capillary electrophoresis-time of flight mass spectrometry; HSQC, heteronuclear single quantum coherence spectroscopy; DQF-COSY, double quantum filtered-correlation spectroscopy; TOCSY, total correlation spectroscopy; HP-COSY, proton phosphorus-correlation spectroscopy.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1122109.
REFERENCES
- Achenbach-Richter L., Gupta R., Stetter K.O., Woese C.R. Were the original eubacteria thermophiles? Syst. Appl. Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- Apostol B.L., Greer C.L. Preferential binding of yeast tRNA ligase to pre-tRNA substrates. Nucleic Acids Res. 1991;19:1853–1860. doi: 10.1093/nar/19.8.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arn E.A., Abelson J.N. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J. Biol. Chem. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- Brown J.R., Douady C.J., Italia M.J., Marshall W.E., Stanhope M.J. Universal trees based on large combined protein sequence data sets. Nat. Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Di Giulio M. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J. Mol. Evol. 2003;57:721–730. doi: 10.1007/s00239-003-2522-6. [DOI] [PubMed] [Google Scholar]
- Englert M., Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Fujishima K., Komasa M., Kitamura S., Suzuki H., Tomita M., Kanai A. Proteome-wide prediction of novel DNA/RNA-binding proteins using amino acid composition and periodicity in the hyperthermophilic archaeon Pyrococcus furiosus . DNA Res. 2007;14:91–102. doi: 10.1093/dnares/dsm011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2006. [Google Scholar]
- Guntert P., Mumenthaler C., Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- Hofmann A., Tarasov S., Grella M., Ruvinov S., Nasr F., Filipowicz W., Wlodawer A. Biophysical characterization of cyclic nucleotide phosphodiesterases. Biochem. Biophys. Res. Commun. 2002;291:875–883. doi: 10.1006/bbrc.2002.6527. [DOI] [PubMed] [Google Scholar]
- Johnson B., Blevins R. NMRView: A computer program for the visualization and analysis of NMR data. J. Biol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kanai A., Oida H., Matsuura N., Doi H. Expression cloning and characterization of a novel gene that encodes the RNA-binding protein FAU-1 from Pyrococcus furiosus . Biochem. J. 2003;372:253–261. doi: 10.1042/BJ20021968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai A., Sato A., Imoto J., Tomita M. Archaeal Pyrococcus furiosus thymidylate synthase 1 is an RNA-binding protein. Biochem. J. 2006;393:373–379. doi: 10.1042/BJ20050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Shirouzu M., Terada T., Yamaguchi H., Murayama K., Sakai H., Kuramitsu S., Yokoyama S. Crystal structure of the 2′-5′ RNA ligase from Thermus thermophilus HB8. J. Mol. Biol. 2003;329:903–911. doi: 10.1016/s0022-2836(03)00448-0. [DOI] [PubMed] [Google Scholar]
- Kozlov G., Denisov A.Y., Pomerantseva E., Gravel M., Braun P.E., Gehring K. Solution structure of the catalytic domain of RICH protein from goldfish. FEBS J. 2007;274:1600–1609. doi: 10.1111/j.1742-4658.2007.05707.x. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Marck C., Grosjean H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C., Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Matsuda T., Sakamoto T., Muto Y., Yokoyama S., Kanai A., Kawai G. 1H, 13C, and 15N resonance assignments of the 2′-5′ RNA ligase-like protein from Pyrococcus furiosus . J. Biomol. NMR. 2006;36(Suppl):16. doi: 10.1007/s10858-005-5581-8. [DOI] [PubMed] [Google Scholar]
- Peck M.L., Herschlag D. Adenosine 5′-O-(3-thio)triphosphate (ATPγS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA. 2003;9:1180–1187. doi: 10.1261/rna.2103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehse P.H., Tahirov T.H. Structure of a putative 2′-5′ RNA ligase from Pyrococcus horikoshii . Acta Crystallogr. D Biol. Crystallogr. 2005;61:1207–1212. doi: 10.1107/S0907444905017841. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y., Tanaka N., Ichimiya T., Kurihara T., Nakamura K.T. Crystal structure of the catalytic fragment of human brain 2′,3′-cyclic-nucleotide 3′-phosphodiesterase. J. Mol. Biol. 2005;346:789–800. doi: 10.1016/j.jmb.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Sato A., Kanai A., Itaya M., Tomita M. Cooperative regulation for Okazaki fragment processing by RNase HII and FEN-1 purified from a hyperthermophilic archaeon, Pyrococcus furiosus . Biochem. Biophys. Res. Commun. 2003;309:247–252. doi: 10.1016/j.bbrc.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Sawaya R., Schwer B., Shuman S. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003;278:43928–43938. doi: 10.1074/jbc.M307839200. [DOI] [PubMed] [Google Scholar]
- Schut G.J., Zhou J., Adams M.W. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: Evidence for a new type of sulfur-reducing enzyme complex. J. Bacteriol. 2001;183:7027–7036. doi: 10.1128/JB.183.24.7027-7036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut G.J., Brehm S.D., Datta S., Adams M.W. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 2003;185:3935–3947. doi: 10.1128/JB.185.13.3935-3947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Spencer E., Furneaux H., Hurwitz J. The role of ATP in in vitro vaccinia virus RNA synthesis effects of AMP-PNP and ATP γ S. J. Biol. Chem. 1980;255:5396–5403. [PubMed] [Google Scholar]
- Soga T., Ueno Y., Naraoka H., Ohashi Y., Tomita M., Nishioka T. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2002;74:2233–2239. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- Sugahara J., Yachie N., Sekine Y., Soma A., Matsui M., Tomita M., Kanai A. SPLITS: A new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- Tocchini-Valentini G.D., Fruscoloni P., Tocchini-Valentini G.P. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. 2005a;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini G.D., Fruscoloni P., Tocchini-Valentini G.P. Structure, function, and evolution of the tRNA endonucleases of Archaea: An example of subfunctionalization. Proc. Natl. Acad. Sci. 2005b;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta C.R., Miao F., Arn E.A., Stevens S.W., Ho C.K., Rauhut R., Abelson J.N. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- Usui K., Katayama S., Kanamori-Katayama M., Ogawa C., Kai C., Okada M., Kawai J., Arakawa T., Carninci P., Itoh M., et al. Protein–protein interactions of the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Genome Biol. 2005;6:R98. doi: 10.1186/gb-2005-6-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.K., Shuman S. Structure–function analysis of yeast tRNA ligase. RNA. 2005;11:966–975. doi: 10.1261/rna.2170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S., Martinez J. hClp1: A novel kinase revitalizes RNA metabolism. Cell Cycle. 2007a;6:2133–2137. doi: 10.4161/cc.6.17.4642. [DOI] [PubMed] [Google Scholar]
- Weitzer S., Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007b;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Yakunin A.F., Proudfoot M., Kuznetsova E., Savchenko A., Brown G., Arrowsmith C.H., Edwards A.M. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 2004;279:36819–36827. doi: 10.1074/jbc.M405120200. [DOI] [PubMed] [Google Scholar]
- Zegers I., Haikal A.F., Palmer R., Wyns L. Crystal structure of RNase T1 with 3′-guanylic acid and guanosine. J. Biol. Chem. 1994;269:127–133. [PubMed] [Google Scholar]