Abstract
The double-stranded RNA (dsRNA) transport protein SID-1 enables systemic RNA interference (RNAi) in Caenorhabditis elegans, whereby silencing initiated by local exposure to dsRNA spreads throughout the animal and to its progeny. Previously, we showed that providing dsRNA in the growth medium of Drosophila S2 cells that express C. elegans SID-1 efficiently triggers RNAi. In these experiments long dsRNA proved to be significantly more effective than short dsRNA in silencing the target gene. Here, we show that equivalent masses of long or short dsRNA accumulate in these cells, indicating that size-dependent silencing is not due to size-selective transport through SID-1. Furthermore, using pulse-chase dsRNA uptake experiments, we show that short dsRNA accumulates more rapidly than long dsRNA. We found that import rates are dependent on dsRNA concentration, consistent with energy-independent, diffusion-limited transport through the SID-1 channel. Comparison of silencing efficiencies between Drosophila S2 cells heterologously expressing SID-1 and primary-cultured C. elegans cells shows similar dsRNA concentration and size dependencies, suggesting that C. elegans regulatory proteins do not measurably enhance or restrict dsRNA transport through SID-1. Finally, we find that coexpressing mutant SID-1 with wild-type SID-1 in S2 cells interferes with SID-1 function, indicating that SID-1 may function as a multimer.
Keywords: RNAi, dsRNA, SID-1, systemic RNAi
INTRODUCTION
In Caenorhabditis elegans, RNAi can be initiated by injection, ingestion, or expression of double-stranded RNA (dsRNA) (Fire et al. 1998; Tabara et al. 1998; Timmons and Fire 1998; Tavernarakis et al. 2000). Once dsRNA is introduced, RNA interference (RNAi) silencing spreads throughout the organism and even into its progeny (Fire et al. 1998). This systemic spread of silencing does not occur in sid-1 mutants, although silencing is observed within the cell where dsRNA is injected or expressed (Winston et al. 2002). sid-1 encodes a protein with nine predicted transmembrane domains, and SID-1 reporter expression is detected in all nonneuronal cells (Winston et al. 2002; Feinberg and Hunter 2003). Genetic analysis shows that SID-1 is required to import silencing signals (Winston et al. 2002). This is likely an ancient conserved function as many organisms with SID-1 homologs show systemic RNAi (Bucher et al. 2002; Newmark et al. 2003; Soutschek et al. 2004; Turner et al. 2006). Consistent with such a conserved function, a mammalian homolog of SID-1 has been shown to enhance siRNA uptake in human cells (Duxbury et al. 2005), while knockdown of the SID-1 homolog Sidt1 disrupts the internalization of cholesterol-modified siRNA in human hepatic cells (Wolfrum et al. 2007).
To study the molecular properties of SID-1, a heterologous assay system was developed using Drosophila S2 cells expressing C. elegans SID-1 (Feinberg and Hunter 2003). Drosophila lacks a SID-1 homolog and systemic RNAi (Roignant et al. 2003), yet Drosophila S2 cells can initiate RNAi triggered by dsRNA present in the growth medium. However, S2 cells expressing SID-1 initiate RNAi in response to at least a 100,000-fold lower dsRNA concentration and in contrast to endogenous uptake, internalize dsRNA in a rapid, largely energy-independent manner (Feinberg and Hunter 2003). These results indicate that rather than a pump or a receptor, SID-1 functions as a dsRNA pore or channel, which enables transfer of dsRNA across the cell membrane into the cytoplasm.
Here, we show using the S2 cell system, that long and short dsRNAs accumulate to similar levels indicating that the length-dependent silencing effect is independent of SID-1 function. Further supporting the lack of preferential transport of long dsRNA, analysis of dsRNA transport kinetics shows that shorter dsRNA accumulates inside cells more rapidly than longer dsRNA. We find that SID-1 expressing S2 cells and C. elegans cells have nearly identical sensitivity to dsRNA concentration and that in both systems long dsRNA causes more efficient silencing. These results suggest that C. elegans embryonic cells do not express any proteins that measurably enhance or restrict dsRNA transport by SID-1. Finally, we show that coexpressing wild-type and mutant SID-1 in the same cells inhibits dsRNA transport, suggesting that SID-1 may function as a multimer.
RESULTS AND DISCUSSION
dsRNA size-dependent silencing in Drosophila S2 cells that express SID-1 is not explained by size-dependent dsRNA uptake
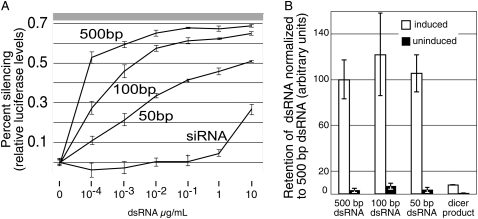
The extent of silencing enabled by SID-1-dependent transport of dsRNA in Drosophila S2 cells shows a striking length dependence (Feinberg and Hunter 2003); long dsRNA (500 base pairs [bp]) silences at much lower concentration than shorter dsRNA (siRNA [19 bp], 50 bp, and 100 bp) (Fig. 1A). To determine whether the size-dependent silencing is caused by preferential transport of long dsRNA or by failure to transport or retain shorter dsRNA, we measured uptake of radiolabeled dsRNA of various sizes (Fig. 1B). The previous analysis of dsRNA uptake used transiently transfected Drosophila S2 cells, thus only 5%–10% of the cells expressed SID-1. To increase the sensitivity of the dsRNA uptake assay, we created a polyclonal S2 cell line whereby all cells contain a metallothionein inducible SID-1 construct. In Feinberg and Hunter (2003) the silencing assays that showed size-dependent silencing were performed with equal masses of dsRNA added to the cells, thus the concentration of the shorter, less efficient dsRNA was present in molar excess to the longer dsRNA. For continuity and to enable direct comparison to these earlier results, we added equal masses of siRNA, 50 bp, 100 bp, and 500 bp dsRNA, and determined the relative mass retained in S2 cells induced to express SID-1 compared to noninduced cells. We found that the mass of 500 bp, 100 bp, and 50 bp dsRNA retained after 30 min were indistinguishable (Fig. 1B). These results suggest that 100 bp and 50 bp dsRNA, although transported and retained at least as well as 500 bp dsRNA, fail to engage the silencing machinery as efficiently as 500 bp RNA. In contrast, the shorter Dicer products, which largely failed to initiate silencing (Fig. 1A; Feinberg and Hunter 2003), were retained poorly in SID-1 expressing S2 cells (Fig. 1B). In related work (J.D. Shih and C.P. Hunter, unpubl.) we find that short dsRNA, including Dicer products, are efficiently transported into SID-1 expressing cells, but that the preferential retention of longer dsRNA requires an intact RNA-induced silencing complex (RISC). Specifically knockdown of Dicer 2 or Argonaute2, which are both required for RISC activity (Lee et al. 2004), reduced long dsRNA retention by 50%–80%. The implication from these results is that SID-1 transported siRNA does not bind efficiently to Dicer, and therefore is not incorporated into RISC. Additional factors, such as the greater coverage of mRNA sequences by longer dsRNA and increased Dicer processing efficiency for longer dsRNA (Elbashir et al. 2001) likely contribute to the size-dependent silencing efficiency. In summary, the results presented here show that the reduced silencing efficiency associated with 100 bp and 50 bp dsRNA cannot be explained by differences in transport via SID-1.
FIGURE 1.
Size-independent transport and retention of dsRNA. (A) dsRNA size-dependent silencing in Drosophila S2 cells expressing SID-1 (data adapted from Feinberg and Hunter 2003). Cells cotransfected with luciferase and SID-1 expressing plasmids were exposed to different concentrations of dsRNA of varying lengths and the extent of silencing was determined. The gray region (top) reflects average luciferase levels at the end of the experiment in samples treated with cycloheximide as a control for protein stability. (B) 10 ng/mL of 500 bp, 100 bp, and 50 bp dsRNA, and siRNA (internally labeled 500 bp dsRNA digested with Dicer) were added to induced and uninduced MT/sid-1 cells for 30 min. The mass of internalized dsRNA was normalized to 500 bp dsRNA. Error bars = 1 standard deviation, n = 2.
dsRNA transport rates through SID-1 are dependent on dsRNA concentration and dsRNA length
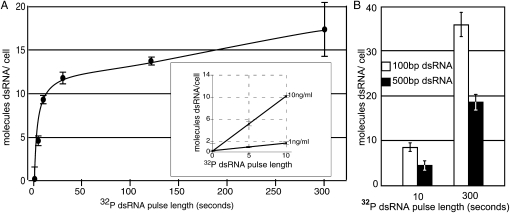
SID-1 mediated dsRNA transport is largely energy independent and thus likely mediated by diffusion-limited processes (Feinberg and Hunter 2003); thus transport of dsRNA into cells is expected to be concentration dependent with internal and external dsRNA concentrations rapidly reaching equilibrium. The above experiments show that at the end of a 30-min uptake experiment that dsRNA length is not a factor in the amount of dsRNA that is transported or retained within cells. Therefore, to determine nonequilibrium accumulation rates we developed a pulse–chase protocol, which allowed us to measure intracellular dsRNA accumulation in the seconds after addition of dsRNA to the cells. We found that the amount of labeled dsRNA retained in cells was stable after the addition of a large excess of unlabeled dsRNA indicating that during the time frame of these experiments there is little exchange between intracellular bound dsRNA and extracellular free dsRNA (data not shown). To estimate dsRNA transport rates, we added 500 bp radiolabeled dsRNA to media containing SID-1-expressing Drosophila S2 cells and then at intervals added a large excess of unlabeled dsRNA. We saw a linear increase in dsRNA uptake in the first ten seconds after radiolabeled dsRNA addition (Fig. 2A). At 10 ng/mL of 500 bp dsRNA the calculated transport rate was approximately one molecule of 500 bp dsRNA/sec/cell. Consistent with expectations for diffusion limited transport mechanisms (Feinberg and Hunter 2003), reducing the concentration of the labeled 500 bp dsRNA to 1 ng/mL, reduced the initial transport rate fivefold (Fig. 2A, inset). That the change in the transport rate is not linear with the amount of dsRNA added may be due to the limited bioavailability of dsRNA in a media solution with many RNA binding proteins, cooperative dsRNA binding, or to intrinsic SID-1 gating properties.
FIGURE 2.
Size- and concentration-dependent dsRNA transport by SID-1. (A) one or 10 ng/mL of radiolabeled 500 bp dsRNA was added to MT/sid-1 cells at time 0 and 100X unlabeled dsRNA was added at the indicated time points. Internalized radiolabeled dsRNA is reported as molecules/cell. Transport of radiolabeled 500 bp dsRNA shows a rapid initial phase (inset), which appears to be linear and concentration dependent in the first 10 sec. Error bars = 1 standard deviation, n = 4. (B) Retention at 10 and 300 sec of 100 bp and 500 bp dsRNA added at equimolar concentrations. Approximately twice as much 100 bp dsRNA was retained at both time points. For data presentation, CPM in uninduced cells were subtracted from the counts measured in induced cells. Error bars = 1 standard deviation, n = 2.
We next sought to compare transport rates between 100 bp and 500 bp dsRNA. We added equimolar amounts (∼30 pM) of 100 bp or 500 bp radiolabeled dsRNA to media containing SID-1-expressing Drosophila S2 cells and then at intervals added a large excess of unlabeled dsRNA. We found that the rate of internalization of the 100 bp dsRNA was approximately double that for the 500 bp dsRNA (Fig. 2B). These data are consistent with the idea that dsRNA is transported lengthwise through a channel or pore, however, if dsRNA length is the rate limiting step for transport, then on average the 500 bp dsRNA should take five times as long to pass through a pore or channel than the 100 bp dsRNA. The observed twofold difference suggests that, at this concentration, transport is not the rate-limiting step.
SID-1 properties in cultured C. elegans cells are similar to those observed in Drosophila S2 cells
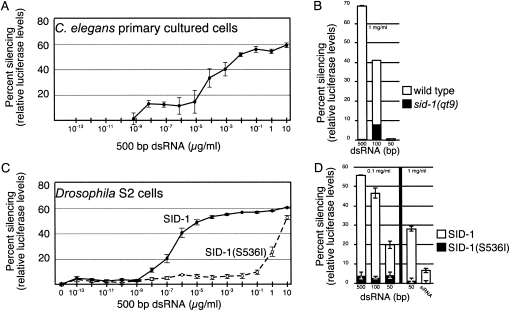
RNAi can be initiated in cultured C. elegans cells by addition of dsRNA to the culture medium (Christensen et al. 2002). We confirmed that soaking-initiated RNAi in these cells was dependent on SID-1 (data not shown). To enable quantitative analysis we then constructed strains with firefly and Renilla luciferase reporters expressed specifically in muscle cells. We found that RNAi of firefly luciferase in C. elegans cells prepared from these strains displayed similar sensitivity to dsRNA concentration in the culture medium as Drosophila S2 cells expressing SID-1 (Fig. 3). Specifically, addition of the same 500 bp firefly luciferase dsRNA to wild-type C. elegans cells and Drosophila S2 cells expressing wild-type SID-1 reduced firefly luciferase levels by 60% at the highest concentration of dsRNA (10 μg/mL). As shown previously, during the limited time of these experiments, a portion of luciferase protein present at the time of dsRNA addition remains active at the end of the experiment, thus the unsilenced 40% likely represents residual stable luciferase protein (Feinberg and Hunter 2003). In both systems, despite the likely differences in SID-1 expression level and the potential for C. elegans regulatory proteins to enhance or restrict dsRNA transport via SID-1, we observed detectable silencing at the same lower limit of 1 pg/mL of dsRNA (Fig. 3A,C). That C. elegans embryonic cells are not more sensitive than Drosophila S2 cells argues that the energy-independent transport observed in the S2 cell system likely reflects the mechanism in C. elegans as well. Attempts to directly measure sid-1-dependent uptake of radiolabeled dsRNA in cultured C. elegans cells have not been successful since we see a persistently high background that prevents clear quantification of data.
FIGURE 3.
Comparison of size and concentration-dependent silencing in C. elegans and Drosophila cells. Size and concentration-dependent silencing triggered by addition of dsRNA to the growth medium of cultured C. elegans and Drosophila cells expressing wild-type or mutant SID-1. (A,B) Cultured C. elegans cells expressing myo-3∷luciferase∷GFP were exposed to the indicated concentration of 500 bp (A) and indicated lengths (B) of luciferase dsRNA and luciferase levels measured 48 h later. The qt9 allele is an early stop codon mutant (Winston et al. 2002). (C,D) Drosophila S2 cells transiently transfected with wild-type SID-1 or missense mutant SID-1(S536I) constructs were exposed to the indicated concentration of 500 bp (C) and indicated lengths (D) of luciferase dsRNA and luciferase levels measured 24 h later. Error bars = 1 standard deviation, n = 3.
C. elegans cells appear to be more discriminating as the response to 50 bp dsRNA is significantly lower than in S2 cells and the response to lower concentrations of dsRNA is similarly less sensitive (Fig. 3B,D). The poor silencing induced by soaking C. elegans cells in 50 bp dsRNA mirrors microinjection experiments in intact worms (Feinberg and Hunter 2003). This difference could reflect the activity of additional proteins in C. elegans that restrict import of small but not longer dsRNA. Alternatively, as described above, in Drosophila S2 cells retention of SID-1-transported dsRNA requires Dicer-2 or Argonaute2, and retention appears to be the limiting step for initiating RNAi. Thus, the reduced effectiveness of dsRNA < 100 bp to initiate silencing in C. elegans may reflect reduced binding to Dicer and or RISC. A restriction on transport of small RNAs may be important for limiting the effect of endogenous regulatory RNAs, such as microRNAs, or for ensuring the specificity of RNAi silencing (Yigit et al. 2006).
SID-1(S536I) acts as a dominant-negative protein in Drosophila S2 cells, suggesting SID-1 may function as a multimer
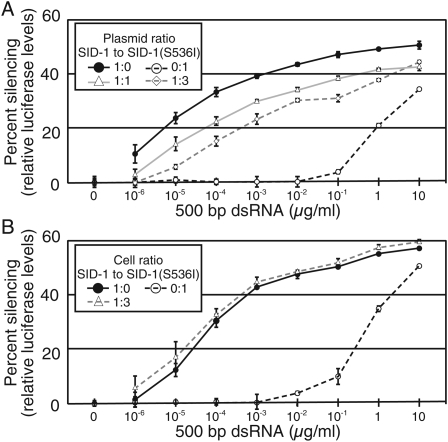
Expression of SID-1 in Drosophila S2 cells is sufficient for dsRNA transport, thus SID-1 functions either as a monomer or a homomultimer. If SID-1 functions as a monomer, then coexpression of the transport defective missense mutant SID-1(S536I) should not affect wild-type SID-1 function. However, if SID-1 forms multimers, then coexpression of SID-1(S536I) may cause a dose-dependent reduction in SID-1 activity. We compared silencing in S2 cells expressing wild-type SID-1 to S2 cells coexpressing wild-type SID-1 and SID-1(S536I). We found that coexpressing wild-type SID-1 and SID-1(S536I) at equimolar ratios reduced silencing efficiency ∼100-fold (Fig. 4A). Specifically, S2 cells expressing only wild-type SID-1 showed half-maximal silencing at ∼50 pg/mL, while cells coexpressing wild-type SID-1 and SID-1(S536I) at a 1:1 ratio showed half-maximal silencing at ∼5 ng/mL. Transfecting the same amount of wild-type SID-1 plasmid, but increasing the amount of SID-1(S536I) expressing plasmid threefold resulted in a further 10-fold reduction in sensitivity (Fig. 4A). These results suggest that SID-1(S536I) forms nonfunctional multimers with wild-type SID-1. Alternatively, SID-1(S536I) may bind and sequester exogenous dsRNA from functional SID-1 monomers. To control for this possibility, we compared silencing between cocultured wild-type SID-1 expressing and SID-1(S536I) expressing cells to wild-type SID-1 expressing cells cultured alone (Fig. 4B). We found no difference in silencing when comparing the cocultured cells to the SID-1 expressing cells demonstrating that SID-1(S536I) does not sequester dsRNA from SID-1. These results suggest that SID-1 functions as a homomultimer.
FIGURE 4.
Coexpression of SID-1(S536I) with wild-type SID-1 interferes with soaking RNAi. (A) Cotransfection of SID-1(S536I) with wild-type SID-1 reduces luciferase silencing. S2 cells were cotransfected with a constant amount of firefly luciferase plasmid and the indicated ratios of plasmids that express wild-type SID-1 and SID-1(S536I). A range of concentrations of 500 bp firefly luciferase dsRNA was then added to the growth medium. (B) SID-1(S536I) does not sequester dsRNA from wild-type SID-1 when expressed in different cells. S2 cells coexpressing firefly luciferase and wild-type SID-1 were cocultured at the indicated ratios with S2 cells expressing SID-1(S536I) with a range of concentrations of 500 bp firefly luciferase dsRNA added to the growth medium. For cells expressing SID-1(S536I) alone (0:1 ratio), firefly luciferase plasmid was cotransfected with the SID-1(S536I) expressing plasmid. Error bars = 1 standard deviation, n = 3.
Although we detected a strong dose-dependent effect of SID-1(S536I) on wild-type SID-1 activity, an effect on systemic RNAi has not been detected in sid-1(qt2)/sid-1(+) heterozygous worms. One explanation for this disparity is that the time allowed for an effect in worms is very large (days) and any reduced transport efficiency in C. elegans may be masked, for example, by RdRP-mediated amplification of transported dsRNA (Sijen et al. 2001). However, because RdRP mutants are RNAi defective (Smardon et al. 2000), this possibility cannot be tested. Confirmation of a functional multimer will require biochemical and biophysical evidence. The formation of functional complexes would be an elegant mechanism of gating SID-1. A channel capable of transporting a dsRNA must have a minimal pore diameter of 3.2 nm, which, if constitutively open, would rapidly dissipate ion gradients and result in cell death. Because heterologous SID-1 expression does not cause acute cell lethality, SID-1 must be ligand gated and such activity is either inherent to SID-1 or can be accomplished by nonspecific protein interactions. Indeed, the constitutively open oligonucleotide channel is rendered oligonucleotide gated by the addition of cytoplasmic malate dehydrogenase (Hanss et al. 2002). Ligand-dependent multimerization could similarly gate SID-1 activity.
CONCLUSIONS
We found that cultured C. elegans embryonic cells and SID-1 expressing Drosophila S2 cells initiate silencing at the same lowest concentration of dsRNA (Fig. 3). This indicates that SID-1, in the context of native C. elegans proteins, is not more efficient at importing dsRNA from the culture medium than heterologously expressed SID-1. Therefore, it is likely that dsRNA transport by SID-1 in C. elegans cells is also passive.
We showed that 100 bp dsRNA enters the cell more quickly than 500 bp dsRNA (Fig. 2). This is consistent with the notion that dsRNA transport is achieved by threading the dsRNA lengthwise through a channel or pore. Consistent with this mechanism of transport, SID-1 is not selective for dsRNA length (Fig. 1); the mass of accumulated 50 bp, 100 bp, and 500 bp dsRNA was indistinguishable. This leads us to conclude, that in Drosophila S2 cells, long dsRNA is simply a more effective RNAi trigger than short dsRNA. In contrast, siRNA was not retained in these cells. This could reflect poor transport and/or poor retention. In results to be reported elsewhere, we find that siRNAs are efficiently transported by SID-1 into cells and that retention of dsRNA requires an intact RISC complex. Together, these results suggest that, in vivo, siRNAs are poor substrates for RISC.
Finally, we show that coexpressing a missense SID-1 mutant protein interferes, in a dose-dependent manner, with the function of wild-type SID-1 (Fig. 4). The simplest interpretation for this dominant-negative effect is that SID-1 functions as a multimer. Although confirmation and functional analysis of multimerization will require the development of additional methods, we suggest two possible roles for multimerization in dsRNA transport. Multimerization may be necessary to create a channel or pore large enough to transport dsRNA. Second, multimerization could play a fundamental role in gating of SID-1 activity; an open channel may only form when dsRNA is being transported. Thus dsRNA, the ligand for SID-1, may participate in the temporary assembly of a functional channel.
MATERIALS AND METHODS
Polymerase chain reaction amplification
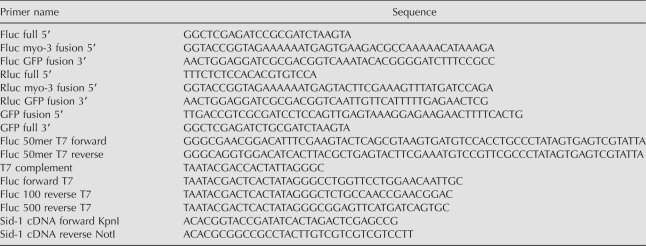
Primer and oligonucleotide sequences are listed in Table 1. Primers were purchased from Integrated DNA Technologies. All PCR amplifications used the EXPAND High Fidelity PCR system (Roche).
TABLE 1.
Oligonucleotides used
DNA constructs
The muscle cell expressed firefly and renilla reniformis luciferase-GFP fusion constructs were assembled by PCR stitching (Thomas et al. 2006). Briefly, firefly and Renilla luciferase were amplified from pGL3-Basic and pRLnull plasmids (Promega) (primers Fluc full 5′ and Fluc GFP fusion 3′, Rluc full 5′ and Rluc GFP fusion 3′) and stitched to GFP (amplified from pPD118.20 with GFP fusion 5′ and GFP full 3′). Firefly luciferase-GFP was then stitched to a myo-3 promoter from pPD118.20 (primers Fluc myo-3 fusion 5′ and GFP full 3′) and used as a PCR stitch construct, while myo-3∷renilla reniformis-GFP was stitched directly to pPD118.20 (primers Rluc myo-3 fusion 5′and GFP full 3′) to create pAJ1(myo-3∷renilla reniformis luciferase∷gfp). To create pMT/sid-1, sid-1(WT)∷FLAG was PCR amplified (primers SID-1 cDNA forward KpnI and SID-1 cDNA reverse NotI) from pHC235 (Feinberg and Hunter 2003) and ligated onto NotI, KpnI digested pMT/lacZ (Invitrogen, SmithKline Beecham).
C. elegans strains
HC246: qtIs23(PCR stitch[myo-3∷firefly luciferase∷gfp], pAJ1[myo-3∷renilla reniformis luciferase∷gfp]). Myo-3∷firefly luciferase∷gfp and pAJ1 were coinjected into Bristol N2 worms, integrated, and out-crossed to produce HC246.
Cell culture
Drosophila S2 and stably transfected MT/sid-1 cells were maintained at 27°C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 50 U/mg penicillin and streptomycin (Invitrogen). For the stable transfected cells 250 μg/mL Hygromycin B was also added. Transfection of Drosophila S2 cells with pHC235 (Act-5∷sid-1-FLAG), pHC247 (Act-5∷sid-1(qt2)-FLAG), and pGL3-basic was as described (Feinberg and Hunter 2003). The sid-1(qt2) allele substitutes isoleucine for serine (S536I). For cotransfection of pHC235 and pHC247 into the same cell, they were cotransfected with pGEM (Promega) as a buffer plasmid in a 1:0:4 pHC235:pHC247:pGEM ratio for SID-1, 0:1:4 for SID-1(S536I), 1:1:3 for a 1:1 SID-1:SID-1(S536I)ratio, and 1:3:1 for a 1:3 SID-1:SID-1(S536I) ratio. To select for stable transfectants pMT/sid-1 was cotransfected with pHygro (Invitrogen) into S2 cells and maintained by Hygromycin B selection (200 μg/mL–250 μg/mL) (Sigma-Aldrich) to produce a polyclonal MT/sid-1 cell line. To induce SID-1 expression MT/sid-1 cells were treated with 0.6 M CuSO4 for 48 h prior to use in the retention assays. C. elegans primary cells were prepared and cultured as described (Christensen et al. 2002) from HC246 embryos.
dsRNA synthesis
Dual T7 PCR DNA templates: 100 bp firefly luciferase (pGL3-Basic [Promega], primers Fluc forward T7 and Fluc 100 reverse T7), 500 bp firefly luciferase (pGL3-Basic [Promgea], primers Fluc forward T7 and Fluc 500 reverse T7), 50 bp firefly luciferase (Fluc 50mer T7 forward, Fluc 50mer T7 reverse, primer T7 complement). RNA synthesized using Ampliscribe T7 kit (Epicentre). Single strands were annealed by heating to 90°C followed by cooling one degree per eight seconds until reaching a temperature of 25°C. dsRNA was purified using Micro Bio-Spin 6 Chromatography Columns (Bio-Rad). dsRNA concentration and purity were verified by agarose gel electrophoresis and spectroscopy.
Luciferase silencing assay
All silencing assays performed on Drosophila S2 cells were performed on transiently transfected cells. Drosophila S2 cells 105 cells per well were plated in triplicate in solid white 96-well plates (Corning). For dsRNA silencing of C. elegans primary cell culture 4 × 105 primary cells (HC246) were plated on each well, three wells per sample. Luciferase activity was measured using the Bright-Glo assay (Promega) for Drosophila S2 cells or Dual-Glo assay (Promega) for C. elegans cells on an AD Analyst (Molecular Devices).
Radiolabeled dsRNA assays
Retention assays were performed as described (Feinberg and Hunter 2003). All retention assays were performed minimally in duplicate on MT/sid-1 cells. For time-dependent assays of SID-1 channel properties 100X competitor dsRNA was added at time intervals after the addition of radiolabeled dsRNA. For the zero time point 100X competitor dsRNA was mixed with radiolabeled dsRNA prior to addition to cells.
ACKNOWLEDGMENTS
We thank Antony Jose, Andrea Hinas, and Jessica J. Smith for critical reading of this paper and all the members of the Hunter laboratory for helpful comments and suggestions. This work was supported by NIH grant GM069891 and NSF grant MCB 0417102.
Footnotes
Article published on line ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1286409.
REFERENCES
- Boulin T., Etchberger J.F., Hobert O. Reporter gene fusions. WormBook. 2006 doi: 10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G., Scholten J., Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr. Biol. 2002;12:R85–R86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- Christensen M., Estevez A., Yin X., Fox R., Morrison R., McDonnell M., Gleason C., Miller D.M., III, Strange K. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron. 2002;33:503–514. doi: 10.1016/s0896-6273(02)00591-3. [DOI] [PubMed] [Google Scholar]
- Duxbury M.S., Ashley S.W., Whang E.E. RNA interference: A mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem. Biophys. Res. Commun. 2005;331:459–463. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg E.H., Hunter C.P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hanss B., Leal-Pinto E., Teixeira A., Christian R.E., Shabanowitz J., Hunt D.F., Klotman P.E. Cytosolic malate dehydrogenase confers selectivity of the nucleic acid-conducting channel. Proc. Natl. Acad. Sci. 2002;99:1707–1712. doi: 10.1073/pnas.022355499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Reddien P.W., Cebria F., Sanchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. 2003;100:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J.Y., Carre C., Mugat B., Szymczak D., Lepesant J.A., Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila . RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke J.M., Stacey S.C., Klein M.E., Mackin N., Maine E.M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans . Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Tabara H., Grishok A., Mello C.C. RNAi in C. elegans: Soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Wang S.L., Dorovkov M., Ryazanov A., Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Turner C.T., Davy M.W., MacDiarmid R.M., Plummer K.M., Birch N.P., Newcomb R.D. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 2006;15:383–391. doi: 10.1111/j.1365-2583.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- Winston W.M., Molodowitch C., Hunter C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charisse K., Ndungo E.M., et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. Analysis of the C. elegans Argonuate family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]