Abstract
MicroRNAs (miRNAs) are sequence-specific negative regulators of gene expression generated by DICER-LIKE1 (DCL1) with the assistance of a double-stranded RNA-binding protein, HYPONASTIC LEAVES1 (HYL1), in Arabidopsis. To achieve a better understanding of miRNA biogenesis, we isolated hyl1 suppressors. Our genetic screening identified a novel semidominant mutation in DCL1 (dcl1-13), which causes an amino acid substitution of Glu-395 with Lys in the ATPase/DExH-box RNA helicase domain. This mutation confers significant restoration from the developmental abnormality and a reduction in the level of miRNA in the loss-of-function mutant of HYL1. However, the dcl1-13 single mutant, exhibiting a decreased number of leaves, showed a slight decrease in miRNA accumulation. Thus, the effect of the dcl1-13 mutation is HYL1 dependent: it promotes miRNA processing in the absence of HYL1, but conversely, impairs it in the presence of HYL1. Our results suggest significant roles of the helicase domain of DCL1, which remain unclear to date, possibly in relation with HYL1.
Keywords: HYPONASTIC LEAVES1, DICER-LIKE1, microRNA, suppressor
INTRODUCTION
MicroRNAs (miRNAs) are small RNAs of around 21–24 nucleotides (nt) in length. They are involved in multiple developmental and physiological processes in various organisms through the negative regulation of gene expression.
miRNAs are produced by Dicer and Drosha proteins, which contain RNase III domains. In Arabidopsis, a miRNA precursor transcript that has a stem–loop structure is processed by DICER-LIKE1 (DCL1) into a miRNA/miRNA* duplex. DCL1 forms a complex with an RNA-binding protein, HYPONASTIC LEAVES1 (HYL1), and a plant-specific zinc finger protein, SERRATE (SE), in the nuclei, possibly at Cajal bodies marked with SmD3/SmB (Fang and Spector 2007; Fujioka et al. 2007; Song et al. 2007). HYL1 belongs to a family of proteins containing double-stranded RNA (dsRNA)-binding domains (dsRBDs). Generally, Dicer proteins and the related RNase III enzymes Drosha proteins interact with specific dsRNA-binding proteins. Mammalian TRBP, a dsRNA-binding protein, is an integral component of a Dicer-containing complex that is required for miRNA biogenesis (Chendrimada et al. 2005). In Drosophila, the dsRNA-binding protein Loquacious (Loqs) associates with Dicer-1 (Saito et al. 2005).
Arabidopsis HYL1 was initially identified as a factor involved in organ morphogenesis and response to phytohormones. A loss-of-function mutant of HYL1 exhibited severely hyponastic rosette leaves and abnormal sensitivities to phytohormones (Lu and Fedoroff 2000). It has since been demonstrated that miRNAs are reduced in abundance and, conversely, that miRNA precursors are overaccumulated, in hyl1 mutants (Han et al. 2004; Vazquez et al. 2004; Kurihara et al. 2006). It was also reported that HYL1 interacts with DCL1 in vitro and in vivo (Hiraguri et al. 2005; Kurihara et al. 2006; Fujioka et al. 2007), has binding activity to dsRNA (Hiraguri et al. 2005), and promotes the accurate processing of miRNA precursors mediated by DCL1 (Dong et al. 2008). Thus, it is assumed that HYL1 associates with DCL1 and supports its processing activity, possibly by binding to RNA substrates.
To achieve a better understanding of miRNA processing, we performed genetic screening of mutants in which the hyl1 phenotype is suppressed. A suppressor of hyl1 had a dominant point mutation (E395K) in the ATPase/DExH-box RNA helicase domain of DCL1. This novel allele was designated dcl1-13. The dcl1-13 mutation led to a recovery of miRNA processing without HYL1. On the contrary, the processing of miRNA in dcl1-13 single mutants was slightly impaired in the presence of HYL1. These results indicate the essential role of the helicase domain of DCL1 possibly in relation with HYL1.
RESULTS
Isolation of hyl1 suppressor
To gain insight into the molecular mechanism underlying miRNA processing, genetic screening was performed to isolate hyl1 suppressors. The hyl1-2 mutation is a presumably null allele with a T-DNA insertion in the first dsRBD resulting in a nonfunctional truncated HYL1 protein. In hyl1-2 mutants, miRNAs are reduced in abundance and the leaves are narrow and small, possibly because DCL1 could not normally recognize or be loaded with miRNA precursor (Vazquez et al. 2004; Kurihara et al. 2006). The hyl1-2 seeds were mutagenized by ethylmethane sulfonate (EMS) and M2 seedlings with rounder, wild-type (WT)-like cotyledons were selected as candidates for hyl1 suppressor. Approximately 38,000 seeds were investigated in total, and 22 individuals were obtained. Out of these 22 lines, 21 were obtained from the same M2 pool that was treated with 0.1% EMS and one line was recovered from a 0.2% EMS-treated M2 pool. Subsequent experiments were carried out using one representative line derived from a 0.1% EMS-treated pool. The obtained suppressor was backcrossed to the original hyl1-2 mutant, and the leaf morphology of the progeny was analyzed. The F1 progeny exhibited leaf morphology similar to the wild type (data not shown), indicating that the hyl1 suppressor mutation is dominant.
An amino acid substitution in the RNA helicase domain of DCL1 causes suppression of the hyl1 phenotype
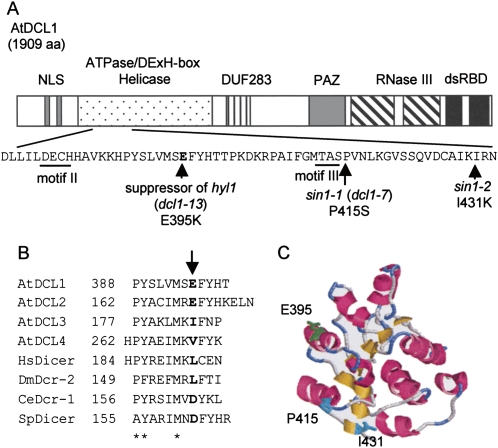
To identify the gene responsible for suppression of the hyl1 phenotype, map-based cloning was performed. The hyl1 suppressor mutation was mapped to within a 3.2 Mbp region between the left end of chromosome 1 and the nga63 polymorphic marker (data not shown). Because DCL1 (At1g01040), which is essential for miRNA biogenesis, exists in this region, the genomic sequence of DCL1 in the hyl1 suppressor mutant was determined. A guanine at nucleotide position 1183 of the coding sequence of DCL1 was substituted to adenine, resulting in an amino acid substitution, glutamic acid (E) to lysine (K), at position 395 in the ATPase/DExH-box RNA helicase domain of DCL1 (Fig. 1A). Introduction of a DCL1 gene containing this mutation into the hyl1-2 mutant significantly suppressed the hyl1 mutant phenotype in T1 plants (Supplemental Fig. S1 and Supplemental Materials and Methods), demonstrating that this mutation causes suppression of the hyl1 phenotype in a dominant fashion. All 22 suppressors screened in this experiment had the same mutation, implying a unique role for this amino acid within the helicase domain. We have designated this new allele of DCL1 as dcl1-13. To date, genetic studies have identified various recessive dcl1 mutants including caf-1, which has pleiotropic defects such as excess stamens and carpels (Jacobsen et al. 1999). In this study we have demonstrated that the dcl1-13 mutation suppresses the hyl1 phenotype in a dominant manner and, to our knowledge, this is the first dominant mutation to be identified in a Dicer protein in any organisms.
FIGURE 1.
Molecular nature of the hyl1 suppressor mutation. (A) Schematic representation of the domain structure of Arabidopsis DCL1 (AtDCL1) and the dcl1-13 mutation in the helicase domain that caused the suppression of the hyl1 mutant phenotype. DCL1 has two nuclear localization signals (NLSs), an ATPase/DExH-box RNA helicase domain, a domain known as DUF238 (domain of unknown function), a PAZ domain, two RNase III domains, and two double-stranded RNA-binding domains (dsRBDs). Between motifs II and III of the helicase domain, the G at position 1183 in the coding sequence was substituted to A in hyl1 suppressor plants, resulting in the amino acid substitution of Glu-395 with Lys (E395K). The transitions of P415S in sin1-1 (dcl1-7) and I431K in sin1-2 were closely located. (B) Alignment of amino acid sequences between motifs II and III in the helicase domains of Arabidopsis DCLs and other Dicer homologs of human, Drosophila, C. elegans, and S. pombe. Sequences that were predicted to form α-helices using the jpred3 program (Cuff et al. 1998) are shown. Arabidopsis DCL1 (AtDCL1; accession no. NP_171612) was aligned with Arabidopsis DCL2 (AtDCL2; NP_566199), DCL3 (AtDCL3; NP_189978), DCL4 (AtDCL4; NP_197532), Homo sapiens Dicer (HsDicer; NP_803187), Drosophila Dcr-2 (DmDcr-2; NP_523778), C. elegans Dcr-1 (CeDcr-1; NP_498761), and S. pombe Dicer (SpDicer; Q09884). Sequences were aligned with the positions of conserved P, Y, and M (letter columns marked with asterisks), which were highly conserved among these proteins. Numbers indicate the positions of the first amino acids. The arrow indicates the position of the hyl1 suppressor mutation in AtDCL1. (C) Homology model of the first RecA-like domain of the AtDCL1 helicase domain (residues 239–433) according to the structure of yeast initiation factor 4A (PDB ID code 1qva). This was constructed using the 3D-JIGSAW program (Bates et al. 2001). The α-helices, β-strands, and turns are indicated with pink, yellow, and blue colors, respectively. Residues E395, P415, and I431 are highlighted.
Conservation of amino acid position 395 among other Dicer proteins
In general, ATPase/DExH-box RNA helicases have eight conserved amino acid sequence motifs designated motif I, Ia, Ib, II, III, IV, V, and VI (Cordin et al. 2006). These motifs are also found in the helicase domain of DCL1 (Jacobsen et al. 1999). The dcl1-13 mutation results in an amino acid change between motifs II and III (Fig. 1A). It was previously reported that short integments1 mutants (sin1-1 and sin1-2), which have defects in female sterility and ovule development, also have point mutations in the RNA helicase domain of DCL1 (Fig. 1A) (Golden et al. 2002). These are recessive, loss-of-function mutations, while dcl1-13 is a dominant mutation, suggesting that dcl1-13 has totally different characteristics from sin1-1 and sin1-2 even though their mutation sites are very close.
Next, we examined the conservation of amino acid sequences and secondary structures between motifs II and III of the helicase domains in four DCL proteins of Arabidopsis and the Dicer proteins of other organisms including human, Drosophila, Caenorhabditis elegans, and Schizosaccharomyces pombe (Fig. 1B). The secondary structures were predicted using the jpred3 program (http://www.compbio.dundee.ac.uk/∼www-jpred/) (Cuff et al. 1998). Although we also examined Drosophila Dicer-1 and yeast initiation factor 4A (yIF4A), both of which contain an RNA helicase domain as well as the proteins listed in Figure 1B, the amino acid sequences in this region were too diverse to obtain an alignment.
While the amino acid sequences between motifs II and III were fairly diverse, the secondary structures of this region were predicted to fold into α-helices, which were well conserved (only amino acid sequences predicted to fold into α-helices are shown in Fig. 1B). The amino acids aligned with Glu-395 of AtDCL1 are acidic (E and D) or hydrophobic (I, V, and L) in all Dicer homologs (Fig. 1B; indicated with bold letters). Although the acidic amino acid Glu-395 is replaced with a basic amino acid Lys by dcl1-13 mutation, this region is still predicted to fold into α-helix (data not shown). These results suggested that an acidic or hydrophobic residue in the position corresponding to Glu-395 of AtDCL1 is essential. A change in the charge at the amino acid position 395 may alter the activity of the helicase domain of AtDCL1.
Members of the DExH-box and related DEAD-box families include two RecA-like domains in their helicase core (Cordin et al. 2006). The homology model of the first RecA-like domain of AtDCL1 was predicted by the 3D-JIGSAW program (http://www.bmm.icnet.uk/servers/3djigsaw/) (Bates et al. 2001) using yIF4A as a template (Fig. 1C). Glu-395 is exposed on the surface of this domain as well as Pro-415 and Ile-431, the mutated residues in sin1-1 and sin1-2, respectively. Generally, RNA helicases convert the energy derived from ATP hydrolysis into structural changes in RNA or RNA/protein complexes. Taken together, these findings suggest that amino acid position 395 of AtDCL1 is involved in recognition of RNA substrate, ATP, or other proteins, via the acidic residue at this position on the surface of the domain.
Semidominant and mild suppressor effect of dcl1-13
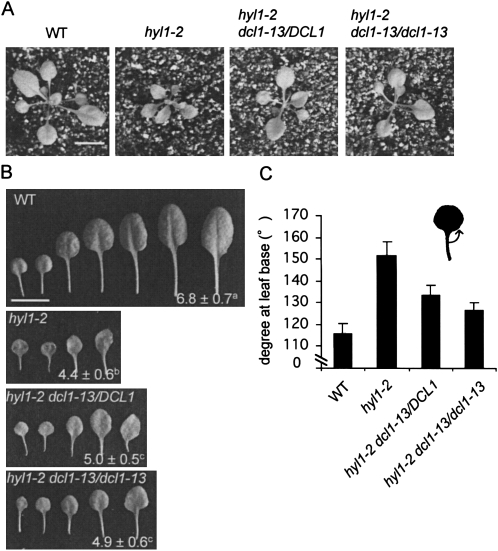
As described above, dcl1-13 dominantly suppressed the hyl1 phenotype. We then examined the phenotypes of heterozygous and homozygous hyl1 suppressors (namely, hyl1-2 dcl1-13/DCL1, and hyl1-2 dcl1-13/dcl1-13, respectively) in detail. Rosette images of WT, hyl1-2 and hyl1 suppressors are shown in Figure 2A. Both heterozygous and homozygous suppressors exhibited reversion from the hyl1 phenotype, although they were still smaller than WT plants. They showed the same level of restoration from the phenotype of hyl1 plants in terms of the numbers of rosette leaves (Fig. 2B). However, their numbers of rosette leaves in these plants were less than those in WT plants. When observing leaf morphology in detail, acuteness of the third and fourth rosette leaves was significantly increased in the order of WT plants, the homozygous suppressor, the heterozygous suppressor, and hyl1 plants (Fig. 2C).
FIGURE 2.
The dcl1-13 mutation suppresses the hyl1 phenotype in a semidominant manner. (A) Rosette images of wild-type (WT) plants, hyl1-2 plants, and heterozygous and homozygous hyl1 suppressors (hyl1-2 dcl1-13/DCL1, and hyl1-2 dcl1-13/dcl1-13, respectively). Bar = 1 cm. (B) Successive rosette leaves and their numbers (average ± SD) from the plants shown in A. n = 17–21. Bar = 1 cm. Values designated by the same superscript letter are not significantly different at the P = 0.01 level in the t-test. (C) Degrees at leaf bases (indicated in illustration) of the third and fourth rosette leaves of plants shown in A. n = 17–20. Bars indicate SD. Values are significantly different between each possible comparison at the P = 0.01 level in the t-test.
These results suggest that the degree of reversion in the homozygous suppressor was more than that in the heterozygous suppressor, and that the dcl1-13 mutation confers suppression of the hyl1 phenotype in a semidominant fashion. The suppressor effect was mild, and it did not completely induce recovery from the hyl1 mutant phenotype, even in homozygous lines.
Recovery of the efficiency of miRNA processing in the hyl1 suppressors
In hyl1-2 mutants, the levels of miRNAs are reduced and, conversely, the levels of miRNA precursor transcripts are increased in abundance (Kurihara et al. 2006). We next examined whether accumulations of miRNAs and their precursors are restored in the hyl1 suppressors (Fig. 3).
FIGURE 3.
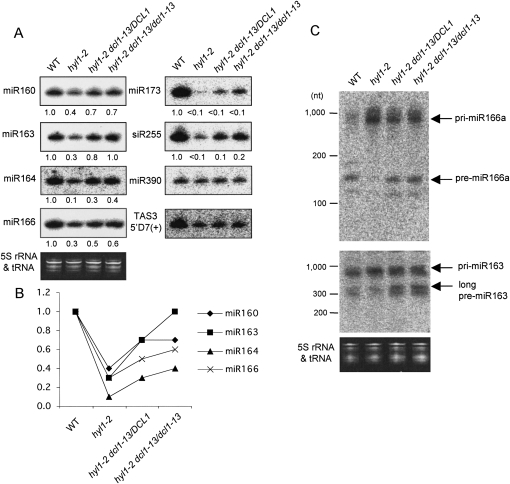
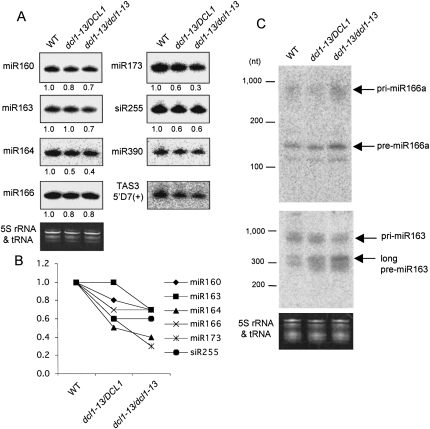
The dcl1-13 mutation restored miRNA processing in hyl1 mutants. (A) Northern blot analyses of miRNAs (miR160, 163, 164, 166, 173, and 390) and tasiRNAs (siR255 and TAS35′D7[+]) in WT plants, hyl1-2 and hyl1 suppressors. Blots are shown with high contrast in order to make the differences clear. Ethidium bromide staining (5S rRNA and tRNA) is shown as a control. Relative RNA levels estimated from band signals are indicated at the bottom of each lane, with small RNA levels in WT plants set to 1.0. The signals for miR390 and TAS3 5′D7(+) were too weak to calculate the levels. (B) The values for the miRNAs (miR160, 163, 164, and 166) in A are graphically shown. (C) Northern blot analyses of miRNA precursors for miR166a and miR163 in WT plants, hyl1-2 and hyl1 suppressors. Ethidium bromide staining (5S rRNA and tRNA) is shown as a control. Bands of pri-/pre-miR166a or miR163 are indicated with arrows. The miR163 precursor has an additional cleavage step to produce its miRNA, resulting in the presence of long and short pre-miR163s (Kurihara and Watanabe 2004). Long pre-miR163 is indicated with an arrow.
Accumulation of six miRNAs (miR160, 163, 164, 166, 173, and 390) was examined by Northern blot analysis (Fig. 3A), and the relative RNA levels estimated from the band signals are shown at the bottom of each lane (Fig. 3A) and represented graphically (Fig. 3B). Most miRNAs examined were recovered in both heterozygous and homozygous suppressors from hyl1 plants, except that the differences in miR390 were unclear because of the weak signals. However, their levels were still lower than those in WT plants except for miR163 in the homozygous suppressor. Similarly to the phenotypes shown in Figure 2, some miRNAs recovered more in the homozygous suppressors than in the heterozygous suppressors, possibly suggesting that the dcl1-13 mutation semidominantly restored miRNA accumulation on a hyl1-2 background.
Two tasiRNAs (siR255 and TAS3 5′D7[+]) were detected in parallel with miRNAs. The generation of tasiRNA is first induced by specific miRNAs, and tasiRNAs are subsequently processed by DCL4, not by DCL1 (Allen et al. 2005; Gasciolli et al. 2005; Yoshikawa et al. 2005; Xie et al. 2005). Therefore, the accumulation of tasiRNA is dependent on miRNA accumulation and could reflect the efficiency of target gene regulation. siR255 comes from the TAS1 transcript, whose cleavage is induced by miR173, and the generation of TAS3 siRNA is induced by miR390. The level of siR255 was clearly increased in the hyl1 suppressors compared with the hyl1 plants, while the level of TAS3 siRNA was almost unchanged. These results indicate that the aberration of target gene regulation was restored to some extent in hyl1 suppressors owing to the restored levels of miRNAs.
In Arabidopsis, miRNAs are produced through the two cleavage steps catalyzed by DCL1 in most cases (Kurihara and Watanabe 2004; Kurihara et al. 2006). First, the dsRNA region of the stem–loop structure in the long miRNA primary transcript (pri-miRNA) is processed resulting in the production of a miRNA precursor (pre-miRNA). Then, the pre-miRNA is cleaved at ∼21 nt from the end to produce a miRNA/miRNA* duplex. Next, we tried to detect the pri- and pre-miRNAs to confirm whether the increase in the levels of miRNAs in the hyl1 suppressors is due to an increase in the efficiency of miRNA precursor processing (Fig. 3C). In the hyl1 suppressors, the levels of pri-miR166a, which was highly accumulated in hyl1-2 plants, were slightly reduced; conversely, the levels of pre-miR166a, which were barely detected in hyl1-2 plants, were increased. A similar tendency was observed for pri-/pre-miR163. These results indicate that the impaired processing from pri-miRNA to pre-miRNA in hyl1 plants was restored by the dcl1-13 mutation.
Taken together, these findings demonstrate that the processing of miRNAs from miRNA precursor transcripts is enhanced in the hyl1 suppressors, possibly leading to the proper regulation of target RNAs by miRNAs.
Phenotype of the dcl1-13 single mutant
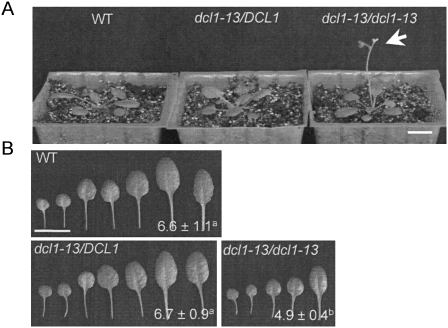
To analyze the effects of the dcl1-13 mutation in the presence of HYL1 activity, the dcl1-13 single mutant was investigated in detail. The hyl1-2 mutation was segregated away from the dcl1-13 mutation by twice crossing the hyl1-2 dcl1-13 double mutant to WT plants. The rosette and leaf morphology of heterozygous dcl1-13 mutant plants (dcl1-13/DCL1) were almost the same as that of WT plants (Fig. 4). Homozygous dcl1-13 plants (dcl1-13/dcl1-13) showed an early-flowering phenotype (Fig. 4A) and a significantly decreased number of leaves (Fig. 4B).
FIGURE 4.
The homozygous dcl1-13 single mutant shows early flowering and decreased number of leaves. (A) Rosette images of WT plants, the heterozygous dcl1-13 mutant (dcl1-13/DCL1) and the homozygous dcl1-13 mutant (dcl1-13/dcl1-13). Bar = 1 cm. The homozygous dcl1-13 shows an early-flowering phenotype (indicated with an arrow). (B) Successive rosette leaves and their numbers (average ± SD) for the plants shown in A. Bar = 1 cm. n = 16–19. Values designated by the same superscript letter are not significantly different at the P = 0.01 level in the t-test.
Next, accumulation of small RNAs and pri-/pre-miRNAs was examined in the dcl1-13 single mutants. Because the dcl1-13 mutation promoted the cleavage of miRNA precursors (Fig. 3), it was predicted that the dcl1-13 single mutants would exhibit increased accumulation of miRNAs compared with WT plants. Contrary to our expectations, the levels of most miRNAs were slightly reduced both in heterozygous and homozygous dcl1-13 mutants (Fig. 5A,B). The levels of some miRNAs were more reduced in the homozygous dcl1-13 plants than in the heterozygous dcl1-13 plants, possibly suggesting a semidominant repression effect of the dcl1-13 mutation in the presence of HYL1 on miRNA accumulation. tasiRNA accumulation was also slightly reduced in dcl1-13 mutant plants (Fig. 5A,B).
FIGURE 5.
miRNA processing is semidominantly impaired by the dcl1-13 mutation in the presence of HYL1. (A) Northern blot analyses of miRNAs (miR160, 163, 164, 166, 173, and 390) and tasiRNAs (siR255 and TAS3 5′D7[+]) in WT plants and dcl1-13 single mutants. Blots are shown with high contrast in order to make the differences clear. Ethidium bromide staining (5S rRNA and tRNA) is shown as a control. Relative RNA levels estimated from band signal intensities are indicated at the bottom of each lane, with small RNA levels in WT plants set to 1.0. The signals for miR390 and TAS3 5′D7(+) were too weak to calculate the level. (B) The values for miRNAs (miR160, 163, 164, 166, and 173) and tasiRNA (siR255) in A are graphically shown. (C) Northern blot analyses of miRNA precursors for miR166a and miR163 in WT plants and dcl1-13 single mutants. Ethidium bromide staining (5S rRNA and tRNA) is shown as a control. Bands of pri-/pre-miR166a or miR163 are indicated with arrows.
Next, the accumulations of miRNA precursors were investigated (Fig. 5C). The levels of pre-miR166a and pre-miR163, intermediates in pri-miRNA to miRNA processing, in the homozygous dcl1-13 plants were clearly higher than those in WT plants. These results, together with the modest reduction in miRNA accumulation in dcl1-13 plants, indicate that the processing of pre-miRNA to miRNA was slightly inhibited by the dcl1-13 mutation in the presence of HYL1. Although the differences in the abundance of miRNAs and their precursors between WT plants and heterozygous dcl1-13 plants were subtle, the repression effect of dcl1-13 on the processing of miRNA precursors seems to be semidominant in the presence of HYL1.
DISCUSSION
In the present study, to analyze in detail the miRNA pathway in Arabidopsis, we screened suppressor mutants of hyl1 and demonstrated that an amino acid substitution of Glu-395 with Lys in the ATPase/DExH-box helicase domain of DCL1, an RNase III enzyme involved in producing miRNAs, could suppress the hyl1 phenotype. In the hyl1 suppressor (hyl1-2 dcl1-13 double mutant), the processing of miRNA precursor was restored, possibly resulting in the restoration of target gene regulation and normal morphogenesis.
Typically, Dicer proteins are ∼200 kDa multidomain proteins that consist of an ATPase/DExH-box helicase domain, a domain of unknown function (DUF283), a PAZ domain, two RNase III nuclease domains (RNase IIIa and RNase IIIb), and one or two dsRBD(s). An intramolecular dimer between RNase IIIa and IIIb domains forms a catalytic center with each RNase III domain cleaving one strand of the dsRNA. The PAZ domain binds the 3′ ends of the dsRNA and the distance between the PAZ domain and the RNase III domains determines the length of the cleavage product (Macrae et al. 2006b).
The requirement of the ATPase/helicase domains of Dicers for the biogenesis of small RNA is different among Dicer proteins in various organisms. Some Dicers, including that of Giardia intestinalis (Macrae et al. 2006b), lack either the helicase domain and/or the dsRBD. In human Dicer with a helicase domain, in vitro activity is ATP independent (Zhang et al. 2002). These Dicers may not require ATPase/helicase domains for their cleavage activities. On the other hand, siRNA generation by Dicer is enhanced by the addition of ATP in Drosophila (Bernstein et al. 2001). The activity of fission yeast Dicer, Dcr1, is dependent on ATP and a mutation in the ATPase motif in the helicase domain disrupts dicing activity (Colmenares et al. 2007). In these Dicers, ATPase/helicase domains may have important roles in the processing of small RNAs. In Arabidopsis, genetic analyses have revealed that sin1-1 and sin1-2 mutants with missense mutations at Pro-415 and Ile -431 in the helicase domain of DCL1, respectively (Fig. 1A), exhibit defects in female fertility and ovule development (Golden et al. 2002). The dcl4-2 mutation converts a well-conserved Glu-583 in the helicase domain to Lys, and reduces the accumulation of siRNAs (Yoshikawa et al. 2005). Recently, the in vitro miRNA processing activity of DCL1 was shown to be enhanced by the addition of ATP (Dong et al. 2008). These results suggest that the ATPase/helicase domains of Arabidopsis DCL1 and DCL4 have important regulatory roles.
It is poorly understood how the RNA helicase domain of Dicer is involved in the process of dsRNA cleavage. In general, RNA helicases participate in various processes involving RNA, such as transcription, pre-mRNA splicing, and translation. They convert the energy from ATP hydrolysis into structural changes in RNA or RNA/protein complexes. The Dicer helicase domain is thought to regulate the loading of dsRNA onto Dicer, promote the translocation of Dicer along dsRNA substrates, and/or facilitate the handoff of RNA substrates or products with other proteins (Macrae et al. 2006a; Colmenares et al. 2007). In this article, we show that an amino acid substitution, E395K in DCL1, promotes miRNA processing in the absence of DCL1's partner HYL1. HYL1 is required for efficient and precise miRNA processing (Kurihara et al. 2006; Dong et al. 2008) probably via the presentation or loading of dsRNA substrates onto DCL1. The dcl1-13 mutation in the helicase domain of DCL1 allows DCL1 to compensate for the loss of HYL1, resulting in the promotion of cleavage of miRNA precursors and the production of miRNAs (Fig. 3). These results support the notion that the DCL1 helicase domain is involved in the recognition and/or presentation of RNA substrates as well as HYL1, and that the mutation promotes its activity.
Curiously, it was found that the effect of this mutation is HYL1 dependent, and opposite in the presence and absence of HYL1. In the absence of HYL1 (hyl1-2 dcl1-13 double mutant), miRNA processing in the hyl1 mutant was restored, indicating that DCL1E395K could function in the place of HYL1, possibly by recognizing/loading RNA substrates. Conversely, in the presence of HYL1 (dcl1-13 single mutant), the processing of miRNA precursors was slightly impaired, suggesting that DCL1E395K has less cleavage activity than WT DCL1, while the RNA recognition/loading was normally performed by HYL1. From these findings, we propose that the DCL1 helicase domain is involved not only in the recognition/loading of RNA substrates as described above, but also in RNA cleavage. DCL1E395K is assumed to have enhanced recognition/loading activity toward RNA substrates, but to have slightly impaired cleavage activity. The helicase domain may function as an intramolecular switch facilitating the cleavage activity of RNase III domains after it recognizes and/or is loaded onto dsRNA.
Recently, it was reported that the helicase domain of human Dicer normally inhibits its catalysis, and that the interaction of Dicer with the dsRNA-binding protein TRBP may induce a structural rearrangement of the helicase domain to trigger catalytic activation (Ma et al. 2008). This model may explain our mutant phenotype; in the absence of HYL1, the DCL1 helicase domain has this inhibitory effect, which is removed by the dcl1-13 mutation. However, a domain of Dicer that is required to bind to a partner dsRNA-binding protein is the helicase domain in humans (Lee et al. 2006), but it is the dsRBD in Arabidopsis (Hiraguri et al. 2005). Therefore, the molecular mechanism underlying how a dsRNA-binding protein promotes Dicer activity might be different among the Dicer proteins of humans and Arabidopsis. To gain some clues into this molecular mechanism, we performed an in vitro ATPase/helicase assay using a recombinant DCL1 helicase domain, but could not determine whether the helicase domain of DCL1E395K is activated or inactivated. An in vitro miRNA processing assay would answer the fundamental questions of whether DCL1E395K has more or less cleavage activity than WT DCL1, and whether this DCL1 activity is altered by HYL1. It will also be intriguing to investigate the conservation of the effects of this mutation in other organisms, and to analyze the molecular mechanism underlying the regulation of Dicer activity by the helicase domain in detail.
MATERIALS AND METHODS
Plant materials and growth conditions
The hyl1-2 (SALK_064863) seeds were obtained from the SALK collection. The Col-0 accession was used as a WT control. Surface-sterilized seeds were plated onto half-strength Murashige-Skoog salts supplemented with 100 mg/L myo-inositol, 1 mg/L nicotinic acid, 1 mg/L pyridoxine hydrochloride, 10 mg/L vitamin B1 hydrochloride, 0.5 g/L 12-(N-morpholino)ethanesulfonic acid (MES), 10 g/L sucrose, and solidified with 0.8% agar. After 3 d at 4°C in the dark, plates were incubated at 22°C with 16 h/8 h of light/dark cycle. After 12 to 14 d, plants were transferred to the mixed soil of Jiffy mix (SAKATA) and vermiculite.
EMS mutagenesis and screening for hyl1 suppressors
To isolate suppressors of the hyl1 phenotype, hyl1-2 seeds were mutagenized with 0.1% or 0.2% ethyl methanesulfonate (EMS) for 16 h at room temperature. Mutagenized M1 seeds were subsequently washed several times with water and grown on soil. M2 seeds were collected separately; five M2 pools were collected from plants treated with 0.1% EMS and one pool was collected from 0.2% EMS-treated plants. Approximately 4000 to 7000 M2 seeds from each pool (38,000 seeds in total) were grown on plates for screening. Seedlings whose cotyledons were larger and rounder than hyl1-2 were selected as suppressors of hyl1. The obtained suppressors were backcrossed to the original hyl1-2 plants to remove excess mutations and to analyze the phenotypes and the nature of the mutations.
Phenotypic analysis
Mutant phenotypes were analyzed using the progeny of heterozygous parents to remove maternal effects. The degrees at leaf bases of third and fourth rosette leaves in WT, hyl1-2, and hyl1 suppressors were measured using ImageJ (http://rsb.info.nih.gov/ij/). The degree and leaf number were statistically compared between each possible pair of genotypes. First, a two-tailed F-test was performed to check the equality of variance at the P = 0.05 level. Next, a two-tailed Student's t-test was performed for groups with equal variances, and a two-tailed Welch's t-test was performed for groups with unequal variances. The Bonferroni correction was applied for multiple comparisons. The tendency was confirmed in three independent experiments.
Northern blot analysis
Northern blot analyses were performed as described previously (Kurihara et al. 2006), with some modifications as follows. Total RNA was extracted from 4-wk-old rosette leaves of Arabidopsis using Trizol (Invitrogen). Two micrograms of total RNA was used for detection of small RNAs, and 5 μg of total RNA was used for detection of pri-/pre-miRNAs. The antisense oligos used for detection of small RNAs are listed in Table 1.
TABLE 1.
Antisense oligo sequences used for small RNA detection
The levels of small RNAs were compared between genotypes by calibrating band signal intensities to RNA levels. First, the calibration curve was made as follows. A twofold dilution series (five steps) of total RNA from WT plants (10 μg in the largest amount) was separated, and miR169 and siR255 were detected as previously described (Kurihara et al. 2006) (Supplemental Fig. S2A: blots). The signal intensities of these bands were determined from the original figures using ImageJ. The background signals were subtracted from band signals and the relative signal intensities were determined with the maximal levels set to 1.00 (Supplemental Fig. S2A: values are indicated at the bottom of each lane). The values were plotted as Supplemental Figure S2B: x-axis is dilution (2x dilution) and y-axis is relative signal intensity. Approximate curves were determined from the plots: y = 0.0949x + 1 (R 2 = 0.9765) for miR169 and y = 0.1079x + 1 (R 2 = 0.996) for siR255. Thus, in this study, an average coefficient (0.101) was used: y = 0.101x + 1. The relative signal intensities (y) for small RNA blots were determined in the same way with levels in WT plants set to 1.00. The relative level of each small RNA was calculated by 2x = 2^{(y − 1)/0.101}. The tendency was confirmed in three independent experiments.
SUPPLEMENTAL MATERIAL
Supplemental Material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Nina Fedoroff (Penn State University) for providing the hyl1-1 seeds and Dr. Munetaka Sugiyama (the University of Tokyo) for giving advice on map-based cloning methods. This work was supported by a Grant-in-Aid from the Japan Society for Promotion of Science for Young Scientists (to Y.T.) and a Grant-in-Aid for Science Research on Priority Areas (Grant No. 20061007) from the Ministry of Education, Culture, Sports, Science and Technology (to Y.W.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1297109.
REFERENCES
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bates P.A., Kelley L.A., MacCallum R.M., Sternberg M.J. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;5(Suppl.):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares S.U., Buker S.M., Buhler M., Dlakic M., Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Cuff J.A., Clamp M.E., Siddiqui A.S., Finlay M., Barton G.J. JPred: A consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Dong Z., Han M.H., Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Utsumi M., Ohba Y., Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis . Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Golden T.A., Schauer S.E., Lang J.D., Pien S., Mushegian A.R., Grossniklaus U., Meinke D.W., Ray A. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis . Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana . Plant Mol. Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Jacobsen S.E., Running M.P., Meyerowitz E.M. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Hur I., Park S.Y., Kim Y.K., Suh M.R., Kim V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E., Macrae I.J., Kirsch J.F., Doudna J.A. Autoinhibition of human Dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I.J., Li F., Zhou K., Cande W.Z., Doudna J.A. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol. 2006a;71:73–80. doi: 10.1101/sqb.2006.71.042. [DOI] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006b;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:1202–1212. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crete P., Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]