Abstract
Lin28 is highly expressed in human and mouse embryonic stem (ES) cells. Here, we show that in mouse ES cells, specific repression of Lin28 results in decreased cell proliferation, while overexpression of Lin28 accelerates cell proliferation. Further, Lin28 associates specifically with ribonucleoprotein particles containing mRNAs for cyclins A and B and cdk4. Importantly, changes in Lin28 levels lead to corresponding changes in the levels of these proteins, and sequences from the 3' untranslated regions of cyclin B and cdk4 mRNAs exhibit stimulatory effects on translation of reporter genes in a Lin28-dependent fashion. Thus, we postulate that Lin28 may play a role in the regulation of translation of genes important for the growth and maintenance of pluripotent cells.
Keywords: Lin28, cell cycle, RNP, embryonic stem cell
INTRODUCTION
Lin28 is an evolutionarily conserved RNA-binding protein originally identified as a key regulator of developmental timing in Caenorhabditis elegans (Moss et al. 1997). In mammals, Lin28 is widely expressed in early-stage embryos, with expression decreasing and becoming restricted to a limited number of tissues as embryonic development proceeds. The skeletal and cardiac muscles are among the few adult tissues in which Lin28 expression persists (Yang and Moss 2003). In human and mouse embryonic stem (ES) cells, Lin28 is abundantly expressed, and expression is down-regulated upon induction of differentiation (Richard et al. 2004; Polesskaya et al. 2007). The functional significance of Lin28 is further highlighted by its ability to enhance the efficiency in reprogramming human fibroblasts to pluripotency (Yu et al. 2007).
Predominantly cytoplasmic, Lin28 can be found in a variety of complexes including messenger ribonucleoprotein particles (mRNPs), polysomes, P-bodies, and stress granules (Balzer and Moss 2007; Rybak et al. 2008). Functionally, Lin28 associates with RNPs containing IGF-2 mRNA and promotes its translation in differentiating muscle cells (Polesskaya et al. 2007). It also inhibits the processing of let-7 microRNAs that have been implicated in the regulation of cell growth and differentiation in embryonic cells (Heo et al. 2008; Newman et al. 2008; Piskounova et al. 2008; Roush and Slack 2008; Rybak et al. 2008; Viswanathan et al. 2008). Thus, Lin28 can regulate gene expression through multiple mechanisms. Here, we report that Lin28 regulates mouse ES cell proliferation likely in part through direct modulation of the expression of a subset of cell cycle genes including cyclins A and B and cdk4.
RESULTS AND DISCUSSION
In the process of studying RNA-binding proteins in neuronal cells, we noticed that Lin28, which is highly expressed in undifferentiated mouse neuroblastoma cells, appeared to promote cell proliferation in these cells. We thus speculated that Lin28 might perform a similar function in mammalian ES cells. To ask whether inhibition of Lin28 could slow cell proliferation, we reduced Lin28 expression using Lin28-specific siRNAs and observed ∼20% fewer cells in Lin28 siRNA-treated versus control siRNA-treated cells 48 h after the transfection. On the other hand, overexpression of Lin28 by Flag-Lin28 transfection resulted in an ∼35% increase in cell number compared with vector-transfected cells. The observed cell-number changes were not due to altered cell death rate, as caspase activity assays (measurements of cell apoptosis) showed no significant differences between the cells after the various treatments. These results suggest that Lin28 may play a role in ES cell growth. To determine whether Lin28 affects ES cell cycle structure, we performed transfection, BrdU incorporation, and flow cytometric analysis. As shown in Figure 1A, suppression of Lin28 led to a modest but consistent increase in the population of ES cells in S phase (69.15 ± 0.37% in Lin28 siRNA transfected versus 63.94 ± 1.36% in control siRNA transfected). This was accompanied by a decreased population in G2/M phase (16.89 ± 0.04% in Lin28 siRNA transfected versus 20.07 ± 0.8% in control siRNA transfected), consistent with decreased progression from S to G2/M phase. Conversely, in a separate experiment, overexpression of Lin28 resulted in a decreased population of ES cells in S phase (64.20 ± 0.06% in Flag-Lin28-transfected versus 71.2 ± 0.37% in vector transfected) that was accompanied by an increased population in the G2/M phase (18.65 ± 0.18% in Flag-Lin28 transfected versus 11.31 ± 0.98% in vector transfected) (Fig. 1B), consistent with increased progression from S to G2/M phase. Due to a relatively higher nonspecific cytoxicity associated with plasmid DNA transfection, the cell cycle patterns differ somewhat between control siRNA and vector transfected cells. Therefore, the percentages between panels A and B cannot be compared directly. Nonetheless, these results imply that Lin28 in its natural context may function to facilitate the progression from S to G2/M phase, which, in turn, accelerates cell proliferation.
FIGURE 1.
Lin28 expression alters cell cycle structure. (A,B) ES cells were transfected with the indicated siRNAs or plasmid DNAs as previously described (Zhang et al. 2007), respectively. Forty-eight hours later, cells were pulse-labeled with BrdU for 1 h and processed for flow cytometric analysis. The percentages of cells in the G1-, S-, or G2/M phase are shown, together with representative dot plots. Values are the mean of two experiments.
To understand how Lin28 may affect cell proliferation, we set out to identify genes whose expression might be modulated by Lin28 in a way similar to that of IGF-2 in muscle cells (Polesskaya et al. 2007). Thus, mouse ES cells were transfected with Flag-Lin28. Flag-lin28-associated RNPs were isolated by immunoprecipitation (IP) using an anti-Flag antibody. RNA samples extracted from IP complexes were used to generate cDNAs by reverse transcription, followed by quantitative real-time PCR analysis to identify associated mRNAs. Figure 2A is a representative result of multiple independent experiments, showing the abundance of mRNAs present in the anti-Flag, relative to those in control (using pre-immune IgG) IP complexes. Cdk4 and cyclin B mRNAs exhibited the most dramatic enrichment in the anti-Flag IP complexes, followed by cyclin A mRNA. Oct4, cdk6, and cyclin D mRNAs displayed less than fivefold enrichment, while the rest showed no enrichment at all. Given that the relative fold enrichment did not correlate with the steady-state levels of the mRNAs (Fig. 2B), the preferential enrichment of cyclins A and B and cdk4 mRNAs strongly suggests that these may be novel in vivo targets for Lin28 regulation.
FIGURE 2.
Lin28 associates with a specific subset of mRNAs. (A) Flag-Lin28 was transfected into mouse ES cells. Immunoprecipitation (IP) was carried out 24 h later and RNP-associated RNAs analyzed by RT-qPCR. mRNA levels present in the anti-Flag relative to control IgG complexes are shown. Each bar represents mean ± SD (n = 3). (B) Steady-state mRNA levels plotted relative to those of β-actin mRNA. Each bar represents mean ± SD (n = 3).
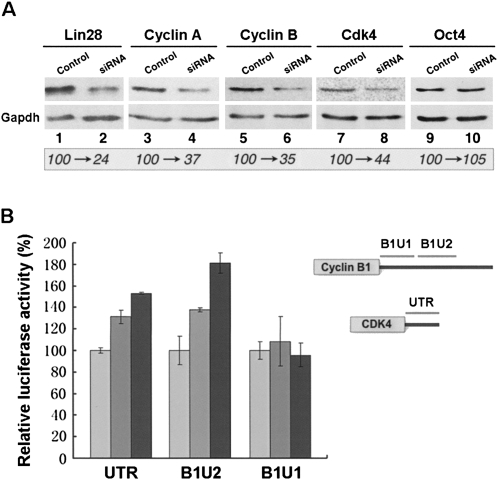
To determine how Lin28 might influence its associated mRNAs, we first examined the steady-state levels of its putative target mRNAs by transient transfection and real-time RT–PCR assays. In repeated experiments, we did not observe significant changes in the levels of its associated mRNAs after modulating Lin28 levels by overexpression and siRNA knock down (data not shown). However, by Western blot analysis, we consistently observed changes in the levels of proteins expressed from cyclins A and B and cdk4 mRNAs. Figure 3A shows representative results of multiple independent experiments having similar outcomes. In cells where Lin28 levels were reduced to 24% (Fig. 3A, cf. lanes 1 and 2), the levels of cyclin A, cyclin B, and cdk4 proteins were reduced to 37% (Fig. 3A, lanes 3,4), 35% (Fig. 3A, lanes 5,6), and 44% (Fig. 3A, lanes 7,8) of those in control siRNA-treated cells, respectively. In these experiments, the level of Oct4 protein was not noticeably affected by Lin28 siRNA (lanes 9,10), suggesting that the pluripotency of the ES cells was not compromised by Lin28 siRNA transfection. Reciprocal results were obtained when Lin28 was overexpressed by Flag-Lin28 transfection (data not shown). These results indicate that Lin28 may act to stimulate the translation of its associated mRNAs. To test this possibility, we inserted fragments from the 3′ UTRs of cyclin B and cdk4 mRNAs into a firefly luciferase reporter construct. The constructs were transiently transfected into HEK293 cells that do not express endogenous Lin28, with or without cotransfection of Flag-Lin28, followed by luciferase activity assays. As shown in Figure 3B, a 261-nt fragment derived from the entire cdk4 3′ UTR, as well as a 317-nt fragment from the middle part of the cyclin B 3′ UTR, were able to enhance luciferase activity in a Lin28-dependent manner. In contrast, a 297-nt fragment from the first part of the cyclin B 3′ UTR failed to stimulate luciferase activity. These results imply that the cyclin B and cdk4 mRNAs may contain sequence elements in their 3′ UTRs that enables translational stimulation mediated by Lin28.
FIGURE 3.
Lin28 may affect the translation of its associated mRNAs. (A) Mouse ES cells were transfected with Lin28 siRNA or control siRNA, and cell proteins isolated and subjected to Western blot analysis. (Top) Western blots using antibodies. (Bottom) Western blots of the same membranes but using an antibody specific for gapdh. Numbers in italics at the bottom indicate protein levels as percentage of Lin28 siRNA treated compared with control siRNA treated after normalization using gapdh signals. (B) Luciferase reporter constructs (Vasudevan and Steitz 2007) containing sequences from cdk4 3′ UTR (UTR), the middle (B1U2), or first (B1U1) part of cyclin B 3′ UTR, were transfected into HEK293 cells together with increasing amounts of Flag-Lin28. Luciferase activities were measured 24 h after the transfection and relative luciferase activities plotted. Luciferase activities from cells without Flag-Lin28 cotransfection were arbitrarily set as 100%. Results are representatives of at least three independent experiments. Numbers shown are means of duplicate samples. Shown on the right are schematic mappings of the individual fragments. Figures are not drawn to scale.
Evidence exists that pluripotency may be mechanistically linked to the unique proliferative properties of ES cells. The mouse ES cell cycle is characterized by very rapid proliferation with an elongated S phase and a shortened G1 phase (for review, see White and Dalton 2005; Berthet and Kaldis 2007). While the protein levels of cyclin E, cdk1, and cdk2 in ES cells are similar to those in differentiated cells, those of cyclins A and B are significantly elevated, with levels only decreasing upon ES cell differentiation (Fujii-Yamamoto et al. 2005). Our studies show that cyclins A and B mRNAs are present in Lin28-containing RNPs (Fig. 2), and that the levels of their encoded proteins change in response to those of Lin28 in ES cells (Fig. 3A). Importantly, we show that the 3′ UTR of cyclin B mRNA appears to harbor sequences capable of stimulating translation of reporter genes (Fig. 3B). Thus, we postulate that Lin28 may enhance the translation of cyclins A and B mRNAs in ES cells, hence elevating the protein levels, which in turn accelerates cell cycle progression. It is intriguing to observe that Lin28 also preferentially associates with cdk4 mRNA-containing RNPs and appears to enhance its translation (Fig. 3). Although the function of cdk4 in mouse ES cells is unclear, it has been shown to be a key factor in the regulation of human ES cell cycle progression (Becker et al. 2006).
How Lin28 specifically recognizes its target mRNAs described here is currently under investigation. Although cyclins A and B were shown to be down-regulated by let-7 expression, the effects were rather indirect (Johnson et al. 2007). This is not surprising, because these mRNAs do not contain the predicted let-7-binding sites, as is the case for mouse cdk4 (the human cdk4 has let-7-binding sites) (http://microRNA.org; http://microrna.sanger.ac.uk). It is conceivable that in ES cells it may be important to prevent the production of functional let-7 microRNAs in order to maintain a state of rapid proliferation (Heo et al. 2008; Newman et al. 2008; Piskounova et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008). Our work, on the other hand, suggests that Lin28 may perform additional and complementary tasks in regulating ES cell proliferation. We propose that in addition to its role in microRNA processing, Lin28 may promote the expression of pro-proliferatory genes by directly enhancing their translation, which would provide a positive feedback system in which to maintain tight control over ES cell proliferation.
MATERIALS AND METHODS
Antibodies, siRNAs, and DNA plasmids
The anti-Lin28 (Abcam, ab46020), anti-Flag (Sigma, F3165), anti-cyclin A2 (Chemicon, 06-138), anti-cyclin B1 (Chemicon, MAB3684), anti-cdk4 (Chemicon, MAB8879), and anti-Oct4 (Chemicon, AB3209) antibodies, and mouse pre-immune IgG (Chemicon, PP54), were purchased. Lin28 siRNA (Dharmacon, L-051530-01) and control siRNA (Dharmacon, D-001810-01-05) were purchased. Flag-Lin28 was created by cloning the mouse Lin28 open reading frame (accession number: NM_145833) into pFLAG-CMV-2 (Sigma, E7398) opened at the NotI and BamHI sites. Cdk4-UTR was created by inserting a 261-base-pair (bp) fragment (nucleotides 1086–1346) from the mouse cdk4 (Accession number: NM_009870) mRNA 3′UTR at the 3′UTR of a firefly luciferase reporter construct (Vasudevan and Steitz 2007) opened at the NotI and XhoI sites. Cyclin B1-B1U2 and B1U1, similarly, were made by cloning a 317-bp (nucleotides 1655–1971) or a 297-bp (nucleotides 1378–1674) fragment from the mouse cyclin B (accession number: NM_172301) mRNA 3′UTR.
Cell culture and transfection
Mouse ES cell line ES-C57BL/6 cells (ATCC, SCRC-1002) were maintained on a mitomycin-inactivated MEF (ATCC, SCRC-1040) feeder layer. The culture medium consisted of DMEM (Gibco, 11965) supplemented with 15% fetal bovine serum (ATCC, SCRR-30-2021), 1% nonessential amino acid (Sigma, M7145), 0.1 mM 2-mercaptoethanol (Sigma, M3148), 1000 unit/mL mouse LIF (Chemicon, ESGRO 107 units), and 1× antimycotic (Gibco, 15240-062). Cell transfections were carried out as described previously (Zhang et al. 2007).
Protein extraction and Western blot analyses
To measure protein levels, cells were harvested and cell pellets directly lysed in 10 vol of 3× SDS-sample buffer by heating at 100°C for 5 min with occasional vortexing to break chromosomal DNA. Cell lysates (5–10 μL/well) were resolved on 12% SDS-PAGE, followed by Western blot analyses using indicated antibodies. Band intensities of scanned films were quantified using Bio-Rad Quantity One 4.4.0 software.
BrdU incorporation and flow cytometric analysis
ES cells grown in a 24-well plate were transfected with siRNAs or DNA plasmids as described above. Forty-eight hours after the transfection, cells were in vivo labeled with BrdU using the FITC BrdU Flow Kit (BD Pharmingen, 559619) and processed for staining and FACS analysis as per the manufacturer's instructions. The measurement of cell cycle parameters was performed in a BD FACSCalibur using CellquestPro software and data processed using FlowJo 7.2.5.
Immunoprecipitation and RNA extraction
To prepare antibodies, 20 μL of protein A Sepharose beads were incubated with 50 μg of anti-Flag M2 antibody or 50 μg of mouse preimmune IgG in 500 μL IP buffer (0.5% Triton X-100, 200 mM NaCl, 10 mM Tris-HCl at pH 7.5, and 10 mM EDTA) at 4°C overnight. The next day, the beads were washed three times with IP buffer and kept on ice until used. To prepare cell lysates, ES cells transfected with Flag-Lin28 were harvested and cell pellets resuspended in 30–50 pellet volume of freshly prepared lysis buffer (0.5% Triton X-100, 10 mM NaCl, 10 mM Tris-HCl at pH 7.5, 10 mM EDTA, 0.5 mM PMSF, 1 mM DTT, 1× protease inhibitor cocktail [Calbiochem], and 400 units/mL RNase inhibitor). The suspensions were incubated on ice for 20 min. After removing insoluble materials by centrifugation, lysates were precleared using 10 μL of protein A sepharose (NaCl was added to a final concentration of 200 mM), followed by addition of yeast tRNA (Ambion) to a final concentration of 40 μg/mL. The cleared lysates were transferred to tubes containing antibody or preimmune IgG coated beads, and IP was carried out by rotating the tubes at 4°C for 3–5 h. Following IP, the beads were washed three times with IP buffer containing 10 μg/mL yeast tRNA by adding 1 mL of the buffer and rotating the tube at 4°C for 2 min each time. RNA extraction from the beads using the RNeasy mini kit from Qiagen was carried out according to the manufacturer's instructions. Reverse transcription was performed using the Bio-Rad iscript cDNA synthesis kit and cDNAs were purified using the Qiagen QIAquick PCR purification kit.
Quantitative real-time PCR
Real-time PCR was carried out on cDNA (5–50 ng) using iQ SYBR Green (Biorad) with a Biorad iCycler. All reactions were done in 25 μL volume. Primers for the individual mouse genes were as follows:
gapdh forward: 5′-TTAGCACCCCTGGCCAAGG-3′;
gapdh reverse: 5′-CTTACTCCTTGGAGGCCATG-3′;
Beta-actin forward: 5′-GTGGGCCGCTCTAGGCACCAA-3′;
Beta-actin reverse: 5′-CTCTTTGATGTCACGCACGATTTC-3′;
cdk4 forward: 5′-TGTGGAGCGTTGGCTGTATC-3′,
cdk4 reverse: 5′-TGGTCGGCTTCAGAGTTCC-3′;
Oct4 forward: 5′-TGGAGAAGGTGGAACCAACTCCC-3′;
Oct4 reverse: 5′-ACACGGTTCTCAATGCTAGTTCGC-3′;
Nanog forward: 5′-GAGATGCTCTGCACAGAGGCTG-3′;
Nanog reverse: 5′-GAGGCAGGTCTTCAGAGGAAGG-3′;
Sox2 forward: 5′-GCACATGAACGGCTGGAGCAACG-3′;
Sox2 reverse: 5′-TGCTGCGAGTAGGACATGCTGTAGG-3′;
Cyclin E (E1) forward: 5′-GCGCGCGCAGAGGTTTCATAG-3′;
Cyclin E (E1) reverse: 5′-TGCAAGGGCTGATTCCTCCAGA-3′;
Cyclin B (B1) forward: 5′-TCCCTCGGTGGGATTCAAGTGC-3′;
Cyclin B (B1) reverse: 5′-CAGGAGTGGCGCCTTGGTATGG-3′;
Cyclin D (D1) forward: 5′-GCTGGAGGTCTGTGAGGAGCA-3′;
Cyclin D (D1) reverse: 5′-CTCCTCGGGCCGGATAGAGTT-3′;
cdk6 forward: 5′-GGCCCTTACCTCGGTGGTCGTC-3′;
cdk6 reverse: 5′-GGCCACGTCCCTAGGCCAGTCT-3′;
cdk2 forward: 5′-CCGGCTCGACACTGAGACTGAAG-3′;
cdk2 reverse: 5′-GAGGAATGCCCGTGAGAGCAGA-3′;
cdk1 forward: 5′-TTGGAGAAGGTACTTACGGTGTGGTG-3′;
cdk1 reverse: 5′-CCAGGAGGGATGGAGTCCAGGT-3′;
Cyclin A (A2) forward: 5′-GCTCAAGACTCGACGGGTTGC-3′; and
Cyclin A (A2) reverse: 5′-GCTGCATTAAAAGCCAGGGCATC-3′.
PCR was performed by initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C. PCR using water instead of template was used as a negative control. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis.
Luciferase assays
Firefly reporter constructs were transfected into HEK293 cells, along with increasing amounts of Flag-Lin28 DNA. In addition, a Renilla reporter was included in all transfections to normalize for extract concentrations, transfection efficiency, and overall translation efficiency. Transfection was carried out in a 48-well plate scale. The amount of total plasmid DNA per well was 400 ng that included 40 ng of firefly reporter DNA, 2 ng of Renilla DNA, and 0, 15, or 30 ng of Flag-Lin28. Further increasing in the amount of Flag-Lin28 DNA in the transfection mix did not lead to further increase in the firefly luciferase activity under the current transfection condition. Luciferase activities were determined using a TD 20/20n (Turner BioSystems) and the Dual Luciferase Assay System (Promega) according to the manufacturer's instructions.
ACKNOWLEDGMENTS
We thank Joan Steitz for the firefly and Renilla reporter constructs. This work was supported by a stem cell grant from the Technology Initiatives for Connecticut Innovations, and funds from the Department of Obstetrics, Gynecology and Reproductive Sciences at Yale University School of Medicine to Y.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1368009.
REFERENCES
- Balzer E., Moss E.G. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Becker K.A., Ghule P.N., Therrien J.A., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. Self- renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Berthet C., Kaldis P. Cell-specific responses to loss of cyclin-dependent kinases. Oncogene. 2007;26:4469–4477. doi: 10.1038/sj.onc.1210243. [DOI] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J., Kim N.V. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Fujii-Yamamoto H., Kim J.M., Arai K., Masai H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J. Biol. Chem. 2005;280:12976–12987. doi: 10.1074/jbc.M412224200. [DOI] [PubMed] [Google Scholar]
- Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Chin L., et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:15399–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Viswanathan S.R., Janas M., Lapierre R.J., Daley G.Q., Sliz P., Gregory R.I. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes & Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Tan S.P., Tan J.H., Chan W.K., Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Roush S., Slack F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Steitz J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- Yang D.H., Moss E.G. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang M., Guller S., Huang Y. Method to enhance transfection efficiency of cell lines and placental fibroblasts. Placenta. 2007;28:779–782. doi: 10.1016/j.placenta.2007.01.012. [DOI] [PubMed] [Google Scholar]