Abstract
In animals, microRNAs (miRNAs), typically, pair to sites of partial complementarity in the 3′-untranslated regions (3′UTRs) of target genes. Regulation by miRNAs often results in down-regulation of target mRNA and protein expression by mechanisms that are yet to be fully elucidated. Additionally, changes in environmental conditions have been shown to influence miRNA function in some cell culture systems. Here, we report the effect of nutrient deprivation on regulation of an endogenous miRNA target in developing worms. In Caenorhabditis elegans, the lin-4 miRNA recognizes multiple sites in the lin-14 3′UTR and directs mRNA degradation and translational repression, but it is unclear how these processes are coupled. In this study, we demonstrate that nutrient deprivation results in loss of lin-14 mRNA, but not protein, repression. In worms removed from feeding conditions, lin-14 mRNA reaccumulates despite the continued expression of lin-4 miRNA. The relative increase in lin-14 mRNA levels during nutrient deprivation is less pronounced in genetic mutants lacking lin-4 miRNA or the lin-14 3′UTR target sites. In conclusion, regulation of lin-14 at the mRNA and protein levels can be uncoupled by changes in culture conditions, indicating that miRNA function can be modulated by environment in multicellular organisms. The awareness that endogenous miRNA pathways can be sensitive to environment is an important consideration for elucidating the mechanism used by miRNAs to regulate target mRNA and protein expression.
Keywords: lin-14, lin-4, miRNA, C. elegans, nutrient deprivation
INTRODUCTION
The lin-4 miRNA and its lin-14 target were discovered as genes essential for developmental timing in Caenorhabditis elegans. Accumulation of lin-4 miRNA toward the end of the first larval stage (L1) is associated with down-regulation of lin-14 protein and mRNA levels, and this decrease is dependent on the miRNA and its target sites in the lin-14 3′-untranslated region (3′UTR) (Lee et al. 1993; Wightman et al. 1993; Ha et al. 1996; Feinbaum and Ambros 1999; Olsen and Ambros 1999; Bagga et al. 2005; Chendrimada et al. 2007). As is the case for lin-4 and its lin-14 target, the molecular mechanism employed by miRNAs to control gene expression is yet to be fully resolved. Current evidence indicates that miRNAs can direct mRNA degradation, particularly through deadenylation, as well as inhibit translation at the initiation or post-initiation steps, including the possibility of proteolysis of nascent peptides encoded by target mRNAs (Filipowicz et al. 2008). The effect of altered cell culture conditions on the regulation of some miRNA targets further complicates the elucidation of miRNA function. For example, amino acid deprivation in Huh7 hepatoma cells relieves translational repression by miR-122 of the cationic amino acid transporter mRNA (Bhattacharyya et al. 2006). Thus, miRNA function can be influenced by extrinsic factors, and understanding the potential sensitivity of miRNA function to environment is important for determining a coherent model of target gene regulation in vivo.
In this study, we investigated the effect of nutrient deprivation on regulation of endogenous lin-14 by lin-4 miRNA in C. elegans. We observed that the coordinated down-regulation of lin-14 mRNA and protein levels becomes uncoupled when worms are removed from feeding conditions. After 4–10 h of food deprivation, lin-14 mRNA, but not protein, was derepressed. The increase in lin-14 mRNA levels occurred despite the maintenance of lin-4 miRNA. Thus, changes in culture conditions can differentially affect expression of target mRNA and protein in a living organism.
RESULTS AND DISCUSSION
lin-14 mRNA and protein levels decrease as lin-4 miRNA accumulates
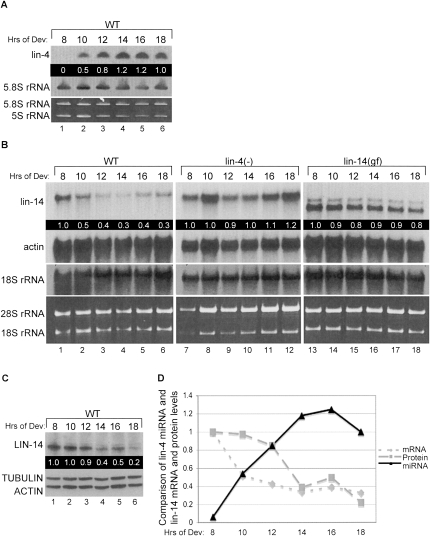
To investigate the coordination of lin-4 miRNA expression with lin-14 mRNA and protein down-regulation, we synchronized wild-type (WT) worms and collected them at 2-h intervals midway through the first (8–14 h) to the beginning of the second (16–18 h) larval stages as they developed on standard worm plates seeded with bacteria. Appearance of lin-4 miRNA correlates with a twofold decrease in lin-14 mRNA levels between 8 and 10 h of development (Fig. 1A,D). In contrast, no significant decrease in lin-14 mRNA levels was detected in staged samples from worm strains null for lin-4 expression [lin-4(e912)] [lin-4(−)] (Lee et al. 1993) or containing a lin-14 mutation [lin-14(n355)] [lin-14(gf)] that deletes the lin-4 target sites (Fig. 1B; Wightman et al. 1993). Western blotting showed a twofold decrease in LIN-14 protein by 14 h and a fivefold drop by 18 h of development (Fig. 1C–D). Taken together, we observed tight temporal coordination between lin-4 miRNA accumulation and lin-14 mRNA decrease followed closely by down-regulation of LIN-14 protein levels (Fig. 1D).
FIGURE 1.
lin-14 mRNA and protein are down-regulated by lin-4 miRNA. (A) Total RNA samples from N2 wild-type (WT) staged larvae developing on food for the indicated hours were probed for lin-4 miRNA and control 5.8S rRNA by Northern blot analysis. Quality of the RNA samples was assessed by ethidium bromide staining of the small ribosomal RNAs. Mature lin-4 miRNA levels were normalized to 5.8S rRNA and values relative to the 18-h time point are indicated in the black box. (B) The WT RNA used in (A) as well as RNA collected from lin-4(e912) [lin-4(−)] and lin-14(n355) [lin-14(gf)] strains staged for the same time course were subjected to Northern blotting to detect lin-14 and control actin mRNA and 18S rRNA. Quality of the RNA samples was assessed by ethidium bromide staining of the large ribosomal RNAs. The lin-14 mRNA levels were normalized to actin, and values relative to the 8-h time point for each strain are indicated in the black box. (C) Protein extracts from WT worms staged as in A were analyzed by Western blotting with antibodies against LIN-14 and control TUBULIN and ACTIN proteins. The LIN-14 protein levels were normalized to TUBULIN, and values relative to the 8-h time point are indicated in the black box. (D) The graph depicts the changes in lin-4 miRNA (triangles), lin-14 mRNA (diamonds), and LIN-14 protein (squares) over the 10-h time course in WT worms, using values calculated as described in A, B, and C.
Down-regulation of lin-14 mRNA but not protein is disrupted upon nutrient deprivation
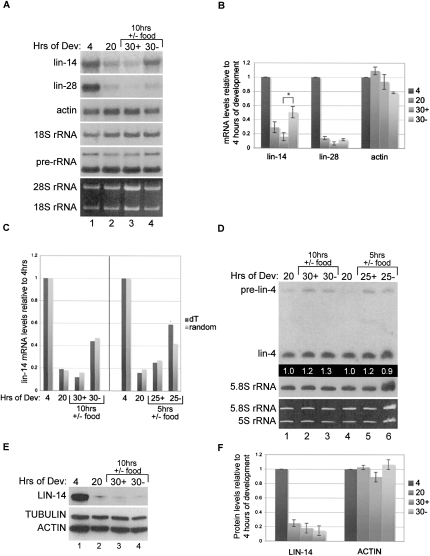
Having established the time course and relative magnitude of lin-14 repression by lin-4 miRNA, we next asked if this regulation was reversible. Since feeding is required for expression of lin-4 miRNA (Feinbaum and Ambros 1999), we chose to test the effect of starvation conditions on regulation of lin-14 (see Materials and Methods). After 10 h of nutrient deprivation, we observed a significant increase in lin-14 mRNA levels compared to samples from continuously fed worms (Fig. 2A–C; Supplemental Fig. 1A). Moreover, the levels of lin-14 mRNA in worms transferred from food are approximately twofold greater than the levels detected at the time of the shift (Fig. 2A–C; Supplemental Fig. 1A). The absence of food for this duration did not have an appreciable effect on rRNA processing, as assessed by the levels of 5.8S containing large ribosomal precursor RNAs, or steady-state actin mRNA levels (Fig. 2A–B; Supplemental Fig. 1A). A starvation period of 5 h similarly resulted in misregulation of lin-14 mRNA levels (Fig. 2C). The lin-28 gene is also regulated by lin-4 miRNA at the mRNA and protein levels (Moss et al. 1997; Seggerson et al. 2002; Bagga et al. 2005), as well as by additional mechanisms (Morita and Han 2006). Although we did not detect reaccumulation of lin-28 mRNA to levels greater than those observed before transfer to plates without food, there appears to be a slightly higher level of mRNA in worms deprived of food for 10 h compared to the control fed worms (Fig. 2A–B; Supplemental Fig. 1A). No significant change in lin-4 precursor or mature levels was detected in worms starved for 5 or 10 h compared to fed worms (Fig. 2D).
FIGURE 2.
lin-14 mRNA but not protein is mis-regulated in worms removed from food. Wild-type worms were cultured on food and collected for RNA or protein at 4 and 20 h of development, and a portion of the 20-h population was replated with (+) or without (−) food for another 5 or 10 h and then collected at the 25-h or 30-h time points, respectively. (A) Total RNA was used for agarose Northern blot analyses to detect lin-14, lin-28, actin, 18S rRNA, and preribosomal RNAs. (B) The mRNA levels of lin-14, lin-28, and actin were normalized to 18S rRNA, and the levels relative to the 4-h time point were graphed. Quantification is the average from three independent experiments. Error bars represent SEM and *P < 0.05. (C) RNA samples from experiments with worms deprived of food for 10 h (left panel) or 5 h (right panel) were used for reverse transcription reactions using oligo dT (dark bars) or random (light bars) primers, and the resulting cDNA was used for quantitative PCR analyses. Samples were normalized to actin and lin-14 mRNA levels are graphed relative to the 4-h time point for each experiment. (D) The RNA used in C was subjected to PAGE Northern blot experiments to detect lin-4 miRNA and control 5.8S rRNA levels. Mature lin-4 miRNA levels were normalized to 5.8S rRNA, and levels relative to the 20-h time point for each experiment are indicated. (E) The same worm populations analyzed in C for the 10-h food deprivation experiment were also used to isolate protein and perform Western blotting to detect LIN-14, ACTIN, and control TUBULIN protein levels. (F) The protein levels of LIN-14 and ACTIN were normalized to TUBULIN, and the levels relative to the 4-h time point were graphed. Quantification is the average from three independent experiments. Error bars represent SEM.
LIN-14 protein does not recover despite the up-regulation of lin-14 mRNA levels when worms are removed from food. Western blot analysis of extracts from staged worms subjected to starvation for 10 h showed continued repression of LIN-14 protein and no significant effect on steady-state ACTIN protein levels (Fig. 2E–F; Supplemental Fig. 1B–C).
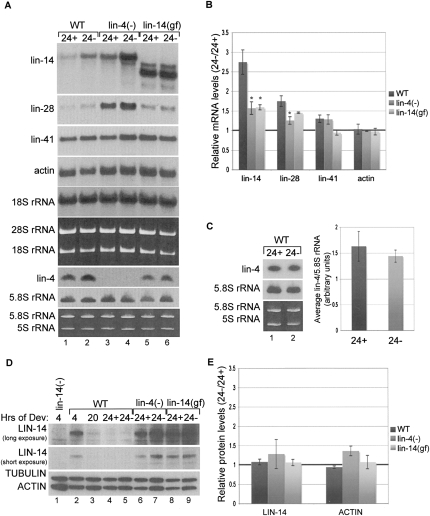
The lin-14 mRNA levels in strains defective in lin-4 miRNA-mediated regulation were less sensitive to nutrient deprivation conditions. Staged WT, lin-4(−) and lin-14(gf) strains were shifted at 20 h of development to plates with or without food and cultured for 4 h before sample collection. The time course was limited to 24 h because additional miRNAs may regulate lin-14 at later time points (Reinhart et al. 2000; Chendrimada et al. 2007). Northern blot analyses revealed greater up-regulation of lin-14 mRNA levels in WT compared to either mutant strain (Fig. 3A–B; Supplemental Fig. 2A). Deregulation of lin-14 mRNA cannot be accounted for by down-regulation of lin-4, as mature levels of this miRNA remain constant in fed versus unfed worms (Fig. 3A,C; Supplemental Fig. 2B). We detected a moderate increase in lin-28 mRNA levels in WT worms deprived of food that was diminished in the lin-4 mutant strain (Fig. 3A–B; Supplemental Fig. 2A). We also analyzed lin-41 mRNA, which is subject to degradation by let-7 miRNA activity around 32 h of development during L3 (Bagga et al. 2005). Barely any change in lin-41 mRNA levels was detected in WT worms cultured with or without food from 20 to 24 h of development (Fig. 3A–B; Supplemental Fig. 2A), indicating that substantial up-regulation is not uniformly observed for all mRNAs under these conditions.
FIGURE 3.
Disruption of lin-4 miRNA function contributes to up-regulation of lin-14 mRNA in worms shifted off of food. Wild-type (WT), lin-4(e912) [lin-4(−)], and lin-14(n355) [lin-14(gf)] worm strains were cultured on food for 20 h and then shifted to plates with (24+) or without (24−) food for 4 h (A) RNA was collected at the 24-h time point and used for Agarose Northern blot analyses to detect lin-14, lin-28, lin-41, and actin mRNAs and 18S rRNA and PAGE Northern blot analyses of lin-4 miRNA and control 5.8S rRNA. (B) The mRNA levels of lin-14, lin-28, lin-41, and actin were normalized to 18S rRNA, and the ratio of mRNA levels detected in 24− versus 24+ was calculated for each worm strain. Quantification is the average from three or more independent experiments. Error bars represent SEM and *P < 0.05 is shown for the mutant compared to the WT strains. (C) RNA from WT worms was subjected to PAGE Northern blot experiments to detect lin-4 miRNA and control 5.8S rRNA levels. RNA from four independent experiments was analyzed on the same blot (see Supplemental Fig. 2B). The lin-4 miRNA levels were normalized to 5.8S rRNA and the values from 24+ and 24− were graphed as the average from the four independent experiments and error bars represent SEM. (D) Protein extracts were collected at the 24-h time points and used for Western blot analyses to detect LIN-14, ACTIN, and control TUBULIN proteins. Additionally, 4-h and 20-h time points are shown for WT as well as a 4-h time point from worms null for LIN-14 protein expression [lin-14(−)] [lin-14(n356)(n540)] (Reinhart and Ruvkun 2001). Two different exposures are shown for the LIN-14 Western to demonstrate the absence of a specific LIN-14 band in the lin-14(−) null control worms. (E) The protein levels of LIN-14 and ACTIN were normalized to TUBULIN and the ratio of LIN-14 and ACTIN detected in 24− versus 24+ was calculated for each worm strain. Quantification is the average from three or more independent experiments. Error bars represent SEM.
In WT worms the strong down-regulation of LIN-14 protein detected between 4 and 20 h of development continues in worms cultured an additional 4 h, regardless of the presence or absence of food (Fig. 3D–E; Supplemental Fig. 2C). Also, no difference was detected in the steady-state protein levels of LIN-14 or ACTIN in the mutant worm strains cultured with or without food for 4 h (Fig. 3D–E; Supplemental Fig. 2C).
Concluding remarks
Observations of variable degrees of lin-14 mRNA versus protein regulation by lin-4 could be influenced by differences in culture conditions (Wightman et al. 1993; Olsen and Ambros, 1999; Bagga et al. 2005; Chendrimada et al. 2007). Under limiting food, liquid culture, or other stress conditions, robust inhibition of LIN-14 protein but not mRNA may ensue. Our results show that lin-4-mediated regulation of lin-14 mRNA levels is particularly sensitive to nutrient deprivation, but reaccumulation of this mRNA does not result in reestablishment of detectable LIN-14 protein levels. Up-regulation of lin-14 mRNA is reduced in worms with mutations that remove the lin-4 miRNA or the target sites in the lin-14 3′UTR, but it is presently unclear if the modest change in lin-14 mRNA levels in the mutants is attributable to miRNA, transcriptional, or other pathways during starvation conditions. Although our data are compatible with the model that during nutrient deprivation lin-4 miRNA maintains translational repression of the lin-14 mRNA, additional pathways could also contribute to inhibition of LIN-14 protein expression under these conditions. For example, reduced target of rapamycin (TOR) activity in response to nutrient deprivation can result in general down-regulation of translation (Hay and Sonenberg 2004; Avruch et al. 2006).
In conclusion, our work demonstrates that changes in environment can alter regulation of a miRNA target in a multicellular organism. Following establishment of lin-4 miRNA-mediated inhibition of lin-14 mRNA and protein expression, nutrient deprivation results in substantial reaccumulation of mRNA without change in protein or miRNA levels. Thus, stress conditions may be an important consideration in analyzing the contribution of mRNA degradation and translational repression to miRNA target regulation in different cell types or organisms.
MATERIALS AND METHODS
Worm strains and culture conditions
The following C. elegans strains were used: N2 Bristol, lin-4(e912)II (DR721), and lin-14(n355)X (MT355). Worms were cultured at 20°C and synchronized by standard hypochlorite treatment. Development was initiated by plating arrested L1 hatchlings on NGM plates seeded with OP50 bacteria. For nutrient deprivation experiments, 20-h L2 stage worms cultured on food were collected and replated on either seeded or unseeded plates and cultured for an additional 4–10 h.
Northern blotting
Polyacrylamide gel electrophoresis (PAGE) Northern methods were used to detect small RNAs (<200 nt) (Pasquinelli et al. 2003). The 5′-radiolabeled DNA oligo probe for lin-4 (A250: AAGCTCCACCCAATCAGAG) was hybridized at 50°C. The control 5.8S probe was generated by Prime-It II Random Primer Labeling Kit (Stratagene) using a gel purified PCR template amplified with the primers:
A479: CTAGCTTCAGCGATGGATCGGTTGC; and
A480: GAACCAGACGTACCAACTGGAGGCCC.
To detect larger RNAs, Northern analyses were performed by separating 5 μg of RNA in 1% agarose gels as previously described (Bracht et al. 2004). Radiolabeled DNA probes were generated from PCR fragments using the following primers:
-
lin-14:
A445: AACAGTTGAAACAGCTCCACCACTC; and
A455: GGATAAGATGGGTGAAGAGACTGATG.
-
lin-28:
A494: GACGGTAGTATCGGAGGGAAGGAATGATG; and
A427: GCGTTCGCCCGCAATAGCGGAACTTACG.
-
actin:
A810: GTGTTCCCATCCATTGTCGGAAGAC; and
A811: GCACTTGCGGTGAACGATGGATGGG.
-
lin-41:
A465: CGAGCGCTTCAGCCAAATCCCC; and
A410: GTTGTAAAATTGGGTGCGCAAG.
-
18S rRNA:
A839: GCGTACGGCTCATTAGAGCAGATATCAC: and
A840: GGTCAGAACTAGGGCGGTATCTAATCG.
Storage phosphor screens exposed to Northern blots were scanned on a Typhoon Trio PhosphorImager (GE Healthcare) and band signals were quantified using ImageQuant software. The mean and SEM were calculated from three or more independent experiments where the control sample was set to 1. A two-tailed Student's t-test was used to determine if relative mRNA levels were significantly different between conditions or worm strains.
Quantitative PCR
Quantitative PCR (qRT-PCR) analyses were performed by the TaqMan method (Applied Biosystems) as previously described (Bagga et al. 2005). Normalization was to actin mRNA levels. Primer sequences are as follows:
ceLin14-1164F: CGAGGAAGACTTTTGGGACAAA;
ceLin14-1236R: ATGCCTAACTCTGCGACATCTG;
ceLin14-1187T: TCCGCGAGGCAATGCGCA;
Ce-act1-790F: CAGCCATCCTTCTTGGGTATG;
Ce-act1-859R: CGCACTTCATGATGGAGTTGTAA; and
Ce-act1-812T:AGTCCGCCGGAATCCACGAGACTT.
Western blotting
To prepare protein for Western blotting, frozen worms were boiled in 2× SDS buffer (Reinhart and Ruvkun 2001) and concentrations were determined by Qubit (Invitrogen). Samples were run on denaturing 4%–20% gradient gels and electroblotted to Immun-Blot PVDF membranes (Bio-Rad). Western analysis for LIN-14 protein was done using 1:1000 affinity purified polyclonal anti-LIN-14 N-terminal antibodies specific for a peptide (PVAQTPTVPQPECPGQIR) (Open Biosystems) present in all isoforms (Reinhart and Ruvkun 2001). Anti-Tubulin (1:1,000, Sigma) and anti-Actin (1:20,000, MP Biomedical) were used as controls. Protein levels were quantified using Quantity One (Bio-Rad) software.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank G. Ruvkun and members of the Pasquinelli laboratory for suggestions and critical reading of the manuscript. qRT-PCR experiments were performed at the UCSD Center for AIDS Research, Genomics Core (5P30 AI36214). This work was supported by NIH (GM071654-01) and the Keck Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1258309.
REFERENCES
- Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A.E. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Feinbaum R., Ambros V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans . Dev. Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Ha I., Wightman B., Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Morita K., Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans . EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., McCoy A., Jimenez E., Salo E., Ruvkun G., Martindale M.Q., Baguna J. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: A role in life history evolution? Evol. Dev. 2003;5:372–378. doi: 10.1046/j.1525-142x.2003.03044.x. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Ruvkun G. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics. 2001;157:199–209. doi: 10.1093/genetics/157.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Seggerson K., Tang L., Moss E.G. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Post-transcriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]