Abstract
Histone mRNAs are the only eukaryotic cellular mRNAs that are not polyadenylated. Synthesis of mature histone mRNA requires only a single processing reaction: an endonucleolytic cleavage between a conserved stem–loop and a purine-rich downstream element to form the 3′ end. The stem–loop binding protein (SLBP) is required for processing, and following processing, histone mRNA is transported to the cytoplasm, where SLBP participates in translation of the histone mRNA and is also involved in regulation of histone mRNA degradation. Here we present an analysis of histone mRNA metabolism in cells with highly reduced levels of SLBP using RNA interference. Knocking down SLBP in U2OS cells results in a reduction in the rate of cell growth and an accumulation of cells in S-phase. Surprisingly, there is only a modest (twofold) decrease in histone mRNA levels. Much of histone mRNA in the SLBP knockdown cells is properly processed but is retained in the nucleus. The processed histone mRNA in SLBP knockdown cells is not rapidly degraded when DNA replication is inhibited. These results suggest a previously undescribed role for SLBP in histone mRNA export.
Keywords: 3′-end formation, stem–loop binding protein, histone mRNA, mRNA export
INTRODUCTION
The replication-dependent histone mRNAs are the only eukaryotic cellular mRNAs that are not polyadenylated, ending instead in a conserved stem–loop (SL) (Marzluff 2005). These mRNAs lack introns and undergo a single processing reaction cleaving the nascent pre-mRNA from the DNA template and producing the mature mRNA. The cleavage reaction requires the stem–loop 5′ of the cleavage site and a purine-rich region, the histone downstream element (HDE), located ∼10 nucleotides (nt) 3′ of the cleavage site. The HDE base pairs with the U7 small nuclear RNA, which is a component of the U7 snRNP (Mowry and Steitz 1987). The stem–loop binding protein (SLBP) binds to the SL and helps stabilize the interaction of U7 snRNP with the pre-mRNA. Several lines of evidence support this model. Pull-down of biotinylated histone pre-mRNA from nuclear extracts results in the concomitant pull-down of the U7 snRNA (Dominski et al. 1999, 2001), which is enhanced by SLBP. In vitro processing of histone pre-mRNA is absolutely dependent on SLBP if the HDE does not interact strongly with U7 snRNA (Dominski et al. 1999). The factor ZFP100 interacts with both the SLBP/SL complex (Dominski et al. 2002) as well as the U7 snRNP specific Sm-like protein Lsm11 (Azzouz et al. 2005; Wagner et al. 2006) and is limiting for histone pre-RNA processing in vivo (Wagner and Marzluff 2006). Mutations in the stem–loop that abolish binding of SLBP result in no expression of processed histone mRNA in vivo (Pandey et al. 1994).
The requirement of SLBP for in vitro processing of histone pre-mRNA in nuclear extracts from mammalian cells on many substrates is not absolute. Extracts depleted of SLBP are incapable of processing the H1t histone pre-mRNA but are competent to process the H2A-614 histone pre-mRNA (Dominski et al. 1999). The difference between these two substrates is that U7 snRNP base pairs more extensively with the HDE of H2A-614 than with the H1t HDE.
Once the U7 snRNP is bound to the HDE, a cleavage factor is recruited containing at least CPSF73 (Dominski et al. 2005) and Symplekin (Kolev and Steitz 2005). CPSF73 is likely the endonuclease that cleaves the histone pre-mRNA from the DNA template (Dominski et al. 2005). The other components of the cleavage factor and the mechanism of recruitment of the cleavage factor to the U7 snRNP/SLBP/SL complex have not been elucidated.
The molecular events that take place following processing in the nucleus and prior to the translation of histone mRNA in the cytoplasm are less well understood. What is clear is that the histone mRNA is rapidly exported (within 5 min) (Schochetman and Perry 1972), and export in Drosophila S2 cells (Erkmann et al. 2005a) and Xenopus oocytes (Huang and Steitz 2001) is dependent on TAP. SLBP is present in the cytoplasm only when histone mRNA is present (Erkmann et al. 2005b), where it is specifically associated with the histone mRNP (Whitfield et al. 2004; Townley-Tilson et al. 2006). In Xenopus oocytes, expression of SLBP stimulates the translation of reporter genes ending in a stem–loop (Sanchez and Marzluff 2002). Recently it has been reported that SLBP interacts with a novel protein, SLIP1, which also interacts with EIF4G and plays a role in translation of histone mRNA (Cakmakci et al. 2008). These data are all consistent with a role of SLBP in the translation of histone mRNA.
In late S phase or when DNA replication is inhibited, histone mRNAs are rapidly degraded. The rapid decay of existing histone mRNA likely requires SLBP. Upf1 binds to SLBP at the 3′ end of histone mRNA immediately after inhibition of DNA replication and is required for histone mRNA degradation (Kaygun and Marzluff 2005a). Recently, we reported that an oligo(U) tail is added to the 3′ end of histone mRNA shortly after inhibiting DNA replication (Mullen and Marzluff 2008). This oligo(U) tail provides a binding site for Lsm1-7, which binds to the oligouridylated histone mRNA together with SLBP. The Lsm1-7 complex likely recruits the enzymes that degrade histone mRNA as it does for poly(A) mRNAs (Parker and Song 2004). SLBP is a cell-cycle regulated protein that accumulates just before entry into S-phase to activate histone pre-mRNA processing, and SLBP is degraded at the end of S-phase. Stabilization of SLBP by mutating the phosphorylation sites required for degradation does not affect degradation of histone mRNA (Zheng et al. 2003), indicating that SLBP degradation is not required for histone mRNA degradation.
In order to understand the critical roles of SLBP in cells, we knocked down SLBP using RNA interference in U2OS cells. Previously, we, and others, had shown that reduction of SLBP levels results in a reduction in the rate of cell growth and an accumulation of cells in S-phase (Zhao et al. 2004; Wagner et al. 2005). However, a thorough molecular analysis of the effect of SLBP knockdown on histone mRNA metabolism has not been reported. Here we show that the major lesion in histone mRNA metabolism as a result of knocking down SLBP is not in histone pre-mRNA processing, but in the export of histone mRNA to the cytoplasm. Properly processed histone mRNA accumulates in the nucleus. Together, these results implicate SLBP as having a critical histone mRNA export function in mammalian cells, as well as its known roles in histone pre-mRNA processing, histone mRNA translation, and regulation of histone mRNA half-life.
RESULTS
SLBP is required for cell growth
We determined the effect of knocking down SLBP in U2OS cells using a double-stranded siRNA that specifically targets SLBP mRNA. Previously we have shown that knockdown of SLBP with this siRNA results in an increase in S-phase cells, and that we can rescue this phenotype by expressing an RNAi-resistant SLBP. Moreover, Muller and colleagues (Zhao et al. 2004) demonstrated that reduced expression of SLBP also resulted in a defect in S-phase progression and a reduced expression of histone mRNAs. Both studies demonstrated S-phase defects but did not elucidate the specific molecular defects underlying this phenotype.
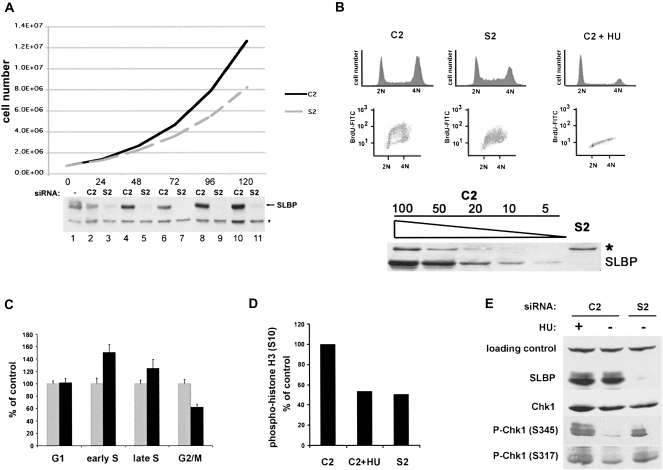
Since we initially observed variable results, which were dependent on the extent of SLBP knockdown, we optimized the knockdown, both for extent and duration, allowing us to assay for histone mRNA metabolic defects while still maintaining efficient SLBP knockdown. We used Western blotting to monitor SLBP protein levels over various times throughout the RNAi transfection (Fig. 1A). Reproducible phenotypes required a knockdown of SLBP >95%, as determined by comparing Western blot intensities from SLBP knockdown cells and a series of dilutions of the control lysates (Fig. 1B, bottom panel). U2OS cells were plated in 24 well dishes and, the following day, transfected with either control (C2) or SLBP (S2) siRNA. We determined the levels of SLBP every 24 h. The day after the first transfection (24 h) (Fig. 1A, lanes 2,3), SLBP protein levels were slightly lower in C2 treated cells than in untransfected cells (Fig. 1A, lane 2). This decrease is likely due to the high cell density used to achieve effective transfection, resulting in a reduction in the number of S-phase cells that contain SLBP. After 24 h, we split the cells out of the 24 well dishes and replated them into 6-well tissue culture dishes. Twenty-four hours later (48 h after first siRNA treatment), we transfected the second dose of siRNA and monitored SLBP levels for the next 4 d. Between 72 and 120 h following the initial hit of siRNA, SLBP protein levels were down-regulated >95%, and experiments assaying histone mRNA metabolism were done during this time. Note that there is clearly some residual SLBP present in the cells at all times. By 144 h, SLBP levels had started to recover (data not shown).
FIGURE 1.
SLBP knockdown cells are viable but grow at a slower rate as a result of various S-phase defects. (A) Growth curve (total cell number) of control (C2) and SLBP (S2) RNAi populations. Lysates from the time points shown in the graph were probed for levels of SLBP by Western blotting; (*) cross-reacting band. (B) SLBP knockdown results in S-phase accumulation and a reduced rate of DNA replication. C2 and S2 treated (72 h) U2OS cells were pulsed for 1 h with 10 μM bromodeoxyuridine (BrdU) and harvested and analyzed by FACS for DNA content and DNA synthesis. Hydroxyurea (HU; 5 mM) was added to a parallel culture of C2 cells 1 h prior to the addition of BrdU. Cells were fixed and BrdU incorporation detected with an anti-BrdU antibody conjugated to FITC and for DNA content with propidium iodide. Cells were analyzed using a FACScan device and Summit software for DNA content and intensity of BrdU incorporation. The levels of SLBP in a typical experiment are shown below the graph. They were determined by Western blotting compared to a dilution series of a control extract (1:2 to 1:20). (C) The cell cycle distribution (G1, early and late S phase, and G2/M) was quantified from the data in B ([gray bars] C2; [black bars] S2) from three independent experiments. Quantities are expressed as a percentage of the C2 control cells. (D) Parallel cultures from B were fixed and stained with an antibody to the mitotic marker phospho-histone H3, and the antibody was detected with a secondary antibody conjugated to FITC. Cells were analyzed similarly as in B, and the percentage of cells with fluorescent signal in the G2 population was quantified. The number of phospho-histone positive cells is expressed as a percentage of the C2 control cells. (E) SLBP knockdown results in the activation of a Chk1-dependent cell cycle checkpoint. Protein lysates from SLBP knockdown cells were analyzed by Western blotting for SLBP, total Chk1 protein, and two phosphorylation sites (serines 317 and 345) using phospho-specific Chk1antibodies. Cells treated with HU for 1 h were used a positive control for Chk1 activation.
As previously shown, knockdown of SLBP to this level resulted in an increase in the number of cells in S-phase (Wagner et al. 2005). We determined the rate of growth of the cells by measuring cell number at the various time points in Fig. 1A. The SLBP knockdown cells grew more slowly, resulting in 30%–40% fewer cells after 5 d of growth. All the cells were viable, and there was no evidence of apoptosis, suggesting that the cells were growing at a slower rate.
Reducing the levels of SLBP decreases the rate of DNA synthesis and activates an S-phase cell cycle checkpoint
To confirm that the SLBP knockdown cells were accumulating in S-phase, we analyzed these cells by flow cytometry. We also asked whether these cells were still synthesizing DNA by pulsing the C2 or S2 siRNA treated U2OS cells with bromodeoxyuridine (BrdU), a thymidine analog, for 30 min followed by analysis of incorporation by FACS, using a FITC coupled anti-BrdU antibody. As a control, we treated cells transfected with C2 (control) siRNA with 5 mM hydroxyurea (HU) for 1 h to abolish BrdU incorporation. The DNA content was measured by propidium iodide staining, and the incorporation of BrdU was measured by FACS. As demonstrated previously, the SLBP knockdown cells accumulated in S-phase of the cell cycle (Fig. 1B, top panel). From the FACS analysis of BrdU incorporation (Fig. 1B, center panel), it is clear that SLBP knockdown cells are still synthesizing DNA (all the S-phase cells incorporated BrdU), but they were synthesizing DNA at about one-half of the rate as control cells, as judged by the intensity of BrdU staining. In the control RNAi (C2) cells (Fig. 1B, left panel), there is efficient incorporation of BrdU as detected by a FITC-conjugated antibody to BrdU. The characteristic arc (Hoek and Stillman 2003) of BrdU incorporation is present in the control cells, showing the high rate of incorporation in early S-phase, and only a small percentage of cells under the arc not incorporating BrdU. In contrast, the SLBP knockdown cells (S2) have a markedly lower BrdU-FITC signal overall and lack the characteristic arc observed in control cells (Fig. 1B, center panel), suggesting that their rate of DNA synthesis is reduced. C2-treated cells treated with HU for 1 h prior to addition of BrdU did not incorporate BrdU (Fig. 1B, right panel).
We also used the BrdU FACS analysis to quantify the cell cycle phase distribution at a higher resolution in the SLBP knockdown cells compared to the C2 control (Fig. 1C). We quantified the G1- and G2-phases as the populations at 2N and 4N, respectively, that did not incorporate BrdU. We separated S-phase into two phases, early and late, by quantifying the two populations that were between 2N and 3N, and 3N and 4N. It is clear from this representation (Fig. 1C) that S2-treated cells accumulate both in the early part of S-phase and also in late S-phase, indicating that the cells have difficulty completing S-phase. The number of cells in G2-phase was reduced by 40%, consistent with cells progressing through S-phase more slowly but through G2/M and G1 normally.
We also determined the mitotic rate in the SLBP knockdown cells using an antibody to phosphorylated histone H3 (serine 10) as a mitotic marker. Positive cells were measured by FACS and were quantified as a percentage of the control (Fig. 1D). It is clear from the mitotic index data that SLBP knockdown cells are going through mitosis at approximately half the rate of C2 control cells, consistent with the finding that G2 cells are reduced by 40%. This is in agreement with the interpretation that there is not a block in progression through G2, but rather a block in completing S-phase (and, hence, in entering G2).
We next determined whether the reduced rate of DNA replication resulted in activation of the S-phase checkpoint, a consequence of defective chromosome replication, damaged DNA, or environmental stress (Sancar et al. 2004). It has been shown previously that overexpression of a dominant-negative chromatin assembly factor 1 (CAF1) or RNAi to CAF1 produces a similar DNA replication phenotype as knocking down SLBP (Hoek and Stillman 2003; Ye et al. 2003). We hypothesized that reducing the levels of SLBP might result in impaired histone protein biosynthesis and hence mimic the defective chromatin assembly phenotype since the chromatin assembly machinery would not have enough histone proteins to properly assemble chromatin.
We assessed the phosphorylation of Chk1, a kinase that is activated by the ATR kinase when DNA replication stress is sensed in S-phase cells. As a control, we again used C2 control cells treated with HU for 1 h since HU treatment is known to activate the DNA replication checkpoint (Sancar et al. 2004). There was increased Chk1 phosphorylation on both serine 345 (S345) and serine 317 (S317) in response to SLBP knockdown (Fig. 1E). The levels of Chk1 phosphorylation on each of these serine residues were higher than that in control cells but less than that in the HU-treated C2 siRNA population. This result supports the notion that reducing the levels of SLBP results in a partial inhibition of DNA replication phenotype that is directly sensed by the S-phase checkpoint sensor and effector molecules.
Processed histone mRNA levels are only reduced 50% in SLBP knockdown cells
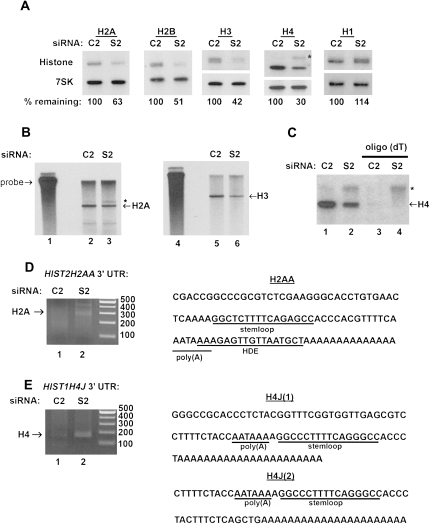
Since SLBP is involved in histone pre-mRNA processing, we determined the levels of histone mRNA, both by Northern blotting and by S1 nuclease mapping to assess whether the 3′-end was formed properly. We used Northern blots to determine histone mRNA levels for all five histone proteins, and each band represents mRNAs from five to 15 genes since the nucleotide sequences of the ORFs are very conserved (Marzluff et al. 2002). We used radiolabeled probes generated by PCR targeting the open reading frame (ORF) to detect histones H2A, H2B, H3, H4, and H1 and used 7SK snRNA as a loading control (Fig. 2A). The abundance of histone H2A, H2B, H3, and H4 mRNAs is reduced to 37% to 70% of control levels in the SLBP knockdown cells when compared to the C2 control. This was a surprisingly high level, but it is consistent with previous reports (Zhao et al. 2004) that also reported only a relatively small reduction in core histone mRNA in HeLa cells.
FIGURE 2.
Histone mRNA levels are reduced in SLBP knockdown cells and a portion of the mRNA is misprocessed and polyadenylated. (A) Northern blot analysis was performed on 2 μg of total RNA from C2 and S2 treated cells. 7SK snRNA is used as a loading control. The percentage of histone mRNA present in the S2 treated cells was quantified using PhosphorImager analysis and ImageQuant software and normalizing mRNA to the 7SK snRNA levels. These results are representative of several different experiments. Note that a slowly migrating product reacting with the histone H4 probe (*) is present in the S2 cells. (B) 3′ S1 nuclease protection assays were used to map the 3′ ends of (lanes 2,3) histone H2A (HIST2H2AA) and (lanes 5,6) histone H3 (HIST2H3A/C) mRNAs. (Lane 3, *) The protected fragment indicates a longer mRNA that is polyadenylated (panel D). (C) Total RNA was selected on oligo(dT) cellulose, and equal proportions of (lanes 1,2) total RNA and (lanes 3,4) poly(A)+ RNA were analyzed by Northern blotting for histone H4 mRNA. (*) The longer polyadenylated H4 mRNA. (D,E) Total RNA from C2 and S2 U2OS cells was primed for reverse transcription with oligo(dT) fused to a T7 adaptor sequence and subjected to PCR using (D) a pan-H2A ORF oligo or (E) a pan-H4 oligo together with a T7 adaptor sequence. The products were analyzed by gel electrophoresis, the products were cloned, and multiple clones from each reaction were sequenced. A single polyadenylation site in histone H2a was found, and two sites in the histone HIST1H4J gene were identified.
We observed a slower migrating band in the histone H4 Northern blot (Fig. 2A, panel 4), which was not present in the control cells. We hypothesized this represented misprocessed histone mRNA since if there were polyadenylation signals 3′ of the genes, the polymerase might read through to that site. We therefore performed both S1 nuclease protection assays and Northern blots using total cell RNA and oligo(dT) selected total cell RNA to assess whether there were read-through mRNAs produced in the SLBP knockdown cells. We detected a small amount of misprocessed histone HIST2H2AA mRNA using S1 nuclease mapping (Fig. 2B), which mapped to a cryptic poly(A) site that is utilized when an intron is inserted into this gene (Pandey et al. 1990). There were no read-through transcripts detected from the HIST2H3A gene. We used oligo(dT) selection to confirm that the slower migrating histone H4 mRNA was polyadenylated (Fig. 2C). We cloned the RNA from both the H2A and H4 histone genes, where we observed misprocessing by oligo(dT)-primed RT-PCR to identify (1) the particular gene that was misprocessed and (2) the location of the polyadenylation site. Cloning of the histone H2A band using a pan-H2A primer targeting the ORF revealed that this mRNA was derived from the HIST2H2AA gene and the polyadenylation occurred after the HDE (Fig. 2D). The AATAAA in this H2A gene is conserved in the mouse and hamster (Debry and Marzluff 1994), and the ortholog of this gene is polyadenylated using this canonical polyadenylation signal in rodent testis (Moss et al. 1994). It has been previously reported that this poly(A)+ mRNA is present when NELF1 or SLBP is knocked down (Narita et al. 2007). We cloned the poly(A)+ histone H4 mRNA, and this mRNA is derived from the HIST1H4J gene. Two different polyadenylation sites were found both of which use a polyadenylation signal present immediately 5′ of the stem–loop sequence (Fig. 2E), which overlaps with the binding site for SLBP. This signal is not conserved in the mouse HIST1H4J gene. Interestingly, the first polyadenylated species resulted from cleavage and polyadenylation 5 nt 3′ of the SL (21 nt from the AAUAAA), the same site where cleavage would occur in the presence of SLBP. The second species of polyadenylated HIST1H4J resulted from cleavage and polyadenylation 34 nt downstream from the AAUAAA signal, just prior to the HDE.
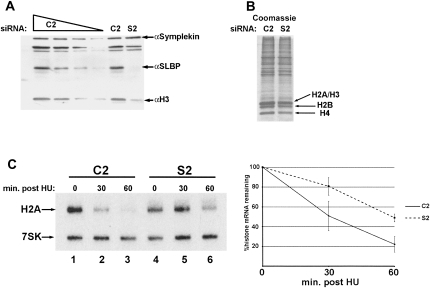
Histone protein levels are reduced in SLBP knockdown cells
Since there was still a substantial amount of properly processed histone mRNA in the SLBP knockdown cells for all the core histone proteins, we suspected that there must be a defect in histone protein production other than in histone pre-mRNA processing. Previous structure–function studies on SLBP have shown that there is a critical domain necessary for histone mRNA translation (Sanchez and Marzluff 2002; Gorgoni et al. 2005). Due to the modest decrease in properly processed histone mRNA in SLBP knockdown cells, we hypothesized that reduction of SLBP might have had a more dramatic effect on histone protein synthesis than on histone pre-mRNA processing. We therefore investigated the levels of histone protein in SLBP knockdown cells. Figure 3A demonstrates that SLBP knockdown is >90%. We used Symplekin, a scaffold protein involved in 3′-end formation (Takagaki and Manley 2000), as a loading control. Using an antibody against histone H3, we find that total histone H3 is reduced by ∼50% in the SLBP knockdown cells relative to the control cells.
FIGURE 3.
Histone protein levels are decreased in SLBP knockdown cells, and the normal histone mRNA degradation in response to HU is impaired. (A) A serial dilution Western blot analysis of C2 (1:2–1:16, lanes 1–4) and a direct comparison of (lane 5) C2 and (lane 6) S2 total cell protein lysates. The levels of SLBP, histone H3 protein, and symplekin as a loading control were analyzed. (B) U2OS cells were knocked down for SLBP and nuclear proteins prepared from the control and SLBP knockdown cells. They were resolved on 15% SDS-polyacrylamide gels and stained with Coomassie blue. (Arrows) The core histone proteins. (C) Histone H2A mRNA is not degraded as efficiently in SLBP knockdown cells compared to control cells. HeLa cells were treated with (lanes 1–3) C2 or (lanes 4–6) S2 siRNAs and treated with 5 mM HU, and cells were harvested (lanes 2,5) 30 min or (lanes 3,6) 60 min later. (Left panel) Total cell RNA was analyzed by Northern blotting with probes to H2A mRNA and 7SK snRNA as a loading control. (Right panel) The results from three independent experiments. (Vertical bars) Standard deviations.
To directly test the effect of knockdown of SLBP on the amount of histone protein, nuclear proteins were prepared from nuclei of control cells and cells with SLBP knocked down. The Coomassie stained gel (Fig. 3B) shows a reduction in total protein levels for all four core histones, consistent with the previous Western results for histone H3. Similar results have been found in five independent experiments, and the levels of the nonhistone proteins remain constant. As judged by FACS analysis, the DNA content per cell is the same in SLBP knockdown cells, which have an increased number of cells in S-phase. We conclude that the amount of histone protein produced in the SLBP knockdown cells is not enough to meet the histone protein demands of the replicating U2OS cell, resulting in a decrease in histone protein in the SLBP knockdown cells, and this must, at least in part, contribute to the DNA replication and growth defects we observed.
Regulation of histone mRNA degradation is impaired in SLBP knockdown cells
Histone mRNA half-life is coordinately regulated with the rate of DNA synthesis, and at the completion of S phase or upon inhibition of DNA synthesis, there is a rapid degradation of histone mRNA. Histone mRNA degradation is a regulated process that requires translation of the histone mRNA (Graves et al. 1987; Kaygun and Marzluff 2005b), and involves the addition of an oligo(U) tail to the 3′end in order to recruit specific decay factors for rapid, bidirectional decay of the mRNA (Mullen and Marzluff 2008). Since reducing SLBP protein levels produced defects in DNA synthesis and normal histone gene expression, we asked if down-regulation of SLBP had an effect on histone mRNA degradation in HeLa cells. We inhibited DNA replication with 5 mM HU in both C2 control cells and SLBP knockdown cells for 30 and 60 min. The representative Northern blot analysis of histone H2A mRNA presented in Figure 3C (and graphically represented as the average of three independent experiments) reveals that the rate of histone mRNA degradation is decreased when SLBP protein levels are down-regulated by siRNA. 7SK snRNA was used as a loading control to normalize the percentage of histone H2A mRNA remaining. Histone mRNA in the C2 control has been significantly reduced at the 30-min time point and is barely detectable at the 60-min time point (Fig. 3C, lanes 2,3). In the SLBP knockdown cells, histone mRNA degradation is stabilized roughly 1.6-fold on average at the 30-min time point and roughly 2.3-fold at the 60-min time point (Fig. 3C). The reduced rate of histone mRNA degradation could reflect the inefficient translation of histone mRNA with low levels of SLBP and/or a requirement for SLBP in histone mRNA degradation.
Processed histone mRNA is retained in the nucleus in SLBP knockdown cells
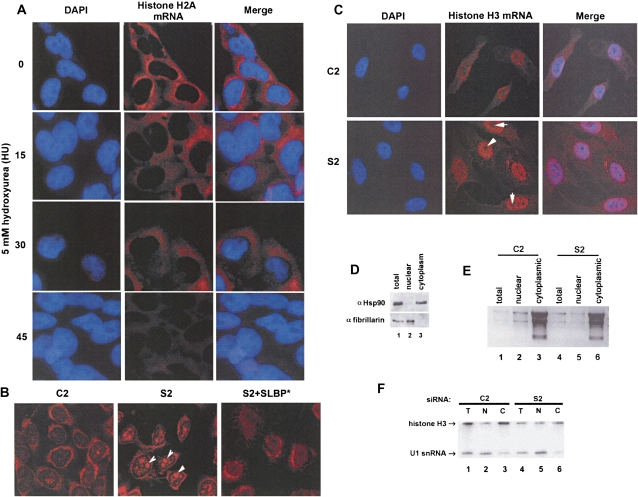
Another possibility for the low rate of histone protein synthesis is that the histone mRNA may be mislocalized. We determined the subcellular localization of the histone mRNA in SLBP knockdown cells. As a control experiment to localize histone mRNA, we synchronized HeLa cells using double thymidine block and released them into S phase for 3 h and performed in situ hybridization with a probe to histone H2A. Histone H2A mRNA is localized strongly around the nucleus in a halo-like appearance, although some signal is present throughout the cytoplasm (Fig. 4A, time 0). To assess probe specificity, we treated the S-phase cells with 5 mM HU for 15, 30, or 45 min since inhibition of DNA synthesis results in rapid degradation of histone mRNA (Fig. 4A, 15, 30, and 45 min time points). As expected, histone H2A signal diminished over time, and this provided confidence that our probe specifically detects the replication-dependent histone mRNAs.
FIGURE 4.
Histone mRNA in SLBP knockdown cells is retained in the nucleus. (A) HeLa cells synchronized by double-thymidine block were released into S phase for 3 h and treated with 5 mM hydroxyurea (HU) for 0, 15, 30, and 45 min. Cells were fixed and prepared for in situ hybridization with antisense DIG-labeled H2a mRNA. Cells were stained with DAPI and anti-DIG antibodies, (blue) DAPI, and (red) histone H2A mRNA. (B) Logarithmically growing HeLa cells were treated with the indicated siRNAs, and in situ hybridization was performed with antisense DIG-labeled H3 mRNA. Note that histone mRNA accumulates in (arrows) nucleoli of HeLa cells in S2-treated cells. (C) U2OS cells were treated with the indicated siRNAs, and in situ hybridization was performed with antisense DIG-labeled H3 mRNA as in B. Cells were stained with DAPI. Note that there is no accumulation of histone mRNA in (arrows) nucleoli in U2OS cells. (D) U2Os cells were fractionated into nuclear and cytoplasm and protein or (E,F) RNA from total and subcellular fractions analyzed. (Lane 1) Total, (lane 2) nuclear, and (lane 3) cytoplasmic protein fractions were separated by 12% polyacrylamide gel electrophoresis and probed for Hsp90 (top, as a cytoplasmic marker) and fibrillarin (bottom, as a nuclear marker by Western blotting. (E) Total (10%) input, nuclear, and cytoplasmic RNA fractions from (lanes 1–3) C2 and (lanes 4–6) S2-treated cells were separated on a 1% agarose gel, and rRNA (as a cytoplasmic marker) was visualized by ethidium bromide staining. (F) Total, nuclear, and cytoplasmic RNAs were analyzed by Northern blot analysis, and histone H3 mRNA and U1 snRNA (nuclear marker) were simultaneously detected using a mixture of radiolabeled probes.
We performed in situ hybridization to histone H2A mRNA in HeLa and H3 mRNA in U2OS cells in which SLBP has been knocked down. In the control HeLa cells, histone H2A mRNA was present in the cytoplasm, with the halo-like appearance in the perinuclear region with little to no staining in the nucleus (Fig. 4B). In contrast, in the SLBP knockdown HeLa cells, in situ hybridization revealed that the histone H2A mRNA was concentrated in large foci in the nucleus, although some signal persisted in the cytoplasm. This phenotype was rescued by expressing an siRNA-resistant form of SLBP, SLBP* (Wagner et al. 2005) in HeLa cells (Fig. 4B). We also performed in situ hybridization to histone H3 mRNA in U2OS cells where SLBP was knocked down (Fig. 4C). In U2OS cells, the phenotype is even more dramatic than in HeLa cells (Fig. 4C). In U2OS cells, there is the characteristic halo-like staining of histone H3 mRNA in the C2 control treated cells, but when SLBP is knocked down (S2), the histone H3 mRNA was concentrated in the nucleus (Fig. 4C). A similar result was also observed in SLBP knockdown U2OS cells when in situ hybridization to histone H2A was performed (data not shown). Since the misprocessed RNA that is polyadenylated is presumably exported via the conventional poly(A) RNA export pathway, the histone mRNA that accumulates in the nucleus is properly processed mRNA.
As an alternative approach to confirm the presence of histone mRNA in the nucleus, we fractionated C2 control and S2 knockdown cells into nuclear and cytoplasmic fractions. We harvested protein and total RNA from these fractions as well as from a portion of the total cell lysate. Hsp90 and fibrillarin were used as cytoplasmic and nuclear markers, respectively, and they were present almost exclusively in the appropriate fraction (Fig. 4D). The first lanes in Fig. 4D represent 100% of the total input from unfractionated cells. Similarly, the ribosomal RNA was largely in the cytoplasmic fraction (Fig. 4E, lanes 2,3).
We also analyzed the histone mRNA in each of the subcellular fractions from the same experiment, and simultaneously probed for histone H3 mRNA and U1 snRNA as a control (Fig. 4F). Consistent with the nuclear localization of spliceosomal snRNAs, we observed that U1 was concentrated in the nuclear fraction of both the C2 control and S2 knockdown (Fig. 4F, lanes 2,5). A small amount of U1 snRNA was detected in the cytoplasm (Fig. 4F, lanes 3,6). Histone H3 mRNA was concentrated in the cytoplasm of C2 control cells (Fig. 4F, lane 3), but in the SLBP knockdown cells, ∼50% of the histone mRNA was in the nucleus. Since the nuclear volume is ∼10% of the volume of the cytoplasm (S2) (Fig. 4F, cf. lanes 3 and 6), this result is consistent with the high concentration of histone mRNA in the nucleus detected by in situ hybridization. Sequestration of histone mRNA in the nucleus upon loss of SLBP is consistent with the decrease in histone protein synthesis due to decreased translation, as well as the failure to rapidly degrade histone mRNA when DNA replication was inhibited.
Taken together these results demonstrate that the major lesion in histone mRNA metabolism when SLBP is knocked down more than 95% is in export of the histone mRNA to the cytoplasm. Processed histone mRNA accumulates in the nucleus, and is relatively stable. Thus the primary molecular defect in histone mRNA metabolism when SLBP is knocked down to 2%–5% of normal levels is the retention of processed histone mRNA in the nucleus.
Tethering of SLBP to nonspecific mRNA results in nuclear export
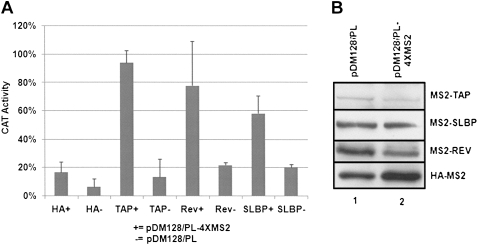
Many general mRNA export factors, including TAP and Rev, promote export to the cytoplasm when tethered to nonspecific mRNAs (Yi et al. 2002). To determine if SLBP facilitates nuclear export of mRNA, we utilized a well-characterized CAT (chloramphenicol acetyl transferase) reporter system. The pDM128/PL reporter contains a CMV-driven bacterial CAT gene located within an intron derived from HIV-1. Under standard conditions, the CAT mRNA is spliced out of the reporter and not expressed due to its nuclear retention. Robust CAT activity can be achieved by the insertion of four MS2 binding sites adjacent to the CAT gene coupled with the expression of a heterologous fusion protein containing the MS2 coat protein and a factor capable of mRNA export (i.e., TAP). The activity results from the export of the CAT mRNA prior to it getting spliced out of the reporter. 293T cells were cotransfected with either pDM128/PL or pDM128/PL-4XMS2 and HA-MS2 fused to TAP, Rev, or SLBP. Expression of MS2-TAP and MS2-Rev resulted in increased levels of CAT activity from the pDM128/PL-4XMS2 reporter compared to the reporter lacking MS2 sites (Fig. 5A), as has been reported for other transport factors (Yang et al. 2001; Wiegand et al. 2002). Cotransfection of MS2-SLBP with the MS2 reporter results in CAT activity levels ∼50% of those observed for TAP and Rev, while coexpression of HA-MS2 had no effect on either reporter. Levels of MS2-TAP, MS2-SLBP, MS2-Rev, and HA-MS2 were determined by Western blotting (Fig. 5B). Note that levels of MS2-TAP expression were lower as reported previously (Wiegand et al. 2002). These data indicate that tethering SLBP to a nonspecific mRNA is sufficient to promote nuclear export, albeit not as efficiently as TAP or Rev.
FIGURE 5.
SLBP promotes mRNA export. (A) 293T cells were transfected with 25 ng of reporter plasmid and 500 ng of HA-MS2 fusion protein plasmid. Forty-eight hours post-transfection, cells were harvested in TEN buffer and a CAT assay was performed on the lysates. CAT activity was normalized to untransfected lysates. Values are for three independent experiments with error bars representing 1 standard deviation. (B) The cell lysates from cells transfected with the two reporters were analyzed by Western blotting using antibody against the HA tag, except for SLBP where the anti-SLBP antibody was used.
DISCUSSION
As the only mRNAs that are not polyadenylated, metazoan histone mRNAs require a distinct set of factors for pre-mRNA processing, translation, and regulation of stability. A common factor in all these processes is SLBP, which binds the stem–loop in the histone pre-mRNA during transcription, remains associated with the mRNA during translation, and is involved in histone mRNA degradation (Marzluff et al. 2008). Here we have ascertained the effect of knocking down SLBP in mammalian cells to <5% of the normal level. The cells remain viable and continue growing at a slow rate. Surprisingly, there is only a modest effect on the accumulation of processed histone mRNA, but a substantial fraction of the histone mRNA is retained in the nucleus, suggesting a previously unrecognized role of SLBP in histone mRNA export in vivo.
We, and others, have previously shown that knocking down SLBP results in slow growth of cells and an accumulation of cells in S-phase (Zhao et al. 2004; Wagner et al. 2005). This effect is due to the SLBP knockdown since normal growth can be restored by expression of an RNAi-resistant SLBP (Wagner et al. 2005). Progression through S-phase was slowed as judged by the release of cells from either nocodazole or HU blocks (Zhao et al. 2004). This is in contrast to knocking down components of the U7 snRNP, which result in an arrest of cells in mid-G1 (Wagner and Marzluff 2006). This evidence suggests that the cell monitors the ability to synthesize histone mRNA by monitoring the presence of an active U7 snRNP, but that cells enter S phase normally when SLBP is reduced.
SLBP is a relatively unstable protein, being degraded at the end of S-phase and not accumulating again until just prior to entry into the next S-phase (Whitfield et al. 2000; Zheng et al. 2003). Thus, it is relatively easy to obtain effective knockdown of the SLBP protein, although for the phenotypes observed here SLBP must be knocked down >95%, and with long exposures of the Western blots, we detect small amounts of SLBP in the knockdown cells. Thus, the results reported here reflect the phenotypes observed when SLBP is severely limiting but not absent from cells.
The most obvious effect of knocking down SLBP is a slow growth rate and a change in cell cycle distribution of the cells, with an accumulation of cells in S-phase. The cells continue dividing, and we see no evidence of apoptosis during the five to six day course of the experiments. The accumulation of cells in S-phase is due to a slower progression through S phase as a result of a reduced rate of DNA replication (Fig. 1). We suggest that the reduced rate of replication is due to a failure to provide replication-dependent canonical histones to the chromatin assembly machinery at a sufficient rate. This likely results in defects at the replication fork, leading to activation of the S-phase checkpoint and slowing the rate of DNA replication. Similar phenotypes have been observed with knockdown of CAF1, the major chromatin assembly factor for the canonical but not the replication-independent histones (Ye et al. 2003).
Surprisingly, the overall level of histone mRNAs is only reduced about twofold in SLBP knockdown cells, and the great majority of the histone mRNA is accurately processed. This implies that the residual SLBP in the cell must be preferentially used for histone pre-mRNA processing. One possibility is that the residual SLBP might be concentrated in the Cajal body, where it could function in histone pre-mRNA processing, but we were not able to detect a specific localization by immunofluorescence. There is a small amount of misprocessed polyadenylated mRNA that accumulates from some of the histone genes, suggesting that the efficiency of processing is reduced in the SLBP knockdown mutants. The HIST2H2AA gene has been previously shown to have a cryptic polyadenylation site in the HDE that is used when histone processing is disrupted by insertion of an intron (Pandey et al. 1994), and in round spermatids (Moss et al. 1994). This mRNA is also polyadenylated when NELF, a transcription elongation inhibitor, is knocked down (Narita et al. 2007). There is also a polyadenylated histone H4 mRNA that accumulates when SLBP is knocked down. This mRNA is derived from a single histone H4 gene, HIST1H4J, which contains a polyadenylation signal in the 3′-UTR, just before the stem–loop. Although the position of the poly(A) tail is at the same site as the cleavage site for histone mRNA, it is likely that this polyadenylation signal, which overlaps the SLBP binding site, directs the cleavage of the HIST1H4J pre-mRNA to yield the polyadenylated mRNA. Each of these polyadenylation signals overlaps a cis-element required for histone mRNA processing, suggesting that normally the histone processing machinery preferentially associates with these signals.
Unlike the situation in Drosophila (Lanzotti et al. 2002; Godfrey et al. 2006), there is not extensive read-through of histone transcripts resulting in polyadenylated mRNAs in mammalian cells when SLBP is reduced. This may reflect a need in Drosophila for efficient termination between closely spaced histone genes, or there may in fact be a role for polyadenylated histone mRNAs in particular cell types during development. The role of SLBP in the processing of histone pre-mRNA is “catalytic,” allowing cells to perform this function even under limiting concentration of SLBP, and then SLBP can be released for processing more histone mRNAs. However the requirement for SLBP in other steps in histone mRNA metabolism (e.g., export or translation) is likely stoichiometric, and the RNAi-mediated reduction in SLBP levels therefore interferes with these functions.
Transport of histone mRNA
Most mRNAs are transported through a pathway that requires TAP (yeast mex67p) as an adaptor protein for export (Erkmann and Kutay 2004). TAP can be recruited to mRNAs derived from spliced pre-mRNAs by binding to components of the exon–junction complex deposited near the exon–exon border after splicing. Unspliced mRNAs such as histone mRNAs must recruit TAP by a different mechanism. There have been several studies relevant to histone mRNA export from the nucleus. In one study the role of processing was assessed by forming the histone 3′ end either by a normal processing reaction or by ribozyme cleavage (Eckner et al. 1991). They found that mature histone mRNAs were cytoplasmic regardless of the mechanism of 3′ end formation. However, if the histone ORF was replaced with the chloramphenicol acetyl transferase ORF, then the mRNA formed by the processing reaction was cytoplasmic, and the mRNA formed by the ribozyme was retained in the nucleus.
Huang and Steitz (2001) studied elements in the histone ORF that were capable of directing transport of an heterologous mRNA in Xenopus oocytes and identified a binding site for the SR protein 9G8 responsible for this activity. The binding site is contained within a region of the histone ORF capable of conferring cytoplasmic localization on other unspliced mRNAs (Huang and Carmichael 1997). These studies suggest a role for 9G8 protein in recruiting TAP and provide a partial explanation for the observed conservation in the nucleotide sequence level in histone ORFs at that site (Debry and Marzluff 1994; Friend et al. 2007). In a previous study of histone mRNA export in Xenopus oocytes, we found there was not a requirement for any specific nucleotide sequence, including the 3′ end of histone mRNA or SLBP, for histone mRNA export (Erkmann et al. 2005a) in Xenopus oocytes. This study suggested there are multiple potential binding sites for proteins that could recruit TAP and that in Xenopus oocytes there is a large supply of these proteins, which is in agreement with other studies that showed the length of the RNA was a major determinant of its transport efficiency in Xenopus oocytes (Masuyama et al. 2004).
In mammalian cells with active RNA metabolism, there is likely to be competition among the mRNAs for the binding proteins and export factors, just as there is for factors involved in RNA processing. Thus it is not surprising to find there might be more stringent requirements for export in mammalian cells than in Xenopus oocytes. Our results suggest there is a critical role for SLBP in histone mRNA export in mammalian cells.
Overall, these results are consistent with a model wherein multiple factors may need to bind to a RNA for efficient export. The processing reaction may facilitate recruitment of some of these factors (Eckner et al. 1991), and there are clearly sequence elements in the histone ORF that promote export (Huang and Carmichael 1997; Huang and Steitz 2001) of mRNAs. Our results suggests that association of SLBP is necessary to promote efficient transport of histone mRNA but not to stabilize the histone mRNA in the nucleus.
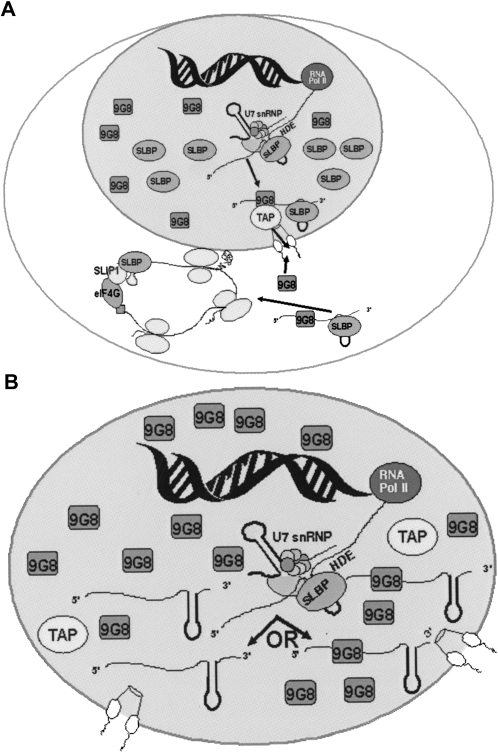
When SLBP levels are greatly reduced, mature histone mRNAs accumulate in the nucleus. A striking aspect of this result is that these mRNAs accumulate to substantial levels. When mutations are introduced into the histone stem–loop that alter SLBP binding (Pandey et al. 1994) or the HDE is deleted preventing U7 snRNP binding (Chodchoy et al. 1991), or when the HDE is naturally suboptimal (Liu et al. 1989), then the unprocessed mRNAs do not accumulate, but rather, must be rapidly degraded. The same is true of improperly processed mRNAs from polyadenylated transcripts. In the SLBP knockdown cells, histone mRNAs are properly processed, and direct sequencing of the mRNAs after circular RT-PCR (Mullen and Marzluff 2008) did not reveal any alterations at the 3′-end of the mRNA (E.J. Wagner, unpubl.). Thus these mature mRNAs, which are not bound to SLBP, are still stable in the nucleus. They may be stable in the nucleus since they have gone through the normal processing pathway, despite the fact that they cannot be exported. A model for histone mRNA export in normal cells and for the nuclear retention of histone mRNA in SLBP knockdown cells is shown in Figure 6.
FIGURE 6.
Model of SLBP-dependent histone mRNA export. (A) Histone mRNA metabolism in normal S-phase cells. SLBP remains associated with the histone mRNA in the nucleus, and the mRNA is efficiently exported associated with multiple proteins that recruit TAP. (B) Histone mRNA metabolism in cells depleted of SLBP. Histone mRNA is processed normally by the residual SLBP, but that SLBP dissociates from the processed mRNA before it can be exported, and the processed message remains in the nucleus, where it is stable.
Decreased levels of histone protein in SLBP knockdown cells
The second feature of the SLBP knockdown cells is that they have a lower amount of histone protein, consistent with a defect in histone protein biosynthesis. Initially we found this surprising, since one might think that reducing histone protein:DNA ratios twofold might have a profound effect on cell physiology and viability. However, in at least one situation, infection of human T lymphocytes with HTLV1, there is an approximately twofold reduction in histone:DNA ratios and mediated by a reduction in core histone mRNA expression without affecting DNA replication (Bogenberger and Laybourn 2008). In yeast one can vary the histone:DNA ratio by changing the dosage of the histone genes (Gunjan and Verreault 2003) or in some mutants by chromatin modification (Xiao et al. 2007; B.D. Strahl, pers. comm.) without affecting viability.
Conclusions
SLBP is essential for the proper accumulation of histones for assembly of newly replicated DNA into chromatin. Our results suggest that a major consequence of SLBP depletion is the failure to export properly processed histone mRNA. This suggests that SLBP must be associated with histone mRNA for export and that SLBP may dissociate from histone mRNA after processing and additional nucleoplasmic SLBP may then reassociate with the histone mRNA prior to export. Understanding and identifying the components of the nuclear histone mRNP, as well as the cytoplasmic histone mRNP, will help elucidate the pathway between processing of the mRNA at the site of transcription and subsequent export and translation of the histone mRNA.
MATERIALS AND METHODS
RNA interference
RNAi was performed using a two-hit method as previously described using Lipofectamine 2000 (Wagner and Garcia-Blanco 2002). Briefly, 110,000 U2OS or 95,000 HeLa cells were plated into 24-well plates, and the following day, siRNAs were transfected into cells. A day following the first hit of siRNA, cells were replated into six well dishes, allowed to grow an additional day, and then transfected with the second hit of siRNA. Cells were harvested 48–72 h following the second hit for analysis. The siRNA sequences of the C2 (control) and S2 (SLBP) have been previously described (Wagner et al. 2005).
DNA synthesis and mitotic index measurements by FACS
We quantified the percentage of bromodeoxyuridine (BrdU) labeled cells in S phase and phospho-histone H3 in mitosis as previously described (Doherty et al. 2003). As controls for these assays, cells were treated with 5 mM HU for 1 h prior to addition of the BrdU. To measure DNA synthesis in S phase, RNAi-treated cells were pulsed with a 1 mM stock solution of BrdU (10 μM final concentration) for 1 h. Acid-treated nuclei were resuspended with 100 μL of a solution containing α-BrdU antibody conjugated to FITC (Becton Dickinson) diluted 1:5 in IFA (10 mM HEPES at pH 7.4, 150 mM NaCl, 4% FBS, 0.1% NaN3) plus 0.5% Tween-20 and incubated in the dark for 2 h at room temperature. Following the incubation, 2 mL of IFA plus 0.5% Tween-20 was added, and nuclei were recovered by centrifugation. Nuclei were resuspended in 500 μL of the IFA plus 0.5% Tween solution containing 5 μg/mL RNase A and 50 μg/mL propidium iodide. Nuclei were incubated overnight at 4°C and analyzed on a Becton-Dickinson (Becton Dickinson) FACScan using Summit Software (Dako Cytomation).
To measure the mitotic index using phospho-histone H3 as a marker, cells were fixed with 1% formaldehyde in PBS for 30 min. Fixed cells were pelleted at 1200 rpm for 5 min at 4°C and the supernatant removed. Cells were resuspended in 1.5 mL of PBS, and then 3 mL of cold 100% ethanol was added and cells were stored overnight at 4°C. The cells were resuspended in 100 μL of a solution containing 0.5 μg of phospho-histone H3 (serine 10) antibody (Upstate Biotechnology) and 5 μg/mL RNase A in IFA plus 0.5% Tween-20 for 2 h at room temperature. The cell pellet was resuspended in 100 μL of the IFA plus 0.5% Tween-20 solution containing an anti-rabbit secondary antibody conjugated to FITC (Santa Cruz Biotechnology) for 1 h at room temperature. The cells were resuspended in 500 μL of the IFA plus 0.5% Tween solution containing 5 μg/mL RNase A and 50 μg/mL propidium iodide and analyzed by FACS.
Northern blot analysis and S1 nuclease protection assays
Northern blot analysis to replication-dependent histone genes and 7SK snRNA was performed as described previously (Mullen and Marzluff 2008). Briefly, 2 μg of total RNA was resolved on 6% urea–polyacrylamide gels, transferred to Hybond N+ nitrocellulose (Amersham Biosciences), and probed in QuickHyb (Stratagene) overnight at 60°C. Probes were generated by random primed, α32P-dCTP labeling (PrimeIt II Kit; Stratagene) of PCR products generated to the complete ORF of cloned human histone genes and to 7SK RNA (Mullen and Marzluff 2008).
S1 nuclease protection assays to map the 3′ ends of histone H2a (HIST2H2AA) and H3 (HIST2H3A/C) were performed as previously described (Kaygun and Marzluff 2005a). Probes for 3′-end labeling were generated by digestion of the HIST2H2AA gene with AscI and the HISTH3A/C gene with BssHI. The digested plasmid DNA was 3′-end labeled with α32P-dCTP and Klenow (New England BioLabs) and hybridized with total cell RNA. Protected fragments were resolved on 6% urea–polyacrylamide gels.
Western blot analysis
Protein lysates from RNAi-treated cells were prepared as previously described (Mullen and Marzluff 2008) and the concentrations quantified using Bradford reagent. The primary antibodies used in this study include rabbit anti-SLBP (Wang et al. 1996), mouse anti-symplekin (BD Biosciences), rabbit anti-fibrillarin (Abcam), rabbit anti-Hsp90 (Cell Signaling Technology), and rabbit anti-histone H3 (Abcam). Protein were resolved on 12% SDS–polyacrylamide gels and transferred to nitrocellulose before detection using appropriate primary and secondary antibodies.
In situ hybridization
In situ hybridizations were performed as described (Erkmann et al. 2005a). RNAi-treated cells were grown on coverslips and washed with PBS and fixed in 3.7% PFA in PBS for 10 min at room temperature. Cells were then washed again and permeabilized in 0.5% Triton X-100 in PBS. Pre-hybridization was performed for 10 min at 37°C using a solution of 4× SSC and 50% formamide preheated to 37°C. Coverslips were inverted onto a 5–10-μL drop of hybridization solution (50% deionized formamide, 10% dextran sulfate, 1× Denhardt's solution, 4× SSC, 1 mg/mL Escherichia coli tRNA) containing 1 ng/μL DIG labeled H2A or H3 probe. Probes were denatured for 5 min at 80°C and cooled on ice prior to addition to hybridization solution. Hybridizations were performed overnight at 37°C in a humidified chamber. Cells were washed once with 2× SSC at 37°C, 3× in 60% formamide in 0.2× SSC at 37°C, once in 2× SSC at room temperature, and 3× with 0.5% Tween-20 in 4× SSC. Coverslips were inverted onto 8-μL drops of 0.5% Tween-20 in 4× SSC containing mouse Cy3 α-DIG antibody diluted 1:200 and incubated for 1 h at 37°C in a humidified chamber. Coverslips were washed three times with 0.5% Tween-20 in 4× SSC and inverted onto 8-μL drops of 0.5% Tween-20 in 4× SSC containing Cy-3 α-mouse secondary antibody diluted 1:200. Secondary incubation was performed for 45 min at 37°C in a humidified chamber. Coverslips were washed 3× with 0.5% Tween-20 in 4× SSC before being fixed with 3.7% PFAin PBS for 5 min at room temperature. After a PBS wash, coverslips were DAPI-stained and mounted onto slides using Fluorsave reagent (Calbiochem). Images were acquired using a Zeiss LSM 510 confocal microscope and analyzed using ImageJ software.
CAT assay
CAT assays were performed essentially as described (Cullen 2004). 293T cells (3 × 105) were cotransfected with 25 ng of reporter plasmid and 500 ng of MS2-fusion plasmid using Lipofectamine-2000. Forty-eight hours after transfection, cells were harvested in TEN buffer (100 mM NaCl, 10 mM EDTA, 10 mM Tris-HCl at pH 7.5), pelleted, and resuspended in 100 μL of 100 mM Tris-HCl (pH7.5). Cells were lysed by freeze–thawing three times, and lysates were pelleted for 5 min at 4°C. Equal amounts of lysates were added to 300 μL of CAT assay buffer (5 mg/mL chlorampenicol in 100 mM Tris-HCl at pH 7.5 containing 1 μL/mL [3H]acetyl coenzyme A). Three milliliters of Econofluor (Packard) scintillation cocktail was added to each sample, and counts were determined in a scintillation counter.
ACKNOWLEDGMENTS
We thank the members of the Marzluff laboratory for critical discussions during the preparation of this manuscript and Hal Boegerd and Heater Karnowski from Brian Cullen's laboratory (Duke University) for advice on the transport assays. This research was supported by NIH grant GM29832 to W.F.M. K.D.S. was supported from NIH training grant T32GM008581, and E.J.W. is funded by NIH fellowship F32GM070101 and Pathway to Independence Award K99GM080447.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1205409.
REFERENCES
- Azzouz T.N., Gruber A., Schumperli D. U7 snRNP-specific Lsm11 protein: Dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 2005;33:2106–2117. doi: 10.1093/nar/gki516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenberger J.M., Laybourn P.J. Human T lymphotropic virus type 1 protein Tax reduces histone levels. Retrovirology. 2008;5:9–15. doi: 10.1186/1742-4690-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmakci N.G., Lerner R.S., Wagner E.J., Zheng L.-X., Marzluff W.F. SLIP1, A factor required for activation of histone mRNA translation by the stem–loop binding protein. Mol. Cell. Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N., Pandey N.B., Marzluff W.F. An intact histone 3′ processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell. Biol. 1991;11:497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. Assaying nuclear messenger RNA export in human cells. Methods Mol. Biol. 2004;257:85–92. doi: 10.1385/1-59259-750-5:085. [DOI] [PubMed] [Google Scholar]
- Debry R.W., Marzluff W.F. Selection on silent sites in the rodent H3 histone gene family. Genetics. 1994;138:191–202. doi: 10.1093/genetics/138.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty S.C., McKeown S.R., Kelvey-Martin V., Downes C.S., Atala A., Yoo J.J., Simpson D.A., Kaufmann W.K. Cell cycle checkpoint function in bladder cancer. J. Natl. Cancer Inst. 2003;95:1859–1868. doi: 10.1093/jnci/djg120. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Zheng L.-X., Sanchez R., Marzluff W.F. The stem–loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Erkmann J.A., Greenland J.A., Marzluff W.F. Mutations in the RNA binding domain of stem–loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol. 2001;21:2008–2017. doi: 10.1128/MCB.21.6.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Erkmann J.A., Yang X., Sanchez R., Marzluff W.F. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ellmeier W., Birnstiel M.L. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann J.A., Kutay U. Nuclear export of mRNA: From the site of transcription to the cytoplasm. Exp. Cell Res. 2004;296:12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Erkmann J.A., Sanchez R., Treichel N., Marzluff W.F., Kutay U. Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA. 2005a;11:45–58. doi: 10.1261/rna.7189205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann J.A., Wagner E.J., Dong J., Zhang Y.P., Kutay U., Marzluff W.F. Nuclear import of the stem–loop binding protein and localization during the cell cycle. Mol. Biol. Cell. 2005b;16:2960–2971. doi: 10.1091/mbc.E04-11-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K., Lovejoy A.F., Steitz J.A. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7 snRNP dependent 3′ end formation. Mol. Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A.C., Kupsco J.M., Burch B.D., Zimmerman R.M., Dominski Z., Marzluff W.F., Duronio R.J. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and block oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B., Andrews S., Schaller A., Schumperli D., Gray N.K., Muller B. The stem–loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R.A., Pandey N.B., Chodchoy N., Marzluff W.F. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Gunjan A., Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae . Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Hoek M., Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Q., Carmichael G.G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc. Natl. Acad. Sci. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Q., Steitz J.A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005a;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 2005b;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N.G., Steitz J.A. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes & Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti D.J., Kaygun H., Yang X., Duronio R.J., Marzluff W.F. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ processing in vivo. Mol. Cell. Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-J., Levine B.J., Skoultchi A.I., Marzluff W.F. The efficiency of 3′-end formation contributes to the relative levels of different histone mRNAs. Mol. Cell. Biol. 1989;9:3499–3508. doi: 10.1128/mcb.9.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F. Metazoan replication dependent histone mRNAs: A unique class of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Marzluff W.F., Gongidi P., Woods K.R., Jin J.P., Maltais L. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., Duronio R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K., Taniguchi I., Kataoka N., Ohno M. RNA length defines RNA export pathway. Genes & Dev. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S.B., Ferry R.A., Groudine M. An alternative pathway of histone mRNA 3′ end formation in mouse round spermatids. Nucleic Acids Res. 1994;22:3160–3166. doi: 10.1093/nar/22.15.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K.L., Steitz J.A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Mullen T.E., Marzluff W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Pandey N.B., Chodchoy N., Liu T.-J., Marzluff W.F. Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res. 1990;18:3161–3170. doi: 10.1093/nar/18.11.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N.B., Williams A.S., Sun J.-H., Brown V.D., Bond U., Marzluff W.F. Point mutations in the stem–loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell. Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sanchez R., Marzluff W.F. The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schochetman G., Perry R.P. Early appearance of histone messenger RNA in polyribosomes of cultured L cells. J. Mol. Biol. 1972;63:591–596. doi: 10.1016/0022-2836(72)90450-0. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Manley J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley-Tilson W.H., Pendergrass S.A., Marzluff W.F., Whitfield M.L. Genome-wide analysis of mRNAs bound to the histone stem–loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Marzluff W.F. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S-phase. Mol. Cell. Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Berkow A., Marzluff W.F. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem. Soc. Trans. 2005;33:471–473. doi: 10.1042/BST0330471. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Ospina J.K., Hu Y., Dundr M., Matera A.G., Marzluff W.F. Conserved zinc fingers mediate multiple function of ZFP100, a U7 snRNP associated protein. RNA. 2006;12:1206–1218. doi: 10.1261/rna.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-F., Whitfield M.L., Ingledue T.I., Dominski Z., Marzluff W.F. The protein which binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Whitfield M.L., Zheng L.-X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., Kaygun H., Erkmann J.A., Townley-Tilson W.H.D., Dominski Z., Marzluff W.F. SLBP is associated with histone mRNA on polyribosomes as a component of histone mRNP. Nucleic Acids Res. 2004;32:4833–4842. doi: 10.1093/nar/gkh798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Coburn G.A., Zeng Y., Kang Y., Bogerd H.P., Cullen B.R. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Shibata Y., Rao B., Laribee R.N., O'Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol. Cell. Biol. 2007;27:721–731. doi: 10.1128/MCB.01628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bogerd H.P., Wang P.J., Page D.C., Cullen B.R. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell. 2001;8:397–406. doi: 10.1016/s1097-2765(01)00303-3. [DOI] [PubMed] [Google Scholar]
- Ye X., Franco A.A., Santos H., Nelson D.M., Kaufman P.D., Adams P.D. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Yi R., Bogerd H.P., Wiegand H.L., Cullen B.R. Both Ran and importins have the ability to function as nuclear mRNA export factors. RNA. 2002;8:180–187. doi: 10.1017/s1355838202012384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Killop-Smith S., Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 2004;117:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- Zheng L.-X., Dominski Z., Yang X., Elms P., Raska C.S., Borchers C.H., Marzluff W.F. Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol. Cell. Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]