Abstract
In this work we used micro-array experiments to determine the role of each nonessential subunit of the conserved Ccr4-Not complex in the control of gene expression in the yeast Saccharomyces cerevisiae. The study was performed with cells growing exponentially in high glucose and with cells grown to glucose depletion. Specific patterns of gene deregulation were observed upon deletion of any given subunit, revealing the specificity of each subunit's function. Consistently, the purification of the Ccr4-Not complex through Caf40p by tandem affinity purification from wild-type cells or cells lacking individual subunits of the Ccr4-Not complex revealed that each subunit had a particular impact on complex integrity. Furthermore, the micro-arrays revealed that the role of each subunit was specific to the growth conditions. From the study of only two different growth conditions, revealing an impact of the Ccr4-Not complex on more than 85% of all studied genes, we can infer that the Ccr4-Not complex is important for expression of most of the yeast genome.
Keywords: Ccr4-Not complex, yeast, micro-arrays, mRNAs
INTRODUCTION
Expression of a functional protein in eukaryotic cells depends upon many steps. For many years it was believed that these steps occurred in a linear manner, and that the major step of the control of gene expression occurred at the beginning, namely, at the level of transcription initiation. Nowadays, it has become clear, however, that the different steps involved in the synthesis of a protein are linked both physically and functionally and that they are all highly regulated (discussed by Guthrie and Steitz 2005, and references therein). Several mRNA processing events occur cotranscriptionally, polyadenylation and transcription termination are coupled, and appropriate packaging of the newly synthesized transcript, necessary for efficient export from the nucleus, is also cotranscriptionally controlled. Coincident with these findings, the importance of mRNA processing, stability, export, and degradation of the messenger, in the appropriate expression of a gene, has become apparent.
The Ccr4-Not complex, conserved from yeast to human and composed of nine core subunits in yeast, Not1-5p, Caf1p, Caf40p, Caf130p, and Ccr4p, is a global regulator of gene expression that contributes to regulate several of the steps, ultimately leading to the stable production of a functional protein (for reviews, see Collart 2003; Collart and Timmers 2004). Indeed, this complex, initially described as a transcriptional regulator, is important for the appropriate distribution of the general transcription factor TFIID on promoters across the genome (Lenssen et al. 2005) but also for the post-translational modification and transcriptional activity of some stress transcription factors (Lenssen et al. 2005, 2007). Furthermore, the Ccr4-Not complex contributes to mRNA degradation, since two of its subunits, Ccr4p and Caf1p, are the major yeast deadenylase (Tucker et al. 2001). Finally, one subunit of the complex, Not4p, was identified as an E3 ligase (Albert et al. 2002), and a role for protein ubiquitination in vivo was demonstrated (Panasenko et al. 2006). The roles of the nonenzymatic subunits of the complex are largely not understood, except for Not1p, which is the scaffold of the complex (Maillet et al. 2000).
The identification of so many different functions dependent upon a single complex has raised questions over the years. To try and understand this functional complexity, we set out to characterize the importance of each nonessential subunit of the Ccr4-Not complex for the expression of the yeast genome by microarray experiments. Through these experiments, we found that each subunit had a specific function in the expression of the genome, function defined also by the growth conditions. These results support the idea that the functional diversity of the Ccr4-Not complex results from specific functions of its various subunits.
RESULTS
The subunits of the Ccr4-Not complex have specific roles in gene expression
To gain a better understanding of the role of the Ccr4-Not complex in regulation of total cellular mRNA levels and, furthermore, to dissect the role of the individual subunits in this control, we performed micro-array experiments with total cellular RNA extracted from cells lacking individual subunits of the Ccr4-Not complex (Table 1). For analysis of gene expression in exponentially growing cells, we kept cells in all cases 24 h in exponential growth to ensure that in all strains, even in the slowest growing strains, genes activated in the precultures grown to saturation had returned to basal levels of expression. For analysis of gene expression in the post-diauxic phase, after 24 h of growth in exponential phase, we let the cells continue to grow until glucose was depleted from the growth medium and collected them 30 min later. Gene expression analysis was performed under these two different growth conditions, because previous studies have suggested that the Ccr4-Not complex is important for transcription in fermenting cells, but also in cells during respiratory growth (for review, see Collart 2003).
TABLE 1.
Strain list
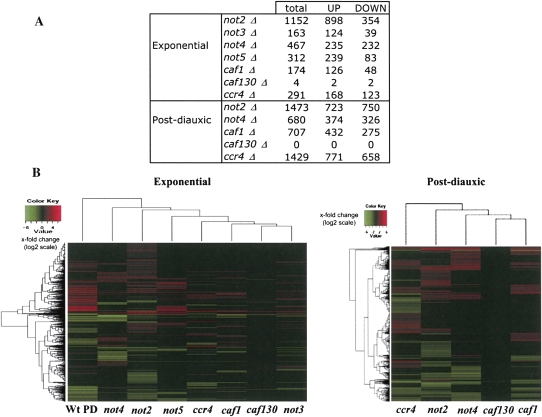
For cells grown in the exponential phase, of the 4283 genes kept for analysis, 1782 were expressed significantly differently in at least one mutant compared to the wild type. This corresponds to a little more than 40% of the analyzed RNAs and 30% of the yeast genome. When compared to wild-type cells, mutant cells lacking one or the other component of the Ccr4-Not complex displayed both increased or decreased levels of specific transcripts (Fig. 1A). Loss of Not2p had the greatest impact on gene expression under these growth conditions, affecting just <20% of the yeast genome, followed by loss of Not4p, Not5p, Ccr4p, Caf1p, Not3p, and finally Caf130p. Clustering of the mutants with regard to their profiles of gene expression revealed that caf130Δ was most similar to the wild type and closest to not3Δ, followed by caf1Δ close to ccr4Δ, whereas not2Δ, not4Δ, and not5Δ were quite different from each other and from the others (Fig. 1B). Interestingly, while these experiments do not distinguish between direct or indirect effects of the different deletions and between effects on transcription or mRNA degradation, the profile of gene deregulation was very specific to each mutant and was also very different from the profile of gene expression changes that occur upon glucose depletion in wild-type cells (Fig. 1B).
FIGURE 1.
Genome wide analysis of gene expression reveals specific roles for subunits of the Ccr4-Not complex. Total cellular RNA was extracted from the indicated wild-type or mutant cells growing exponentially or 30 min after glucose depletion (post-diauxic) and was analyzed by micro-array experiments using the Affymetrix system. (A) The number of target genes defined in each case is indicated. (B) Hierarchical clustering of gene expression in exponentially growing cells (left panel) or cells grown to the post-diauxic phase (right panel). Each line represents a gene, and the color value indicates the fold change (in log2 scale) between the mutant and the wild type in the same growth condition, except in the left lane of the left panel, where we show changes in expression of the genome in wild-type cells after the diauxic shift.
For cells grown to glucose depletion, of the 4283 genes analyzed, 2740 were expressed significantly differently in at least one mutant compared to the wild type, either up-regulated or down-regulated (Fig. 1A). This corresponds to half of the genes analyzed and more than one third of the yeast genome. Thus, the Ccr4-Not complex plays a greater role in gene expression changes that occur upon glucose depletion than in exponentially growing cells (2740 genes affected versus 1782). Under these growth conditions, clustering of the mutants with regard to gene expression revealed again that all mutants were very different (Fig. 1B), but in this case the absence of Ccr4p had as great an impact as the absence of Not2p on gene expression, just >20% of the yeast genome (Fig. 1A).
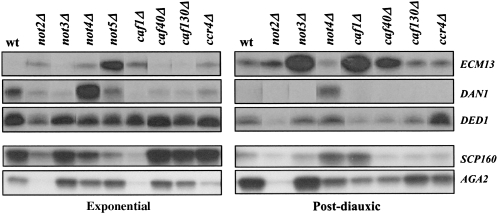
Without defining the mechanism by which each subunit is important for gene expression, these results nevertheless strikingly demonstrate that each subunit of the Ccr4-Not complex plays a specific role in gene expression. To confirm this specific role for each subunit by a different approach, we chose at random five genes (ECM13, DAN1, DED1, SCP160, and AGA2) that were deregulated in one or more cases in the micro-array experiments and analyzed the mRNA levels for these genes, in the wild type and in the mutants by S1 digestion. The mRNA levels were very different in the different mutants of the Ccr4-Not complex, either in high glucose or in the post-diauxic phase (Fig. 2). This analysis confirmed the very specific pattern of gene deregulation for each mutant.
FIGURE 2.
S1 analysis of specific transcripts confirms specific roles for the Ccr4-Not complex subunits in expression of mRNAs. Total cellular RNA was extracted from the indicated wild-type and mutant strains grown to exponential phase or 30 min after glucose depletion (post-diauxic) as indicated, and 30 μg were analyzed by S1 digestion for the levels of the indicated mRNAs in the different indicated strains. All hybridizations were internally controlled by cohybridization with DED1 and all hybridizations with a given probe were analyzed on the same gel even though not always in the same order, such that in the figure signals for each strain have been in some cases cut and pasted such that the order for each of the strains is in relatively the same position.
The Ccr4-Not complex is important for expression of most of the yeast genome and genes related to specific functions depend upon the presence of specific Ccr4-Not subunits
In the analysis presented above, we tested two different physiological conditions, namely, cells growing exponentially in rich medium and high glucose and cells grown to glucose depletion. Our results revealed that the genes dependent upon the presence of each subunit were defined by the growth conditions. Indeed, the target genes for the five mutants analyzed in both growth conditions were generally different in the two physiological conditions tested, and only 847 target genes were found in both growth conditions in one or the other mutant. Thus, we observed that a total of 3675 of the 4283 genes kept in the analysis, namely, >85% (and covering 70% of the genome), were affected significantly in at least one mutant and one growth condition tested. Since only two different growth conditions were tested, one can expect that if more conditions were tested, one would find that the Ccr4-Not complex, in fact, is required, directly or indirectly, for the appropriate expression of most of the yeast genome.
As mentioned above, in general these arrays revealed that the function of the individual subunits in gene expression was very specific, since the patterns of gene deregulation in the absence of each individual subunit were very different. For exponentially growing cells, only 78 genes were significantly deregulated in at least four mutants (see Supplemental Table 1). Clearly, the number of target genes shared by the different mutants is very small relative to the total amount of genes affected in the mutants.
Specific effect of each Ccr4-Not complex subunit for complex integrity
We were interested in light of these findings to analyze the importance of each subunit for Ccr4-Not complex integrity. We made use of a strain expressing Tap-tagged Caf40p from which we have previously successfully purified the Ccr4-Not complex (Lenssen et al. 2007). We deleted each nonessential subunit of the complex from this strain to make new strains. Tagged Caf40p was expressed at similar levels in all strains except in not2Δ, where its levels were reduced (shown in Supplemental Fig. 1A), and was purified by tandem affinity purification. The eluates from the final affinity column were analyzed by separation on SDS-PAGE (shown in Supplemental Fig. 1B) followed by mass spectrometry of all bands visible by Coomassie staining, and also by Western blotting with polyclonal antibodies against Caf40p, Not1p, Not3p, Not4p, Not5p, Caf1p, and Ccr4p. This allowed us to determine which subunits of the Ccr4-Not complex copurified with Caf40p in each of the strains as summarized in Table 2. In cells lacking Ccr4p, Caf40p probably dissociates from the Ccr4-Not complex, as no other subunit of the complex copurified with Caf40p, and this is likely to be somewhat the case also in cells lacking Caf1p in which no Ccr4p and only low levels of Not1p and Not5p were detected (Table 2). Thus for these two mutants it was not possible to assess the integrity of the Ccr4-Not complex with our approach, which was nevertheless conclusive for the other mutants. For instance, the deletion of Caf130p, which has only a modest effect on gene expression, had no effect on the association of the other subunits with Caf40p. In contrast, deletion of Not2p, which leads to important gene deregulation, resulted in dissociation of Caf130p, Not4p, and Not5p from the remaining Caf40p complex. In general, this study showed that each deletion had a very specific effect on the integrity of the Ccr4-Not complex, a finding that is in good correlation with the very specific effect of each subunit on gene expression.
TABLE 2.
List of Ccr4-Not subunits that copurify with Tap-tagged Caf40p
Specific cellular functions are affected in cells lacking specific subunits of the Ccr4-Not complex
We next determined whether genes defining specific cellular functions might be enriched upon deletion of specific subunits of the Ccr4-Not complex in given growth conditions. For this we used Funcassociate, a web-based tool that statistically tests the difference of Gene Ontology categories between two lists of genes. Our analysis revealed that, indeed, certain functions were overrepresented in the target genes defined for specific mutants of the Ccr4-Not complex (Fig. 3). For instance, exosome genes (GO term 0000178) were significantly up-regulated in exponentially growing cells lacking Not2p specifically, and this could be verified by S1 analysis of total cellular RNA extracted from wild-type, not2Δ, and not3Δ strains (shown in Supplemental Fig. 2). Genes involved in oxidative phosphorylation and respiratory-chain phosphorylation (GO term 0006119) were significantly up-regulated in exponentially growing cells lacking Caf1p, whereas genes encoding histones were significantly down-regulated in exponentially growing cells lacking Ccr4p, genes involved in vacuolar function were significantly up-regulated in cells lacking Not3p, and finally genes encoding cell wall were up-regulated in cells lacking Not5p. Again, this analysis confirmed very specific functions for each subunit of the Ccr4-Not complex.
FIGURE 3.
Genes involved in specific functions are overrepresented in the pool of genes deregulated in specific mutants of the Ccr4-Not complex. This figure lists a selection of subpopulations of genes identified as families that are significantly overrepresented in target genes for given mutants. Black shows genes that are not affected, red shows genes that are overexpressed, and green shows genes that are underexpressed.
DISCUSSION
The Ccr4-Not complex consists of subunits with very specific cellular functions
In this work we have tried to understand how the conserved multisubunit Ccr4-Not complex can regulate the very many different cellular functions with which it has been associated. Our study leads to the conclusion that despite the association of all of the subunits in a single complex, each subunit has a very specific role in gene expression. Indeed, a different pattern of gene deregulation was observed when each nonessential subunit of the complex was deleted. In addition, genes involved in different cellular functions were significantly overrepresented in the different mutants. Besides the families mentioned above and provided in Figure 3, several other families were identified for each mutant (Supplemental Table 2). This suggests that not only does each subunit have a specific role, but, moreover, each subunit appears to be important for the expression of genes involved in more than one cellular function. Furthermore, under different growth conditions, different functions are dependent upon different subunits. Taken together, these findings provide an explanation for the multiplicity of cellular functions that depend upon the complex, considering that the complex is composed of nine core subunits.
The role of the individual subunits of the Ccr4-Not complex has also been addressed using microarrays by the laboratory of Clyde Denis (Cui et al. 2008). They have found, like us, that Ccr4p and Caf1p have a different effect on gene expression than the Not proteins, but they underline in addition that Ccr4p and Caf1p have important shared effects, and the roles of Not3p and Not4p as well as Not5p correlate well with each other. The role of Not2p was not addressed in their work. In general, their study reveals more target genes for each subunit in cells growing in high glucose than our study, suggesting a difference in stringency of analysis between both studies, and neither the strain background nor the growth conditions were identical. This is likely to be very relevant to explain differences since our study underlines the importance of the growth condition for the role of each Ccr4-Not complex subunit in gene expression. Nevertheless, it is apparent in their study that, while they found similar effects of Ccr4p and Caf1p or the Not proteins on gene expression, the roles of the different subunits are not identical, and this is consistent with our findings.
Our study suggests that expression of most of the genome is likely to depend upon one or the other subunit of the Ccr4-Not complex in some growth condition. Indeed, we have analyzed only two growth conditions and determined that two thirds of the genome depends upon at least one subunit of the Ccr4-Not complex. Certainly this finding is very consistent with the knowledge that the Ccr4-Not complex is essential for yeast growth (Maillet et al. 2000) and that the complex is conserved in higher eukaryotes (Albert et al. 2000; Gavin et al. 2002).
An open question is why nine subunits with different functions need to be associated in a complex to mediate their functions. Indeed, surprisingly few genes were found to depend upon many subunits of the Ccr4-Not complex, suggesting that the Ccr4-Not complex is likely to be highly modular in structure. Either the association of the subunits in a complex places the subunits in the appropriate configuration to mediate their functions or the complex acts as a platform for storage of subunits with different functions. The subunits might need to be together, because the functions that they regulate need to be coordinately regulated. In this regard, it is interesting to note that the only subunit of the complex that is absolutely essential for growth is Not1p, which is the scaffold of the complex. This suggests that appropriate gene control by the various subunits does require that the proteins be associated in a complex. How each subunit works in its specific function, namely, whether it functions by transiently dissociating from the complex or by interacting with partner proteins within the complex, are interesting questions to start addressing. The gene families specific for each subunit identified in this study should provide a useful tool to perform such studies in the future.
MATERIALS AND METHODS
Strains, plasmids, and media
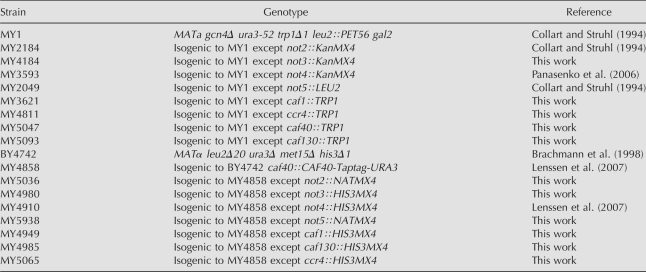
The strains used in this study are listed in Table 1. All media were standard, either YPD for glucose rich medium or synthetic complete media when the presence of a plasmid was selected for. Glucose concentrations in the growth medium were determined using an enzymatic glucose assay (GOD; Roche Diagnostics). Single-step deletions were performed by PCR as described by Longtine et al. (1998). All of the strains were checked with a PCR reaction performed on genomic DNA extracts using a primer localized in the marker gene and a primer localized at the 5′ noncoding sequence of the target gene. All primers used are available upon request. Many new strains were created by crosses followed by sporulation and tetrad dissection (see Table 1).
Microarray analyses
Wild-type and not2Δ, not3Δ, not4Δ, not5Δ, caf1Δ, caf130Δ, and ccr4Δ null strains (Table 1) were grown in glucose rich medium to exponential phase and collected. Wild-type and not2Δ, not4Δ, caf1Δ, caf130Δ, and ccr4Δ null strains were also grown in YPD to glucose depletion and collected 30 min after glucose depletion.
Total RNA was extracted by the acid phenol method and we followed the standard Affymetrix procedure as previously described (James et al. 2007). Description of this procedure and description of the probe sets are presented on the Affymetrix website (www.affymetrix.com/index.affx).
Each sample was prepared twice independently. The hybridized arrays were scanned and the raw image data were processed by Affymetrix software (MicroArraySuite, MicroDB, and DataMiningTool). Samples that were found to be problematic were discarded (more details on the quality control are given below). This is the case for one sample from the caf1Δ strain grown to glucose depletion and one sample from the not5Δ and not4Δ strains growing exponentially. Furthermore, because the not5Δ mutant flocculates, it was difficult to prepare samples from this strain 30 min after glucose depletion, and the attempt was abandoned. Indeed, it is difficult to have a homogenous population with regard to contact with the growth medium in the case of a strain that flocculates. This was true even in not5Δ cells growing in high glucose medium, but to a lesser extent because flocculation of this strain increases when glucose levels decrease. Nevertheless, gene expression changes relative to wild-type cells may have been underestimated in this mutant growing exponentially, because of the heterogeneity of the cells within the population.
The statistical analysis of the arrays was made with the R statistical environment (www.r-project.org) and a number of packages from Bioconductor (Gentleman et al. 2004) (affy, affyPLM, genefilter, limma, and gplots). Chip data quality was controlled through histograms of the signal intensities, RNA degradation plots, and plots of normalized unscaled standard errors (NUSE). The robust multichip average (RMA) algorithm (Irizarry et al. 2003) implemented in the Affymetrix package, affy, was used to correct background, then to quantile-normalize and summarize the perfect match (PM) probes of each probeset into an expression measure (in log base 2 scale).
The probe sets were filtered to discard those that measured too low levels of expression (log2 of 50 in one sample was the minimum) or that were not variable enough across the samples (interquartile range lower than 0.5) or that mapped to deleted genes or to marker genes or, finally, that measured Ty RNAs, tRNAs, or nonannotated RNAs (658 elements). Finally, of the 200 genes represented by more than one probeset (based on the SGD IDs provided by Affymetrix), only the ones showing the highest variability across the samples were kept for analysis. This left 4283 genes for the analysis. The mean of Pearson's correlation coefficients between duplicates was r = 0.96 after these filtering steps.
Differential gene expression of each mutant versus the wild type under the same growth condition and of the wild type in the post-diauxic phase versus the wild type in exponential phase was assessed using an empirical Bayes method after fitting the expression data to a linear model (package limma). The obtained P values were adjusted for multiple testing by Benjamini and Hochberg's method (Benjamini and Hochberg 1995).
Raw data and RMA-calculated signal intensities are available at the NCBI Gene Expression Omnibus repository (GEO series GSE10521). A dendogram of the resulting analysis is provided in Supplemental Figure 3.
S1 analysis
Total cellular RNA extraction and S1 analyses were performed as described previously (Collart 1996). The sequences of the specific oligonucleotides are available upon request.
Tandem affinity purificiation
Tandem affinity purifications from wild-type or mutant cells expressing Tap-tagged Caf40p grown to an OD600 of 1–2 were performed as previously described (Lenssen et al. 2007).
Western blot analysis
For Western blot analysis, proteins isolated by tandem affinity purification were separated on SDS-PAGE, 7% for detection of Not1p, Not3p, Not5p, and Caf1p or 10% for detection of Ccr4p, Not4p, and Caf40p-CBP, transferred to nitrocellulose, and analyzed by Western blotting with polyclonal primary antibodies, followed by secondary antibodies conjugated to alkaline phosphatase.
Mass spectrometry
For mass spectrometry analysis, the sample preparation, gel separation, and gel staining were performed according to the SWISS-2DPAGE protocols. Briefly, the samples were separated on 4%–12% SDS-PAGE gradient gels and stained using Coomassie in keratin free conditions. The visualized bands were excised from the gel and treated for mass spectrometry fingerprint analysis as follows: They were washed 15 min at room termperature with acetonitril 30%, incubated 35 min at 56°C in 10 mM 1,4-dithioerythritol to reduce the proteins, followed by 30 min incubation in 55 mM iodoacetamid at room temperature in the dark. After one wash for 10 min with 50 mM ammonium biocarbonate (pH 8.0) and two other washes of 10 min with 30% acetonitril, samples were then completely dried down in a vacuum centrifuge for 1 h. After sample rehydration in 20 μL of freshly prepared digestion buffer containing 6.25 ng/μL trypsin for 45 min on ice, digestion was allowed to proceed overnight at 37°C. To extract the peptides from the gel, digestion supernatant was recovered and pooled with two successive washes of 20 min using 40 μL 1% trifluoro acetic acid (TFA) and one wash using 0.1% TFA to remove salts. The collected supernatants were lyophilized and washed again with 35 μL 0.1% TFA before sample concentration to 2–5 μL. One microliter was loaded on a Maldi plate, and after matrix addition, sample acquisition was performed using the Maldi-Tof MS (VOYAGEUR CONTROL PANEL program), and peptides were submitted to fingerprinting analysis. The obtained spectra were finally analyzed using the DATA EXPLORER program, and proteins were identified using the MASCOT SEARCH website.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Jacques Schrenzel for the use of his Affymetrix platform and for helpful discussions, Thierry Sengstag and Pierre Farmer for their expert advices about statistical analyzes using R and Bioconductor, and Patrick Linder and Geoffroy Colau for a critical reading of this manuscript. This work was supported by the Leenards Foundation, the Lucie and Ernst Schmidheiny Foundation, and by Grant No. 3100AO-100793 from the National Science Foundation, as well as by IHP/Network European Grant No. HPRN-CTG-2002-00261, supported by the Office Fédéral de l'Education et de la Science, Grant No. 02.0017, to M.A.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1348209.
REFERENCES
- Albert T.K., Lemaire M., van Berkum N.L., Gentz R., Collart M.A., Timmers H.T. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert T.K., Hanzawa H., Legtenberg Y.I., de Ruwe M.J., van den Heuvel F.A., Collart M.A., Boelens R., Timmers H.T. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc [Ser. B] 1995;57:289–300. [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Collart M.A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Collart M.A., Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes & Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- Collart M.A., Timmers H.T. The eukaryotic Ccr4-not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- Cui Y., Ramnarain D.B., Chiang Y.C., Ding L.H., McMahon J.S., Denis C.L. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genomics. 2008;279:323–337. doi: 10.1007/s00438-007-0314-1. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Steitz J. Nucleus and gene expression. Coordinated nuclear events regulate mRNA synthesis, processing, export, and turnover. Curr. Opin. Cell Biol. 2005;17:239–241. [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- James N., Landrieux E., Collart M.A. A SAGA-independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4-Not complex in Saccharomyces cerevisiae . Genetics. 2007;177:123–135. doi: 10.1534/genetics.107.076299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E., James N., Pedruzzi I., Dubouloz F., Cameroni E., Bisig R., Maillet L., Werner M., Roosen J., Petrovic K., et al. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation–via a newly identified Glc7/Bud14 type I protein phosphatase module–and TFIID promoter distribution. Mol. Cell. Biol. 2005;25:488–498. doi: 10.1128/MCB.25.1.488-498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E., Azzouz N., Michel A., Landrieux E., Collart M.A. The Ccr4-Not complex regulates Skn7 through Srb10 kinase. Eukaryot. Cell. 2007;6:2251–2259. doi: 10.1128/EC.00327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maillet L., Tu C., Hong Y.K., Shuster E.O., Collart M.A. The essential function of Not1 lies within the Ccr4-Not complex. J. Mol. Biol. 2000;303:131–143. doi: 10.1006/jmbi.2000.4131. [DOI] [PubMed] [Google Scholar]
- Panasenko O., Landrieux E., Feuermann M., Finka A., Paquet N., Collart M.A. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 2006;281:31389–31398. doi: 10.1074/jbc.M604986200. [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]