Abstract
Dynactin, a large multisubunit complex, is required for intracellular transport by dynein; however, its cellular functions and mechanism of action are not clear. Prior studies suggested that dynactin increases dynein processivity by tethering the motor to the microtubule through its own microtubule binding domains. However, this hypothesis could not be tested without a recombinant source of dynactin. Here, we have produced recombinant dynactin and dynein in Saccharomyces cerevisiae, and examined the effect of dynactin on dynein in single-molecule motility assays. We show that dynactin increases the run length of single dynein motors, but does not alter the directionality of dynein movement. Enhancement of dynein processivity by dynactin does not require the microtubule (MT) binding domains of Nip100 (the yeast p150Glued homolog). Dynactin lacking these MT binding domains also supports the proper localization and function of dynein during nuclear segregation in vivo. Instead, a segment of the coiled-coil of Nip100 is required for these activities. Our results directly demonstrate that dynactin increases the processivity of dynein through a mechanism independent of microtubule tethering.
Keywords: microtubule, motor protein, nuclear segregation, p150Glued, single molecule
Dynactin, a large (≈1.2 MD) complex, was first identified as an activator of dynein-mediated, minus-end-directed vesicle transport (1, 2), and has subsequently been shown to be essential for nearly every cellular function of cytoplasmic dynein (3, 4). Several dynactin alleles have been linked to human neurological disease, which most likely results from a defect in intracellular trafficking (5, 6). Dynactin is composed of a filament of the actin-related protein Arp1, capped at each end by additional subunits. The barbed end subcomplex contains a dimer of the largest subunit, p150Glued (Nip100 in yeast), which binds to dynein directly (4). The N- terminus of p150Glued contains a CAP-Gly domain and a basic region, both of which have been shown to bind microtubules (MTs) (7–9), followed by a coiled-coil that projects as a 24-nm stalk from the Arp1 filament (10, 11).
Two general mechanisms have been proposed through which dynactin could aid the cellular function of dynein. First, many studies have provided evidence that dynactin is important for localizing cytoplasmic dynein to its proper intracellular cargo (4). Dynactin also might modulate dynein motor activity, an idea that has been explored through several in vitro motility assays. Dynactin has been proposed to increase the processivity of dynein, based on findings that dynactin increases the run length of dynein-coated beads in vitro (12). Increased processivity might be important in the cell for uninterrupted transport over long distance or for transport under high load. However, because dynactin and dynein were nonspecifically adsorbed onto beads in this study, it could not be determined whether dynein–dynactin complexes were observed, or if dynactin bound separately from dynein on the bead surface and exerted its effects by interacting independently with MTs.
The MT binding domains of the p150Glued subunit of dynactin have been proposed to have important roles in both dynactin functions. The MT-binding CAP-Gly domain of p150Glued is required for dynactin to localize to MT plus ends in some cell types, and has been implicated in dynein recruitment and MT anchoring at the centrosome (13–15). The initial discovery of the CAP-Gly MT binding domain at the N-terminus of p150Glued also led to the hypothesis that dynactin might increase the processivity of dynein by tethering dynein to the MT, thus suppressing dynein release and the termination of a run (7). Consistent with this idea, antibodies against the CAP-Gly domain were shown to abrogate the ability of dynactin to increase the run length of dynein-coated beads (12), and short polypeptides containing the MT binding basic region of p150Glued increased bead run length when coadsorbed with dynein onto beads (8). However, it was not possible to directly test the MT tethering hypothesis by assaying dynactin lacking its MT binding domains, because these studies were performed with native bovine dynactin, rather than dynactin from a recombinant source.
A recent study suggested that dynactin also might modulate the directionality of dynein. Dynein–dynactin complexes containing a GFP-tagged dynactin subunit were purified from transgenic mice, and their movements along MTs were observed by single-molecule fluorescence microscopy (16). In striking contrast to previous observations, these dynactin-dynein complexes exhibited frequent plus-end-directed excursions (often >1 μm), in addition to the minus-end-directed movement previously observed for dynein. Ross et al. raised the possibility that dynactin might facilitate these reversals in the direction of dynein movement, although the motility of the dynein alone was not observed in this study (16).
Here, we have developed a new system for examining the effects of dynactin on single molecule dynein motility using purified, recombinant dynein and dynactin complexes from Saccharomyces cerevisiae. We show that the dynein–dynactin cocomplex is more processive than dynein alone, but does not exhibit more plus-end-directed motion. Dynactin does not require the MT binding domains of Nip100 (the yeast p150Glued subunit) to increase dynein processivity or for dynein localization and function in nuclear segregation. Instead, our data indicate that a coiled-coil region adjacent to the MT binding regions in Nip100 is critical for these activities. Therefore, dynactin is unlikely to act as a simple MT tether.
Results
Yeast Dynein–Dynactin Is a Unidirectional Motor Complex with Enhanced Processivity.
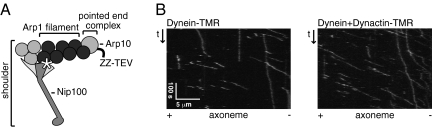
We have used the budding yeast S. cerevisiae as a source of recombinant dynactin for our experiments, using a strategy similar to one previously used to purify dynein [Fig. 1A; purification shown in Fig. S1A] (17). An N-terminal affinity tag was introduced at the genomic locus of ARP10 (which encodes a protein that binds to the pointed end of the Arp1 filament, see Fig. 1A). We also introduced a C-terminal HaloTag (Promega) at the genomic locus of NIP100 (the yeast p150Glued subunit), which allowed us to site-specifically label the dynactin complex with a fluorescent dye [tetramethyl rhodamine (TMR)]. Because the purification tag and fluorescent tag were placed on different subunits at opposite ends of the complex, only intact complexes were purified and labeled with TMR.
Fig. 1.
Purification of dynactin from S. cerevisiae. (A) Diagram of the dynactin complex. Intact dynactin complexes were affinity purified by using a ZZ-TEV tag on the N terminus of Arp10, and the Nip100 subunit was labeled with a TMR-conjugated C-terminal HaloTag (white asterix). (B) Reconstituted dynein–dynactin complexes move processively and unidirectionally along axonemes. Kymographs of dynein-TMR (Left) or unlabeled dynein and dynactin-TMR (Right) moving along axonemes are shown.
Using purified dynactin, we sought to determine the effect of dynactin on the motility of single dynein complexes, using total internal reflection fluorescence microscopy (TIRF). We found that TMR-labeled dynactin molecules displayed no detectable interaction with the MT, even when imaged at a high frame rate (30 per second). The lack of observable binding could reflect a very weak MT binding affinity of yeast dynactin, or that purified dynactin in the absence of dynein is in an inhibited state. However, when coincubated with unlabeled recombinant yeast dynein, TMR-labeled dynactin now bound to MTs and moved processively toward the minus-end of axonemal MTs (Fig. 1B; Movie S1). Because dynactin-TMR alone did not move, and dynein was unlabeled in this assay, these moving particles must be dynein–dynactin complexes.
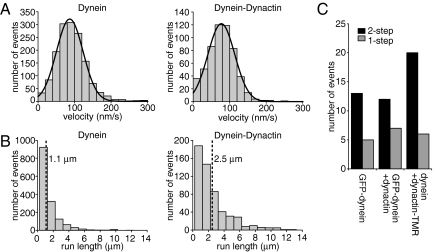
The S. cerevisiae dynein–dynactin-TMR complexes moved at a similar velocity to dynein-TMR alone (77 ± 37 nm/s, mean ± SD), compared with 87 ± 36 nm/s for dynein) (Fig. 2A and Movie S2). Notably, the dynein–dynactin complexes had a significantly longer run length (2.54 ± 0.17 μm, mean ± SE) than dynein alone (1.14 ± 0.04 μm) (Fig. 2B; Fig. S2).
Fig. 2.
Single dynein–dynactin complexes exhibit robust minus-end-directed motility with enhanced processivity. (A) Histograms of dynein and dynein–dynactin velocities. The velocity of dynein is 87 ± 36 nm/s (mean ± SD; n = 1499), and the velocity of dynein–dynactin is 77 ± 37 nm/s (n = 560). Curved lines represent Gaussian fits of the data. A few outlying points are truncated from histograms of run lengths and velocity for display purposes. (B) Histograms of dynein and dynein–dynactin run lengths. The run length of dynein is 1.15 ± 0.04 μm (mean ± SE), and the run length of dynein–dynactin is 2.54 ± 0.17 μm (determined from cumulative probability functions; see Fig. S2). Dashed lines indicate the mean. (C) Histogram of bleaching events. All bleaching events observed occurred in 1 or 2 steps. GFP-dynein, n = 18; GFP-dynein–dynactin, n = 19; dynein–dynactin-TMR, n = 26.
One possible explanation for this longer run length is that dynactin could link 2 or more dynein complexes together; one dynein motor in a multimeric complex might remain bound while another dissociated, leading to an apparent increase in run length. To determine the number of dynein motors within a moving dynein–dynactin complex, we analyzed photobleaching events of GFP-labeled dynein, with and without dynactin. When dynactin was present, we confirmed that GFP-dynein spots were bound to TMR-labeled dynactin by 2-color observation. In both cases, we observed only 1- and 2-step photobleaching of dynein, indicating that the dynein–dynactin complex contains a single dynein dimer (Fig. 2C). To determine the number of dynactin complexes bound to each dynein, we observed photobleaching of moving TMR-labeled dynactin spots. Again, we observed only 1- and 2-step photobleaching events, indicating that each dynein binds 1 dimer of Nip100, and thus a single dynactin complex (Fig. 2C). Therefore, a single dynactin complex can enhance the processivity of a single dynein motor.
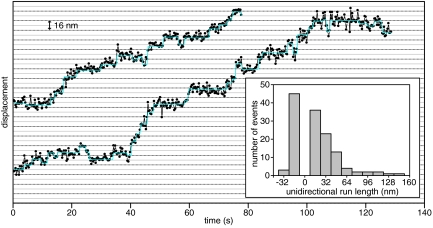
A previous study of dynein–dynactin motility reported plus-end-directed runs exceeding 1 μm (16). From our kymograph analysis, we did not observe any plus-end-directed runs >0.3 μm, the diffraction limit. To characterize the directionality of dynein–dynactin movement with higher spatial precision, we tracked dynein–dynactin complexes using Gaussian fitting, with a mean rmsd of 4.9 nm (Fig. 3) (18). Because this precision is lower than in previous studies of Qdot-labeled dynein, assigning individual steps was difficult, but within some traces, we observed stepwise movement of dynein–dynactin complexes that was grossly similar to the stepping behavior that has been observed for dynein alone (17). To quantify plus- and minus-end-directed components of dynein–dynactin movement, we measured the length of each unidirectional segment of each run. Movement toward the plus-end persisted over much shorter distances than movement toward the minus-end; only 2% of plus-end-directed segments were >24 nm long, compared with 36% of minus-end-directed segments (Fig. 3). Previously, dynein was shown to exhibit short plus-end-directed movement, taking only a few plus end-directed steps in a row (17). We conclude from this that dynactin does not induce bidirectional movement in the yeast dynein motor.
Fig. 3.
High spatial precision measurements of dynactin movement along MTs. Gaussian fits of the position of moving dynein–dynactin-TMR spots at each point in time are displayed as black circles connected by lines, and steps extracted from these fits are displayed as cyan lines. Horizontal lines are separated by 8 nm. (Inset) Histogram of the length of unidirectional segments (distance traveled before a ≥8-nm reversal) of dynein–dynactin movment.
CC1 Region of Dynactin, but Not Its MT Binding Domains, Are Required for Enhanced Processivity.
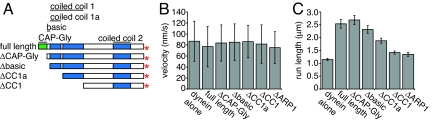
Having shown that dynactin is a dynein processivity factor using a recombinant system, we next performed structure-function studies to investigate the mechanism of this effect. The N-terminal CAP-Gly domain of p150Glued and an adjacent basic region (7–9) have been shown to interact with MTs, and it has been hypothesized that MT tethering by these domains might prevent dynein from dissociating from a MT and terminating a processive run. However, without a recombinant source of dynactin, it was not possible to test whether these domains are responsible for the effect of dynactin on dynein processivity. We truncated the CAP-Gly domain (ΔCAP-Gly) or the CAP-Gly domain and the adjacent basic region (Δbasic) from the single genomic copy of NIP100 and purified dynactin complexes containing these truncated Nip100 subunits (Fig. 4A). These truncated dynactin complexes associated with dynein and moved along axonemes with similar velocities (Fig. 4B) to wild-type dynein–dynactin complexes. Notably, the run lengths were not significantly reduced by these truncations, and remained 2-fold longer than that of dynein alone (Fig. 4C). Therefore, dynactin does not require its MT binding domains to increase the processivity of dynein.
Fig. 4.
The N-terminal coiled-coil (CC1) of Nip100, but not its N-terminal MT binding domains, is required for dynein processivity enhancement. (A) Diagram of truncations of the Nip100 subunit of dynactin. The N-terminally truncated Nip100 subunits lack the following residues: ΔCAP-Gly, 1–68; Δbasic, 1–102; ΔCC1a, 1–182; ΔCC1, 1–377. (B) Mean velocities of dynein–dynactin containing truncations in Nip100 or lacking Arp1, determined from a Gaussian fit. Error bars show SD. Dynein, n = 1,499; dynein plus dynactin, n = 560; dynein plus ΔCAP-Gly-dynactin, n = 899; dynein plus Δbasic-dynactin, n = 487; dynein plus ΔCC1A-dynactin, n = 496; dynein plus ΔCC1-dynactin, n = 939; dynein plus ΔArp1-dynactin, n = 599. (C) Mean run lengths of dynein–dynactin variants, determined from cumulative probability distributions (Fig. S2). Error bars show SE.
To identify a domain that might be required for this activity, we performed further truncations of NIP100. C-terminal to the CAP-Gly domain and basic region is a coiled-coil region that has been reported to bind to the N-terminus of the dynein intermediate chain (19). The first half of this coiled-coil is thought to form the 24-nm projecting stalk seen by EM (10, 11), whereas the second half may fold back into the shoulder of dynactin. We prepared dynactin complexes containing truncations of the first half (ΔCC1A) or the entirety (ΔCC1) of this coiled-coil domain, and examined how these alterations affected dynein–dynactin motility (Fig. 4A). We observed robust movement of dynein–dynactin cocomplexes with these truncations, and dynein coimmunoprecipitated weakly with both WT and ΔCC1A-dynactin, indicating that CC1 truncation does not grossly perturb the interaction of dynactin with dynein (Fig. S1B). Cocomplex formation instead may depend on an interaction between the central region of Nip100 and the dynein intermediate chain (20). Dynein bound to ΔCC1A-dynactin exhibited a longer run length (1.88 ± 0.10 μm) (Fig. 4C) than dynein alone, although this activation was reduced compared with wild-type dynein–dynactin. However, dynein in complex with ΔCC1-dynactin had a mean run length of 1.42 ± 0.06 μm (Fig. 4C), approaching the run length of dynein alone (1.15 ± 0.04 μm) (see SI Methods, showing that shorter run length is not due to premature dynactin dissociation from dynein). Therefore, CC1 is crucial to dynactin's effect on dynein processivity.
We next wished to determine whether the ability of dynactin to enhance dynein processivity is solely contained in the Nip100-containing “shoulder” subcomplex of dynactin, or whether the Arp1 filament contributes to this activity. Deletion of Arp1 or overexpression of p50 (Jnm1 in yeast) separates the shoulder subcomplex of dynactin, containing p150Glued, p50, and p24 (Ldb18 in yeast), from the rest of the complex (21–23), and causes strong perturbations of cellular dynein functions. We purified dynactin lacking the Arp1 filament, using an affinity tag on the Nip100 subunit in a yeast strain harboring a deletion in the ARP1 gene. Although deletion of ARP1 destabilizes p150Glued in some cell types (23–25), we found that Nip100 was stable in an arp1Δ background, as was previously observed in yeast (Fig. S1B) (23). ΔArp1-dynactin moved along axonemes with dynein, but the run length of this cocomplex (1.35 ± 0.08 μm, see Fig. 4C) was similar to dynein alone, indicating that an intact dynactin complex is necessary for increasing dynein processivity.
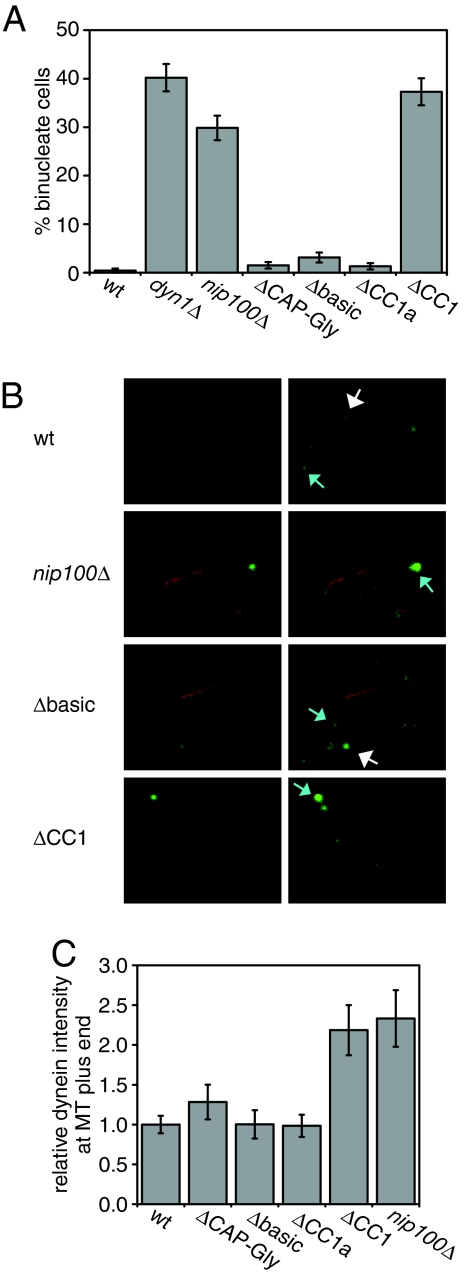
Nip100 CC1, but Not Nip100 MT Binding Domains, Is Required for Dynactin Function in Vivo.
Having determined which parts of the dynactin complex contribute to its in vitro enhancement of dynein processivity, we wished to test whether these domains are required for dynactin function in live cells. Dynactin is essential for the only function of dynein in yeast, pulling the nucleus into the bud neck during early anaphase (26–28). Cells with defects in dynein or dynactin function frequently missegregate their nuclei, retaining both in the mother cell. We found that nuclear segregation occurred normally in yeast harboring truncations of the N-terminal MT binding sites (ΔCAP-Gly and Δbasic) or the first half of CC1 (ΔCC1a) of NIP100, but was strongly perturbed in yeast with a complete CC1 truncation (Fig. 5A). In this ΔCC1-NIP100 strain, a similar percentage of binucleate mother cells were observed as in a nip100Δ strain. Immunoblots revealed that the cellular levels of ΔCC1a-Nip100 and ΔCC1-Nip100 are reduced relative to wild-type Nip100 (≈4-fold, see Fig. S1C). However, because the levels of both CC1-truncated proteins are similar, but only ΔCC1-Nip100 causes a cellular phenotype, this phenotype most likely arises from a specific requirement for CC1.
Fig. 5.
Effect of Nip100 truncations on dynactin function in yeast cells. (A) The fidelity of nuclear segregation in cells containing truncated Nip100, as indicated by the percentage of anaphase cells with binucleate mothers. Error bars show SEM; n > 200 for each strain. (B) Localization of dynein in cells containing truncated Nip100, displayed as Z-projections of confocal stacks. Images on the right are duplicates of those to their left, scaled such that wild-type localization is easily visible. Dyn1–3xGFP is shown in green, and mCherry–Tubl is shown in red. Examples of astral MT plus-end and cortical localization are indicated with blue and white arrows, respectively. (C) Quantification of Dyn1–3xGFP intensity at astral MT plus-ends. Intensities are normalized to intensity in WT cells. Error bars show SEM. WT, n = 22; ΔCAP-Gly-NIP100, n = 8; Δbasic-NIP100, n = 6; ΔCC1A-NIP100, n = 12; ΔCC1-NIP100, n = 11; ΔNIP100, n = 11.
To perform its cellular function, yeast dynein must localize to the bud cortex, where it walks along the astral MTs; thus pulling the nucleus toward the bud. The dynactin components Nip100 and Arp1 are required for this localization; dynein accumulates at the plus-ends of astral MTs in strains bearing deletions of these genes (29, 30). From this evidence, it has been proposed that dynactin may assist in transferring dynein from the plus-end of MTs to the cortex before motility begins. To determine whether the N-terminal domains of Nip100 are required for this transfer step, we observed the localization of Dyn1–3xGFP in the background of NIP100 truncations. Dyn1–3xGFP localized normally in ΔCAP-Gly-NIP100, Δbasic-NIP100, and ΔCC1A-NIP100 cells; these cells exhibited cortical spots of dynein and wild-type levels of dynein at astral MT plus-ends (Fig. 5 B and C). In contrast, ΔCC1-NIP100 cells lacked cortical dynein, and exhibited a 2-fold increase in dynein at astral MT plus-ends, similar to that observed in nip100Δ cells (Fig. 5 B and C). Thus, the first Nip100 coiled-coil, but not the N-terminal MT binding domains, is needed for proper localization of dynein during cell division.
Discussion
Dynactin is essential to dynein-mediated intracellular transport, but its size and complexity have hindered the elucidation of its function. Using dynactin purified from a recombinant source in a direct, single molecule assay, we have shown that dynactin increases the processivity of single dynein motors. Previous studies of mammalian dynactin and dynein (8, 12) first showed that dynactin could increase the processivity of dynein bound to beads. However, these early experiments were performed by nonspecific adsorption of the complexes to beads, which complicated mechanistic interpretation of these results. For example, dynactin and dynein might have bound separately on the bead and independently interacted with the MT, rather than operating as a dynein–dynactin cocomplex. Here, we show through direct observation that a single dynein with a single bound dynactin moves for longer distances along a MT than dynein alone.
Dynactin has been proposed to increase dynein processivity by acting as a tether to the MT surface (8, 9, 12). This hypothesis is supported by observations that beads coated with mammalian dynactin or N-terminal fragments of 150Glued bound to MTs and diffused 1-dimensionally (8, 9, 12). A monoclonal antibody to the N-terminal MT binding region of dynactin also was found to reduce the processivity of dynein and dynactin-adsorbed beads (12). However, by truncation of the Nip100 subunit within the intact dynactin complex, we find that the N-terminal MT binding regions of Nip100 are not required for dynactin to increase the processivity of yeast dynein. Also, we have not observed dynactin binding to or diffusing along MTs on its own; this lack of MT interaction may reflect an inhibited state of dynactin, as suggested by some studies in yeast and mammalian cells (13, 23). The MT interactions observed for bead-adsorbed dynactin and p150Glued fragments and the often pleiotropic effects of antibody inhibition also may not reflect the behavior of the intact complex. Nevertheless, our results indicate that dynactin increases the processivity of dynein by a mechanism other than MT tethering. The mechanism of this effect remains unknown; however, a possible model is that dynactin alters the coordination of the motor domains of dynein in a manner that reduces its probability of dissociation during processive motion.
A recent study suggested that dynactin can induce large plus end-directed movements of dynein, which the authors proposed were facilitated by MT tethering (16). We did not observe similar micrometer-scale plus-end-directed movements in our preparation of dynein–dynactin by kymograph analysis, and high-precision tracking of dynein–dynactin did not show longer plus end-directed movements than were previously observed for dynein alone (36 nm maximum, compared with 70 nm previously measured) (17). Therefore, our results indicate that the dynein–dynactin complex functions essentially as a unidirectional motor that takes an occasional one or few backwards steps, and no additional mechanism is required to ensure minus-end-directed transport by dynein in the cell. The difference between our observations and the study performed by Ross et al. (16) may reflect different behaviors of dynein–dynactin from yeast and mammals, and will require further investigation.
Our in vivo studies also reveal that the known MT binding regions of dynactin are not required for the functions of dynactin in yeast. This finding is surprising, given the high conservation of the p150Glued CAP-Gly domain across species. However, dynactin lacking the MT binding domains of p150Glued (CAP-Gly and basic regions) has been shown to support normal dynein-driven organelle motility in Drosophila S2 cells and in HeLa cells, indicating that the primary role of dynactin in cargo transport may not be as a MT tether (31, 32). However, mitotic functions of dynactin in S2 cells do require these domains (31). A neuronal isoform of p150Glued that lacks these MT binding domains incorporates into dynein–dynactin cocomplexes, and participates in axonal transport, again implying that some functions of dynactin may not involve MT tethering (33, 34). Thus, the MT binding domains of dynactin may be necessary only for a subset of activities of dynactin.
We also show that CC1, but not the N-terminal MT binding regions of Nip100, is required for the dynactin-mediated transfer of dynein from the plus-ends of MTs to the cell cortex. This requirement may reflect an undescribed role for dynactin as a cargo adapter at the cell cortex. However, as the in vitro and in vivo activity of dynactin are both disrupted by truncation of Nip100 CC1, perhaps dynactin's regulation of dynein motor function is important for the transfer process from the MT plus-end to the cortex. Supporting this idea, a small deletion (8 amino acids) in the catalytic core of dynein also causes dynein to accumulate at astral MT plus-ends (30). The specific requirement for the CC1 domain of Nip100 for in vitro processivity enhancement and nuclear segregation in vivo suggests that dynactin's regulation of dynein motor activity is important for dynein function in living cells.
Materials and Methods
Yeast Strains.
Strains used in this study are listed in Table S1. Deletion, tagging, and truncation of dynein and dynactin subunits were performed at chromosomal loci by using standard homologous recombination techniques. The genomic copy of Dyn1 was tagged with 3 tandem repeats of GFP by using a plasmid provided by John Cooper (Washington University School of Medicine, St. Louis, MO) (29).
Purification and Fluorescent Labeling of Yeast Cytoplasmic Dynein and Dynactin.
Yeast cultures were grown in YPD to an OD600 between 0.8 and 1. Cells were harvested by centrifugation and washed once with water; the pellet then was resuspended in residual water, and frozen by drops in liquid nitrogen. The frozen cell pellet was lysed by grinding in a liquid nitrogen-cooled coffee grinder, and the resulting powder was thawed with 0.3 volumes of 4× lysis buffer (1× lysis buffer: 30 mM Hepes, pH 7.2/50 mM potassium acetate/2 mM magnesium acetate/1 mM EGTA/10% glycerol/1 mM DTT/0.1 mM Mg ATP/1 mM Pefabloc/10 μg/mL leupeptin/10 μg/mL pepstatin A/0.2% Triton X-100). The crude lysate was centrifuged at 290,000 × g for 25 min, and the resulting supernatant was incubated with IgG Sepharose (Amersham Pharmacia) for 1 h at 4 °C. The IgG beads were then washed twice with lysis buffer, and bound Halotag fusion proteins were labeled with 10 μM TMR-conjugated Halotag ligand (Promega). The beads were washed 3 times with TEV cleavage buffer (10 mM Tris, pH 8.0/150 mM KCl/10% glycerol/0.1 mM ATP/1 mM DTT/1 mM Pefabloc), and the beads containing bound dynein or dynactin were incubated with TEV protease for 1 h at 16 °C. The resulting solution of dynein or dynactin was aliquotted and frozen in liquid nitrogen.
Motility Assays.
For single molecule TIRF microscopy, Cy5-labeled sea urchin axonemes were added to a flow chamber, where they adhered tightly to the glass coverslip, and the chamber was washed to remove free axonemes. Dynein, dynactin, or a mixture of the complexes were flowed into the chamber, and incubated for 2 min. If dynein–dynactin complexes were to be observed, dynein and dynactin were first preincubated for 5 min. The chamber was washed again and motility buffer (30 mM Hepes, pH 7.4/100 mM potassium acetate/2 mM magnesium acetate/1 mM EGTA/10% glycerol/1 mM DTT/1 mM MgATP/oxygen scavenger system; see ref. 17) was added. TMR-labeled single molecules and Cy5-labeled axonemes were visualized with a custom built objective-type total internal reflection microscope equipped with a 100×, 1.45 NA objective and a 1.6× optovar, using an argon laser with 514-nm illumination at 3 mW to image TMR and a helium-neon laser with 633-nm illumination to image Cy5. Images were acquired every 2 s for 8 min with a cooled, intensified CCD camera (Mega10 S30Z, Stanford Photonics), controlled with QED software. Velocities and run lengths of moving molecules were determined from kymograph analysis in ImageJ as previously described (17). Mean velocity was determined by fitting a Gaussian function to a histogram of velocities by using Origin software. Mean run length was calculated by evaluating the cumulative probability function of the measured run lengths, correcting for photobleaching, and for the length of the axonemes, as previously described (17). Every construct was assayed from at least 2 separate cell growths and protein preparations.
Photobleaching experiments to determine dynein–dynactin stoichiometry were performed under the above described motility assay conditions, with the exception that the oxygen scavenger solution was not included when observing Nip100-TMR photobleaching. Samples were observed by using an inverted microscope (TE2000U, Nikon) equipped for TIRF with a 100×, 1.45 NA objective. The sample was illuminated and imaged continuously with 491 nm (GFP photobleaching) or 561 nm (TMR photobleaching) diode lasers, and images were captured every 100 ms (GFP photobleaching) or 200 ms (TMR photobleaching) by using a back-thinned electron multiplying CCD camera (iXonEM+, Andor). The system was controlled with μManager (www.micro-manager.org) software. Fluorescence intensities of moving motors were determined from kymographs by using ImageJ.
High spatial precision measurements of dynein–dynactin stepping were performed by using the microscope setup described for photobleaching experiments. To prepare slides for these observations, flow chambers were coated with biotinylated BSA, incubated with streptavidin, and then incubated with biotinylated and Cy5-labeled microtubules. Dynein–dynactin complexes were introduced and exchanged into motility buffer as described above, except ATP was included at 300 nM with an ATP regenerating system (1% pyruvate kinase and 10 mM phosphoenolpyruvate); 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid was added to the motility buffer at 0.5 mg/mL to reduce TMR blinking. A 2D Gaussian fit to determine the centroid of dynactin-TMR spots was performed as described previously (18).
Nuclear Segregation Assay.
To assess the fidelity of nuclear segregation, cells were grown in YPD at 30 °C to OD600 = 0.3–1, diluted to OD600 = 0.3, and grown for 16 h at 16 °C. Cells were fixed and stained with DAPI, and wild-type and binucleate anaphase mother cells were counted.
Live Cell Microscopy.
Dyn1 was C-terminally tagged with 3xGFP by using a previously described plasmid (29). Cells were grown overnight in minimal medium supplemented with 20 mg/L adenine, and plated on agarose pads, as previously described (35). Samples were imaged at room temperature by using a spinning disc confocal scanhead (Yokogawa Electric and Solamere) mounted on an inverted microscope (Axiovert 200M, Zeiss) with a 100×, 1.45 NA objective. Images were captured on a cooled CCD camera (C9100-13, Hamamatsu), and image acquisition was controlled with μManager (www.micro-manager.org) software. Stacks of 0.2-μm slices were collected and collapsed into 2D projections by using ImageJ.
Supplementary Material
Acknowledgments.
We thank Nico Stuurman for advice on microscopy, Nenad Amodaj for MicroManager scripting, Andrew Carter for MATLAB scripting, and Andrew Carter and Niels Bradshaw for thought-provoking discussions. This work was supported by National Institutes of Health Grant P01-AR42895 (to R.D.V.), the National Science Foundation (J.R.K.), and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900976106/DCSupplemental.
References
- 1.Gill SR, et al. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:639–650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1990;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 5.Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 6.Munch C, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- 7.Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc Natl Acad Sci USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Shiroguchi K, Edamatsu M, Toyoshima YY. Microtubule-binding properties of dynactin p150 expedient for dynein motility. Biochem Biophys Res Commun. 2006;340:23–28. doi: 10.1016/j.bbrc.2005.11.145. [DOI] [PubMed] [Google Scholar]
- 10.Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: An actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckley DM, et al. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quintyne NJ, et al. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 17.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 19.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deacon SW, et al. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valetti C, et al. Role of dynactin in endocytic traffic: Effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol. 1999;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- 25.Haghnia M, et al. Dynactin is required for coordinated bidirectional motility, but not for dynein membrane attachment. Mol Biol Cell. 2007;18:2081–2089. doi: 10.1091/mbc.E06-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eshel D, et al. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein PacI during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheeman B, et al. Determinants of S. cerevisiae dynein localization and activation: Implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, et al. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit R, Levy JR, Tokito M, Ligon LA, Holzbaur EL. Regulation of dynactin through the differential expression of p150Glued isoforms. J Biol Chem. 2008;283:33611–33619. doi: 10.1074/jbc.M804840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokito MK, Howland DS, Lee VM, Holzbaur EL. Functionally distinct isoforms of dynactin are expressed in human neurons. Mol Biol Cell. 1996;7:1167–1180. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dillman JF, III, Dabney LP, Pfister KK. Cytoplasmic dynein is associated with slow axonal transport. Proc Natl Acad Sci USA. 1996;93:141–144. doi: 10.1073/pnas.93.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.