Abstract
Background
A large and unsuspected tuberculosis outbreak involving 18.7% of the total of the tuberculosis cases studied, was detected in a population-based molecular epidemiological study performed in Zaragoza (Spain) from 2001 to 2004.
Methods
The Mycobacterium tuberculosis drug-susceptible strain, named MTZ strain, was genetically characterized by IS6110-RFLP, Spoligotyping and by MIRU-VNTR typing and the genetic patterns obtained were compared with those included in international databases. The characteristics of the affected patients, in an attempt to understand why the MTZ strain was so highly transmitted among the population were also analyzed.
Results
The genetic profile of the MTZ strain was rare and not widely distributed in our area or elsewhere. The patients affected did not show any notable risk factor for TB.
Conclusion
The M. tuberculosis strain MTZ, might have particular transmissibility or virulence properties, and we believe that greater focus should be placed on stopping its widespread dissemination.
Background
Mycobacterium tuberculosis is an extremely successful pathogen that kills nearly two million people in the world each year [1]. The study of tuberculosis (TB) epidemiology and transmission, traditionally involving patient contact tracing, has been improved by the use of molecular strain typing [2]. Molecular epidemiological studies have added much-needed accuracy and precision to the study of transmission dynamics, and have allowed previously unresolved issues to be newly addressed, e.g. the classification of recent-versus-reactive disease, the extent of exogenous reinfection and the detection of unsuspected transmission events [2].
Advances in molecular typing have led to the identification of highly transmissible M. tuberculosis complex strains in the last years. In New York City, the W strain caused more than 350 cases [3] and spread to other American states [4]. A Beijing strain imported from Liberia affected 75 patients in the Gran Canaria Island during the 1990s [5]. Currently, the major threat for TB control is the transmission of extensively drug-resistant (XDR) strains [6,7]. However other strains, neither belonging to the W-Beijing family nor being drug-resistant, have shown extensive dissemination in various countries: the C strain in New York City [8,9], the CH strain in the UK [10,11], the Harlingen strain in the Netherlands [12,13] and the Danish Cluster 1 and 2 strains in Denmark [14].
In Zaragoza (Spain), an unsuspected, extensive transmission of a drug-susceptible M. tuberculosis strain was detected during a population-based molecular epidemiological survey from 2001 to 2004 [15]. In this study, 454 M. tuberculosis isolates were analyzed by IS6110-RFLP, and 52.6% were clustered. The largest cluster contained 85 isolates (18.7%); the strain causing this cluster was named MTZ, Mycobacterium tuberculosis Zaragoza.
The reasons for the dominance and widespread of the MTZ strain were unclear. The objectives of the present work were: i) to characterize the MTZ isolates by other genotyping techniques such as Spoligotyping and MIRU-VNTR typing, and include it in a Principal Genotype Group (PGG) ii) to compare the genetic patterns obtained with those included in international databases and iii) to analyze the characteristics of the affected patients, in an attempt to understand why the MTZ strain was so highly transmitted among the population.
Methods
The MTZ cluster was identified in a population-based study conducted in the province of Zaragoza between 1st June 2001 and 31st May 2004. A total of 454 M. tuberculosis complex isolates were typed by IS6110-RFLP: 239 isolates were grouped into 45 clusters, each cluster containing between two and 85 isolates [15].
DNA fingerprinting
IS6110-RFLP
RFLP analysis of the 85 isolates by Southern blotting and DNA hybridization with IS6110 was performed in the previous study according to the standard fingerprinting method [16]. The RFLP pattern was entered into the Spanish Database of the University of Zaragoza. This database includes 5,694 IS6110-RFLP entries from drug-susceptible and drug-resistant M. tuberculosis complex isolates, 4,637 (81%) of which are from Spanish isolates. The MTZ RFLP pattern was compared with the other database entries.
Spoligotyping
Spoligotyping was performed according to the method described by Kamerbeek et al. [17]. The spoligotype obtained was contrasted with entries contained in the SpolDB4 database [18]http://www.pasteur-guadeloupe.fr/tb/spoldb4/spoldb4.pdf and was further compared with the updated SITVIT2 database. SITVIT2 is a proprietary database maintained at the Pasteur Institute of Guadeloupe, which contains both spoligotype and MIRU-VNTR patterns of the M. tuberculosis. At the time of this comparison (30th October 2008), it contained data on about 70,000 strains from 160 countries of origin.
MIRU-VNTR
The 85 isolates were genotyped by PCR amplification of a highly discriminatory subset of 15 loci proposed by Supply et al. [19]. Analyses were performed using five multiplex PCRs (Table 1), PCR mixtures and conditions were as described in reference 19 with some modifications. PCR products were separated on a 48-capillary MegaBACE™ 500 Sequencer (GE Healthcare Life Sciences) using Rox-labeled MegaBACE ET900-R as a size standard. PCR fragments sizes were determined using the MegaBACE™ Fragment Profiler v1.2 software (GE Healthcare Life Sciences). VNTR alleles were assigned according to size offsets, which correct the differences in relative migration between the size standard and the amplicons. Agarose gel electrophoresis with PCR products of known size were used to define size offsets.
Table 1.
PCR mixtures and conditions used for the MIRU-VNTR genotyping
| Multiplex | Locus | Alias | VNTR length (bp) | [MgCl2] (mM) | PCR primer pairs (5' to 3'), with labelling indicated in bracketsa |
| Mix 1 | 580 | MIRU 4 | 77 | 3 | GCGCGAGAGCCCGAACTGC (FAM) GCGCAGCAGAAACGCCAGC |
| 2996 | MIRU 26 | 51 | 3 | TAGGTCTACCGTCGAAATCTGTGAC CATAGGCGACCAGGCGAATAG (HEX) |
|
| 802 | MIRU 40 | 54 | 3 | GGGTTGCTGGATGACAACGTGT (TAMRA) GGGTGATCTCGGCGAAATCAGATA |
|
| Mix 2 | 960 | MIRU 10 | 53 | 2 | GTTCTTGACCAACTGCAGTCGTCC GCCACCTTGGTGATCAGCTACCT (FAM) |
| 1644 | MIRU 16 | 53 | 2 | TCGGTGATCGGGTCCAGTCCAAGTA CCCGTCGTGCAGCCCTGGTAC (HEX) |
|
| 3192 | MIRU 31 | 53 | 2 | ACTGATTGGCTTCATACGGCTTTA GTGCCGACGTGGTCTTGAT (TAMRA) |
|
| Mix 3 | 424 | Mtub04 | 51 | 1.5 | CTTGGCCGGCATCAAGCGCATTATT GGCAGCAGAGCCCGGGATTCTTC (FAM) |
| 577 | ETR C | 58 | 1.5 | CGAGAGTGGCAGTGGCGGTTATCT (HEX) AATGACTTGAACGCGCAAATTGTGA |
|
| 2165 | ETR A | 75 | 1.5 | AAATCGGTCCCATCACCTTCTTAT (TAMRA) CGAAGCCTGGGGTGCCCGCGATTT | |
| Mix 4 | 2401 | Mtub30 | 58 | 3 | CTTGAAGCCCCGGTCTCATCTGT (FAM) ACTTGAACCCCCACGCCCATTAGTA |
| 3690 | Mtub39 | 58 | 3 | CGGTGGAGGCGATGAACGTCTTC (HEX) TAGAGCGGCACGGGGGAAAGCTTAG |
|
| 4156 | QUB-4156 | 59 | 3 | TGACCACGGATTGCTCTAGT GCCGGCGTCCATGTT (TAMRA) |
|
| Mix 5 | 2163b | QUB-11b | 69 | 1.5 | CGTAAGGGGGATGCGGGAAATAGG CGAAGTGAATGGTGGCAT (FAM) |
| 1955 | Mtub21 | 57 | 1.5 | AGATCCCAGTTGTCGTCGTC (HEX) CAACATCGCCTGGTTCTGTA |
|
| 4052 | QUB-26 | 111 | 1.5 | AACGCTCAGCTGTCGGAT (TAMRA) CGGCCGTGCCGGCCAGGTCCTTCCCGAT |
|
a VIC and NED labeling used in reference 19 have been replaced by HEX (5'-hexachloro-fluorescein phosphoramidite) and TAMRA (6'-carboxy-tetramethyl-rhodamine), respectively.
Two MTZ isolates were also genotyped by PCR amplification of the 12 "old" loci initially described [20], six of which were also present in the 15 MIRU-VNTR panel and compared with the updated SITVIT2 database.
The 21 loci analysed were sent to be compared to a web server, MIRU-VNTRplus [21], that includes a collection of 186 strains representing the major MTBC lineages. For 15 different MIRU-VNTR it was also possible the comparison with the patterns included in MLVA database http://minisatellites.u-psud.fr/MLVAnet/.
Assignation of the MTZ strain to one of the three principal genotypic groups
M. tuberculosis MTZ strain was assigned to one of the three PGG delineated by Sreevatsan et al. [22]. Polymorphism at codon 463 of the katG gene was evaluated by PCR amplification of a 620 bp portion of the gene with the forward primer katG904 (5'-AGCTCGTATGGCACCGGAAC) and the reverse primer katG1523 (5'-TTGACCTCCCACCCGACTTG) [23], followed by digestion with MspI. In the presence of the CGG variant of codon 463, a MspI recognition site is formed, and the two alleles are easily differentiated by their restriction patterns.
Polymorphism at codon 95 of the gyrA gene was detected by PCR amplification of a 320 bp fragment using the primers gyrA1 (5'-CAGCTACATCGACTATGCGA) and gyrA2 (5'-GGGCTTCGGTGTACCTCAT) [24] followed by DNA sequencing.
Information and statistical analysis of the patients
Medical and laboratory records of the patients were retrospectively and thoroughly reviewed. The primary routine contact investigation reports of the TB surveillance system in Zaragoza were also collected. The information included demographic data (age, sex, country of origin, place of residence), microbiological data (date of isolation and drug sensitivity), clinical data (site of disease), risk factors for TB (homelessness, injection drug use, alcohol and/or tobacco abuse, presence of HIV infection and history of previous imprisonment), and other information about possible epidemiological links.
The characteristics of the 85 patients in the MTZ cluster were compared with the other 369 TB patients studied in 2001–2004 [15]. We used the chi-square test (Yates-corrected) or Fisher's exact test to compare categorical data. The Mann-Whitney non-parametric test was used to compare the distribution of age as a continuous variable. The SPSS program for Windows (version 11.5; SPSS Inc, Chicago Il) was used for statistical analyses.
Results and discussion
DNA fingerprinting
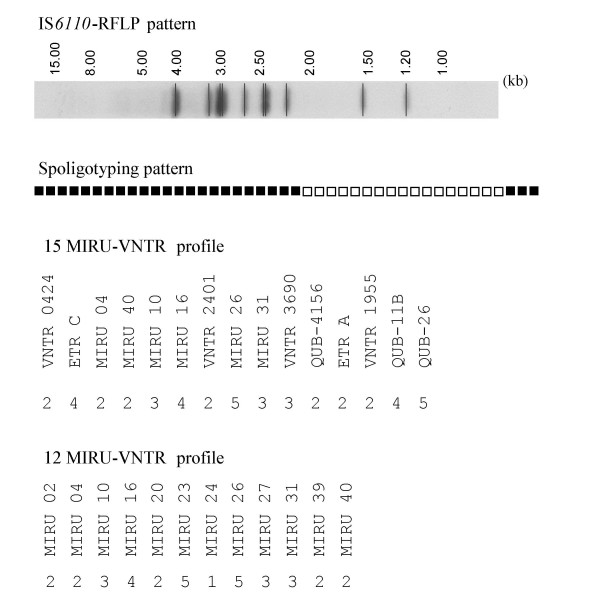
The 85 isolates of the MTZ cluster all showed the same IS6110-RFLP pattern, Spoligotype pattern and MIRU-VNTR pattern. The genetic profiles are shown in Figure 1. RFLP detected ten copies of IS6110 in the MTZ strain; this pattern was previously identified in only two patients from Zaragoza in 1993 and 1995 [25,26] and did not match any other profiles included in the database of the University of Zaragoza. Thus it is likely that this strain spread recently and was not endemic in our region during the nineties.
Figure 1.
IS6110-RFLP pattern, Spoligotyping pattern and MIRU-VNTR profiles of the MTZ strain.
M. tuberculosis MTZ strain was assigned to one of the three PGG (22) trying to provide a better framework for phylogenetic of this M. tuberculosis strain. The SNPs in codon KatG463 (CGG) and codon gyrA95 (AGC) revealed that the MTZ strain belonged to the principal genetic group 3.
The MTZ spoligotyping profile did not belong to any of the predominant spoligotype families from Spain, Europe or any other region of the world and did not match highly transmissible profiles like the W-Beijing [18]. The spoligotyping profile corresponded to the SIT number 773 in the SpolDB4 database (octal number 777777760000031), with no related genetic family identified and only five isolates reported (four from New York City, USA, and one from Jakarta, Indonesia) [18]. Further comparison with the updated database did not show any new additions since the last 5 strains described in SpolDB4. The search for 12-loci MIRU pattern 223425153322 showed that it belonged to a widely reported shared-type MIT157 in the updated SITVIT2 database; it was shared by a total of 61 strains from 9 different settings: Croatia (20/61 or 33%), Belgium (16/61 or 26%), Great Britain (11/61 or 18%), Spain (3/61 or 5%), French overseas departments of the Americas (Guadeloupe, Martinique, French Guiana; 5/61 or 8%), Sweden (2/61 or 3%), South Africa (2/61 or 3%), USA (1/61 or 1.5%), and Italy (1/61 or 1.5%). Spoligotypes were available for only 31/61 strains sharing MIT157, with the following distribution of the genotypic lineages: T lineage (25/31 or 80%), Haarlem (4/31 or 13%), LAM (1/31 or 3%), and Beijing (1/31 or 3%). The signature of the SIT773 contained a very unusual absence of spacers 37 to 40, which does not denote any known lineage-specific signature in SpolDB4 or SITVIT2 databases. The overall pattern of SIT773 has a resemblance to the "Zero copy" lineage described previously (18), nonetheless, the fact that MIRU24 value is equal to "1" together with the assignation at the group 3 of the PGG, suggest that the MTZ strain might be an evolutionary "modern" strain of tubercle bacilli. Further investigations using SNPs and 24-loci MIRU-VNTR would be necessary to determine the exact phylogeographical specificity of the strains containing this SIT773 pattern.
The comparison to MIRU-VNTRplus web server http://www.miru-vntrplus.org/ (21), and with MLVA database in http://minisatellites.u-psud.fr/MLVAnet/, did not report any new information.
The molecular typing of the 85 isolates by Spoligotyping and MIRU-VNTR typing was also useful to confirm the composition of the large MTZ cluster initially identified by IS6110-RFLP.M. tuberculosis isolates from epidemiologically linked patients generally show identical IS6110-RFLP patterns; however, IS6110-based RFLP fingerprints are not always reliable indicators of epidemiological linkage [27-29], and MIRU-VNTR typing can subdivide IS6110-RFLP clusters with no epidemiological connections [19,30]. The epidemiological information on the MTZ cases was limited, because the molecular typing was retrospective and subsequent to the contact tracing and the patients were not re-interviewed. Despite the absence of epidemiological links (only small groups of patients, constituting a total of 26 patients altogether, were clearly linked) (Table 2), the MIRU typing did not separate the cluster of 85 isolates, suggesting that all the patients studied could be connected.
Table 2.
Characteristics of epidemiologically linked patients of the MTZ cluster
| ID n° | Age (years) | Sex | Date of isolation | HIV status | Origin | Epidemiological link | Area of residencea |
| 121 | 43 | Man | 200402 | Negative | Spain | Family contact | 10 |
| 130 | 38 | Woman | 200402 | Negative | Spain | Children attending the same nursery | 10 |
| 140 | 26 | Woman | 200404 | Negative | Spain | Nursery staff b | - |
| 449 | 1 | Man | 200404 | Unknown | Spain | Children attending the same nursery | 1 |
| 443 | 2 | Woman | 200404 | Unknown | Spain | Children attending the same nursery | 3 |
| 437 | 1 | Man | 200404 | Unknown | Spain | Children attending the same nursery | 1 |
| 445 | 1 | Man | 200404 | Unknown | Spain | Children attending the same nursery | 2 |
| 148 | 2 | Woman | 200404 | Unknown | Spain | Children attending the same nursery | 1 |
| 441 | 2 | Woman | 200404 | Unknown | Spain | Children attending the same nursery | 2 |
| 444 | 1 | Man | 200404 | Unknown | Spain | Children attending the same nursery | 1 |
| 442 | 1 | Woman | 200404 | Unknown | Spain | Children attending the same nursery | 4 |
| 180 | 2 | Woman | 200108 | Unknown | Spain | Family contact | - |
| 186 | 20 | Woman | 200109 | Negative | Spain | 18b | |
| 159 | 35 | Man | 200106 | Negative | Spain | - | |
| 269 | 26 | Woman | 200207 | Negative | Algeria | Family contact | 16 |
| 257 | 43 | Man | 200206 | Negative | Spain | 16 | |
| 272 | 51 | Man | 200208 | Unknown | Spain | Family contact | 18c |
| 285 | 19 | Man | 200209 | Negative | Spain | 18c | |
| 160 | 41 | Man | 200106 | Negative | Spain | Family contact | 18d |
| 341 | 5 | Man | 200304 | Unknown | Spain | 18d | |
| 201 | 27 | Man | 200111 | Negative | Spain | Family contact | 17 |
| 196 | 23 | Woman | 200110 | Negative | Spain | 17 | |
| 422 | 24 | Woman | 200402 | Unknown | Spain | Family contact | 7 |
| 419 | 8 months | Woman | 200402 | Negative | Spain | 7 | |
| 36 | 25 | Man | 200205 | Positive | Spain | Friendship contact | - |
| 56 | 25 | Man | 200209 | Unknown | Spain | - |
a The areas of residence are shown in figure 2. The dash (-) means that the patient did not live in any of the 18 areas of residence (see figure 2). b The parents of the woman who worked at the nursery lived close to area n° 5. The woman also worked for 7 years in a bingo hall in area 18d, and for one year in a bingo hall between the areas 7 and 8.
n°: patient identification number.
Characteristics and epidemiological investigation of the patients
Sixty (70.6%) of the 85 cases of the MTZ cluster were men and 25 (29.4%) were women. The median age of the patients was 30 years (25th percentile [P25] to P75, 23 to 41) with a range of 8 months to 68 years. Seventy-three (86%) patients were born in Spain; 12 (14%) were foreign-born. All patients presented with a drug-susceptible isolate, except one patient with history of relapse and non-adherence to the TB treatment, who presented a rifampicin-resistant isolate.
We compared the characteristics of the MTZ-infected patients with the rest of clustered patients from the previous study (154 cases) and with total patients studied (clustered and non-clustered, 369 cases). The median age of MTZ-infected patients was lower (30 years) than that of other clustered patients (median age 38 years; P25 to P75, 29 to 46) or than that of all patients (median age 40 years; P25 to P75, 30 to 62) (P < 0.001 in both comparisons). Almost all (94.1%) of the MTZ-infected patients lived in Zaragoza city; the corresponding values were 81.5% for other clustered patients (P = 0.012) and 76.1% for all patients (P < 0.001). The MTZ-infected patients were less likely to be injection drug users (9.9%) than other clustered patients (22.9%) (P = 0.024), and presented more frequently with pulmonary TB than all other patients (95.3% vs. 85.1%; P = 0.019). In many TB outbreaks, the patients share a common risk factor for TB such as homelessness, HIV infection, injection drug abuse or alcohol abuse [5-9,31]. However, in our study, MTZ cluster patients do not appear to be linked by any common characteristic or risk factors for TB. Other European studies have reported the dissemination of TB strains among low-risk individuals, such as the Harlingen strain in the Netherlands or the Danish cluster 2 strain in Denmark [12-14].
A number of epidemiological links could be established from the data available from patient medical records and contact investigation reports. Eight children (all between one and two years old) and one nursery staff were linked because they attended the same nursery [32]. Familial links were identified involving 15 household contacts in seven different families and a friendship link between two other patients. Patient characteristics are detailed in Table 2.
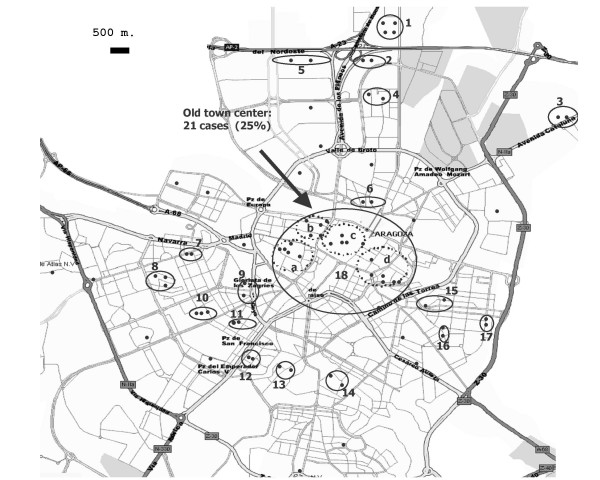
A significant data was the proportion of patients linked by place of residence. The study area included the province of Zaragoza [15], which is composed of both urban and rural areas, but 94.1% of the MTZ-infected patients lived in Zaragoza city. Thirty-nine patients were linked into groups of two or three individuals because they lived in the same street, or in parallel, adjoining or perpendicular streets (Table 3). Seventy-eight cases of the MTZ cluster were situated in the city of Zaragoza, and 21 (25%) of these were concentrated in the old town centre (Figure 2). The other seven patients lived in peripheral districts or in rural areas. Other studies have demonstrated that patients whose residences are geographically aggregated may be clustered [33] and therefore, the location of TB exposure is an important factor to consider in addition to the contact tracing [9,34,35]. TB transmission between sporadic or casual contacts has been documented [29,31]; hypothetically, the transmission of the MTZ strain could have occurred casually in public settings, probably near to the residences of patients.
Table 3.
Characteristics of the MTZ cluster patients linked by the place of residence and/or by common risk factors for TB
| ID n° | Age (years) | Sex | Date of isolation | HIV status | Origin | Area of Residence a | Characteristics in common b |
| 371 | 30 | Man | 200307 | Negative | Guinea | 11 | Neighbours |
| 42 | 25 | Man | 200206 | Negative | Spain | 11 | |
| 177 | 32 | Woman | 200109 | Negative | Spain | 18d | Neighbours |
| 171 | 38 | Man | 200108 | Negative | Spain | 18d | |
| 60 | 44 | Man | 200209 | Negative | Spain | - | Prison, alcoholism |
| 258 | 36 | Man | 200205 | Positive | Spain | - | Prison, alcoholism |
| 167 | 47 | Man | 200107 | Unknown | Spain | - | Prison |
| 246 | 33 | Man | 200204 | Negative | Spain | 18b | Neighbour of 322 |
| 322 | 27 | Man | 200303 | Negative | Egypt | 18b | Prison, neighbour of 246, IDU |
| 69 | 24 | Man | 200211 | Negative | Unknown | 18a | Neighbours, alcoholism |
| 263 | 53 | Man | 200207 | Negative | Spain | 18a | Neighbours, alcoholism |
| 374 | 46 | Man | 200308 | Positive | Spain | 18a | Ex-IDU |
| 217 | 34 | Man | 200112 | Positive | Unknown | - | Ex-IDU |
| 23 | 36 | Man | 200112 | Positive | Spain | - | Ex-IDU |
| 304 | 34 | Man | 200303 | Negative | South America | - | Ex-IDU |
| 79 | 26 | Man | 200212 | Positive | Venezuela | 6 | - |
| 434 | 56 | Man | 200403 | Unknown | Spain | 6 | Alcoholism. Worked next to area 5 |
| 271 | 39 | Man | 200208 | Negative | Spain | - | Alcoholism |
| 178 | 47 | Man | 200109 | Negative | Spain | 3 | Alcoholism |
| 428 | 26 | Woman | 200402 | Positive | Spain | 4 | Lived in the same area than 442 (table 2) |
| 318 | 44 | Man | 200303 | Negative | Gambia | - | 15 years in Spain |
| 312 | 25 | Man | 200302 | Negative | Gambia | 10 | 10 years in Spain. Worked in area 8. |
| 59 | 39 | Woman | 200209 | Negative | Spain | 8 | |
| 76 | 43 | Man | 200212 | Unknown | Ukraine | 8 | |
| 125 | 60 | Woman | 200402 | Negative | Spain | 8 | |
| 302 | 41 | Man | 200301 | Negative | Spain | 15 | |
| 427 | 46 | Man | 200402 | Negative | Spain | 15 | |
| 94 | 50 | Man | 200307 | Negative | Spain | 9 | |
| 194 | 57 | Man | 200109 | Negative | Spain | 9 | |
| 209 | 31 | Man | 200112 | Negative | Spain | 12 | |
| 283 | 26 | Man | 200209 | Negative | Spain | 12 | |
| 174 | 36 | Woman | 200108 | Negative | Spain | 13 | |
| 205 | 26 | Woman | 200111 | Negative | Spain | 13 | |
| 342 | 21 | Man | 200305 | Negative | Spain | 14 | |
| 276 | 35 | Man | 200209 | Negative | Spain | 14 | |
| 237 | 49 | Man | 200204 | Positive | Spain | 5 | |
| 297 | 28 | Man | 200210 | Negative | Unknown | 5 | |
| 247 | 36 | Woman | 200205 | Positive | Spain | 18a | |
| 199 | 63 | Man | 200111 | Unknown | Spain | 18a | |
| 385 | 44 | Man | 200309 | Positive | Spain | 18b | |
| 18 | 68 | Man | 200111 | Unknown | Spain | 18b | |
| 431 | 29 | Woman | 200402 | Positive | Spain | 18c | |
| 214 | 53 | Man | 200201 | Negative | Spain | 18c | |
| 370 | 52 | Man | 200308 | Negative | Spain | 18c | |
| 26 | 31 | Woman | 200201 | Negative | Spain | 18c | |
| 260 | 33 | Man | 200206 | Negative | Spain | 18d | |
| 158 | 20 | Woman | 200106 | Unknown | Spain | 18d |
Figure 2.
Location of 78 cases of the MTZ cluster in Zaragoza City according to the place of residence. The cases grouped inside a circle lived in the same street, or in parallel, adjoining or perpendicular streets. The areas that included two or more cases are numbered from 1 to 18.
In one of the small outbreaks caused by MTZ strain registered among children attending at a nursery, 11.7% showed a positive skin test and 90.9% of them developed the illness [32]. However, the percentage of infected children was surprisingly low in view of the dissemination of this strain among the total population in Zaragoza. Like strains involved in other outbreaks, the MTZ strain might have unique characteristics for virulence and/or transmissibility [36-39]. The transmission of tuberculosis may have been due to the increased virulence of the strain rather than to environmental factors or patient characteristics.
Another possible explanation for the extensive spread of the MTZ strain could be that this strain was endemic in our region, but in a previous molecular study performed in the same area between 1993 and 1995 [25,26], MTZ was only isolated from two Spanish patients who were not apparently connected; one in 1993 and one in 1995. The first patient was an 11 year-old girl living in a rural area, and the second was a 22 year-old woman living in the old town centre in Zaragoza. Surprisingly, both patients presented with rare extra-pulmonary locations of TB (bone TB and intestinal TB respectively), whereas 95.3% of the cases reported from 2001 to 2004 presented with pulmonary TB. The period from 1995 to 2001 has not been studied, and it seems likely that more MTZ cases would be identified during this period. A more detailed epidemiological investigation of the outbreak is needed to elucidate the chain of transmission.
Conclusion
In conclusion, neither the genetic profiles exhibited by the MTZ strain nor the characteristics of the patients affected could explain the reasons for the dominance and widespread of this modern strain. The MTZ strain might have particular transmissibility or virulence properties that need to be studied, and we believe that greater focus should be placed on stopping its widespread dissemination. Molecular typing has been a decisive tool to detect this unsuspected TB outbreak, and demonstrates the importance of the combination of both traditional approaches and molecular epidemiology for TB surveillance.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SS and AILC designed the study, supervised all the experimental work, analyzed the results, wrote the manuscript. AILC and PG performed the experimental assays. MALe and MAV compiled all the isolates and the first clinical and epidemiological data. JG, MALa collect all the clinical and epidemiological data. MJI helped in analysis of results. NR helped in the global analysis of the genotype. CM and MJR critically reviewed the final version of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by Fondo de Investigaciones Sanitarias (FIS 06/1624).
The authors thank Thierry Zozio (Pasteur Institute of Guadeloupe) for data comparison using the SITVIT2 database. Any inquiries regarding the SITVIT2 should be addressed to nrastogi@pasteur-guadeloupe.fr. The authors also would like to thank Carmen Lafoz, Daniel Ibarz, Ana Picó and Ana Belén Gómez for their excellent technical assistance and Alberto Cebollada for his help with the statistical analysis.
Contributor Information
Ana Isabel López-Calleja, Email: ailopezc@aragon.es.
Patricia Gavín, Email: pgavinb.iacs@aragon.es.
Ma Antonia Lezcano, Email: malezcano@salud.aragon.es.
Ma Asunción Vitoria, Email: mavitoria@salud.aragon.es.
Ma José Iglesias, Email: iglesias@unizar.es.
Joaquín Guimbao, Email: jguimbao@aragon.es.
Ma Ángeles Lázaro, Email: malazaro@aragon.es.
Nalin Rastogi, Email: nrastogi@pasteur-guadeloupe.fr.
Ma José Revillo, Email: mjrevillo@salud.aragon.es.
Carlos Martín, Email: carlos@unizar.es.
Sofia Samper, Email: ssamper.iacs@aragon.es.
References
- World Health Organization . (WHO/HTM/TB/2007376) Geneva, World Health Organization; 2007. Global tuberculosis control: surveillance, planning, financing. WHO report 2007. [Google Scholar]
- Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Molecular Epidemiology of Tuberculosis: Current Insights. Clin Microbiol Rev. 2006;19:658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML, Moghazeh SL, Eisner W, Daniel TM, Kaplan MH, Crawford JT, Musser JM, Kreiswirth BN. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–457. doi: 10.1001/jama.275.6.452. [DOI] [PubMed] [Google Scholar]
- Bifani PJ, Mathema B, Liu Z, Moghazeh SL, Shopsin B, Tempalski B, Driscol J, Frothingham R, Musser JM, Alcabes P, Kreiswirth BN. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- Caminero JA, Pena MJ, Campos-Herrero MI, Rodríguez JC, García I, Cabrera P, Lafoz C, Samper S, Takiff H, Afonso O, Pavón JM, Torres MJ, van Soolingen D, Enarson DA, Martin C. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med. 2001;164:1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–80. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- Samper S, Martín C. Spread of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007;13:647–8. doi: 10.3201/eid1304.061329. http://www.cdc.gov/eid/content/13/4/647.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CR, Quinn GC, Kreiswirth BN, Perlman DC, Salomon N, Schluger N, Lutfey M, Berger J, Poltoratskaia N, Riley LW. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis. J Infect Dis. 1997;176:478–84. doi: 10.1086/514067. [DOI] [PubMed] [Google Scholar]
- Macaraig M, Agerton T, Driver CR, Munsiff SS, Abdelwahab J, Park J, Kreiswirth B, Driscoll J, Zhao B. Strain-specific differences in two large Mycobacterium tuberculosis genotype clusters in isolates collected from homeless patients in New York City from 2001 to 2004. J Clin Microbiol. 2006;44:2890–6. doi: 10.1128/JCM.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar K, Shafi J, Smith RJ, Stabler RA, Andrew PW, Modha D, Bryant G, Monk P, Hinds J, Butcher PD, Barer MR. Use of genome level-informed PCR as a new investigational approach for analysis of outbreak-associated Mycobacterium tuberculosis isolates. J Clin Microbiol. 2004;42:1890–6. doi: 10.1128/JCM.42.5.1890-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Moss F. TB in Leicester: out of control, or just one of those things? BMJ. 2001;322:1133–4. doi: 10.1136/bmj.322.7295.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers A, Drost AP, van Soolingen D, Veen J. Use of DNA fingerprinting in international source case finding during a large outbreak of tuberculosis in The Netherlands. Int J Tuberc Lung Dis. 1997;1:239–45. [PubMed] [Google Scholar]
- van Soolingen D, Borgdorff MW, de Haas PE, Sebek MM, Veen J, Dessens M, Kremer K, van Embden JD. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J Infect Dis. 1999;180:726–36. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- Lillebaek T, Dirksen A, Kok-Jensen A, Andersen AB. A dominant Mycobacterium tuberculosis strain emerging in Denmark. Int J Tuberc Lung Dis. 2004;8:1001–6. [PubMed] [Google Scholar]
- López-Calleja AI, Lezcano MA, Vitoria MA, Iglesias MJ, Cebollada A, Lafoz C, Gavin P, Aristimuño L, Revillo MJ, Martin C, Samper S. Genotyping of Mycobacterium tuberculosis over two periods: a changing scenario for tuberculosis transmission. Int J Tuberc Lung Dis. 2007;11:1080–6. [PubMed] [Google Scholar]
- van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, Small P. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ho ML, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39:3563–71. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and user-strategy of MIRU-VNTR plus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–9. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevatsan S, Pan X, Stockbauer K, Connell N, Kreiswirth B, Whittam T, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl JR, Sandhu GS, Kline BC, Cockerill FR., III . PCR-RFLP detection of point mutations in the catalase-peroxidase gene (katG) of M. tuberculosis associated with isoniazid resistance. In: Persing DH, editor. PCR protocols for emerging infectious diseases. ASM Press, Washington, D.C; 1996. pp. 144–149. [Google Scholar]
- Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, Cole ST, Jacobs WR, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–80. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper S, Iglesias MJ, Rabanaque MJ, Lezcano MA, Vitoria LA, Rubio MC, Gómez-Lus R, Gómez LI, Otal I, Martín C. The molecular epidemiology of tuberculosis in Zaragoza, Spain: a retrospective epidemiological study in 1993. Int J Tuberc Lung Dis. 1998;2:281–7. [PubMed] [Google Scholar]
- Iglesias MJ. PhD Thesis. University of Zaragoza, Zaragoza, Spain; 1998. Molecular epidemiology of tuberculosis in Zaragoza 1993–1995. [PubMed] [Google Scholar]
- Braden CR, Templeton GL, Cave MD, Valway S, Onorato IM, Castro KG, Moers D, Yang Z, Stead WW, Bates JH. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–52. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- Gillespie SH, Dickens A, McHugh TD. False molecular clusters due to nonrandom association of IS6110 with Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2081–6. doi: 10.1128/jcm.38.6.2081-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom H, Hoijng SP, de Haas PE, Langendam MW, Horsman A, van Soolingen D, Coutinho RA. Clustered tuberculosis cases: do they represent recent transmission and can they be detected earlier? Am J Respir Crit Care Med. 2004;169:806–10. doi: 10.1164/rccm.200306-856OC. [DOI] [PubMed] [Google Scholar]
- van Deutekom H, Supply P, de Haas PE, Willery E, Hoijng SP, Locht C, Coutinho RA, van Soolingen D. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J Clin Microbiol. 2005;43:4473–9. doi: 10.1128/JCM.43.9.4473-4479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diel R, Meywald-Walter K, Gottschalk R, Rüsch-Gerdes S, Niemann S. Ongoing outbreak of tuberculosis in a low-incidence community: a molecular-epidemiological evaluation. Int J Tuberc Lung Dis. 2004;8:855–61. [PubMed] [Google Scholar]
- Sarrat Torres R, Macipe Costa R, de Juan Martín F, Lezcano Carreras MA, Bouthelier Moreno M, Marín Bravo MC, Esteban Ibarz JA. Epidemic outbreak of tuberculosis in a daycare centre in Zaragoza (Spain) An Pediatr (Barc) 2006;65:219–24. doi: 10.1157/13092157. [DOI] [PubMed] [Google Scholar]
- Bishai WR, Graham NM, Harrington S, Pope DS, Hooper N, Astemborski J, Sheely L, Vlahov D, Glass GE, Chaisson RE. Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA. 1998;280:1679–84. doi: 10.1001/jama.280.19.1679. [DOI] [PubMed] [Google Scholar]
- Barnes PF, el-Hajj H, Preston-Martin S, Cave MD, Jones BE, Otaya M, Pogoda J, Eisenach KD. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. doi: 10.1001/jama.275.4.305. [DOI] [PubMed] [Google Scholar]
- Barnes PF, Yang Z, Preston-Martin S, Pogoda JM, Jones BE, Otaya M, Eisenach KD, Knowles L, Harvey S, Cave MD. Patterns of tuberculosis transmission in Central Los Angeles. JAMA. 1997;278:1159–63. doi: 10.1001/jama.278.14.1159. [DOI] [PubMed] [Google Scholar]
- Beggs ML, Eisenach KD, Cave MD. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2923–8. doi: 10.1128/jcm.38.8.2923-2928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41:179–87. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- Soto CY, Menéndez MC, Pérez E, Samper S, Gómez AB, García MJ, Martín C. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J Clin Microbiol. 2004;42:212–9. doi: 10.1128/JCM.42.1.212-219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, Jones JS, Westmoreland H, Onorato IM. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–9. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]