Abstract
Background
The telomeres of somatic cells become shorter with individual aging. However, no significant change in subtelomeric methylation of somatic cells with aging has yet been reported.
Methods
Telomere lengths of the peripheral blood cells of 148 normal Japanese were analyzed by Southern blotting using methylation-sensitive and -insensitive isoschizomers.
Results
With aging, long telomeres decrease and short telomeres increase, and the contents of the telomeres with methylated subtelomere increase in long telomeres, thus leading us to postulate that telomeres with less methylated subtelomeres tend to become shortened faster.
Conclusions
A telomere length distribution analysis with methylation-sensitive and -insensitive isoschizomer seems to be a useful tool to assess the subtelomeric methylation status of the somatic cell population. The subtelomeric methylation of peripheral blood cells is also indicated to be an indicator for aging-associated genomic changes.
Keywords: Telomere, Aging, DNA methylation, Japanese
TELOMERES are a repetitive DNA structure at the very ends of chromosomes, which stabilize chromosomal structures and protect them from harmful DNA recombination between different chromosomes (1). The replication of normal somatic cells is finite and that after a critical number of cell divisions the cells reach a state in which further division cannot occur (2). This state is called “replicative senescence,” and it occurs in association with shortened telomeres because DNA replication during mitosis is incomplete and results in a loss of 50–200 terminal base pairs (bp) in telomeres per cell division (3). The cells shift to the state of cell senescence without DNA replication and mitosis in order to prevent further telomere shortening, which could lead to an unstable chromosome structure and unfavorable chromosomal recombination, thus possibly leading to carcinogenesis (1). In various organ systems, replicative senescence can result in reduced tissue regeneration with aging (4,5).
The cell loss resulting from replicative senescence is compensated by supplies of young cells derived from tissue-specific stem cells harbored in each tissue, which maintain their telomere length by their telomere-elongating telomerase activity over their lifetime (6,7).
The telomerase activity decreases promptly as cell differentiation proceeds in humans (1). Telomeres undergo attrition in their length with each cycle of somatic cells thereafter (1), and thus, the replicative history of somatic cells is a major determinant of telomere length. In human beings, the telomere length of replicating somatic cells is inversely related to the donor’s age (8,9), although it is highly variable among same-age donors (9,10). Another determinant of this phenomenon is heredity (11). Studies of monozygous and heterozygous twin pairs have indicated a high heritability for mean terminal restriction fragment (TRF) length of blood cell. The individual differences in the mean TRF length in blood seem to a large extent to be genetically determined (12,13). Pathophysiological conditions including mental stress, smoking, obesity, diabetes mellitus, ischemic heart diseases, Alzheimer’s disease, Parkinson’s disease, and sarcoidosis also influence such aging-associated telomere attrition (14–21). Therefore, many factors affect telomeric attrition with aging.
The subtelomeric methylated status was recently suggested to be related to telomere attrition. Telomeres and adjacent subtelomeric regions are packaged as heterochromatin in many organisms. Mammalian telomeres are thought to have properties that are characteristic of heterochromatin, as indicated by the fact that they can transcriptionally silence nearby genes (22–24). Less methylated genomic regions reveal lower content of unmethylated CG dinucleotides, which is associated with euchromatin structure and hypomethylation corresponds to euchromatin associated with a high dimensional genomic DNA structure suppressing transcription and inhibiting recombination between sequences integrated at different locations (25,26). A recent report shows that the shortened telomere region tends to accompany subtelomeric hypomethylation in mice of the fifth generation of telomerase activity–deficient tert−/tert− mutants (27). In humans, subtelomeric DNA is hypomethylated in sperm and ova, and these regions are subjected to de novo methylation during development (28–30). In mice, this activity is carried out by DNA methyltransferase 3b (Dnmt3b). Mutations in DNMT3B in humans lead to the autosomal-recessive ICF (immunodeficiency, centromeric region instability, facial anomalies) syndrome (31). The subtelomeric regions in lymphoblastoid and fibroblast cells of ICF patients are hypomethylated to similar levels as those seen in sperm. In addition, the telomeres in this syndrome are abnormally short, and many chromosome ends lack detectable telomere fluorescence in situ hybridization signals (32). From these recent findings, we hypothesized that the genomic epigenetic status in the somatic cells including subtelomeric DNA methylation in an individual was also being associated with telomeric attrition. We therefore decided to pursue the possibility that the extent of the subtelomeric methylation may thus be a tool to evaluate individual biologic aging. No correlation between subtelomeric methylation and human individual aging has been reported. Subtelomeric methylation status can be associated with aging-related telomere attrition, which is accelerated in various kinds of disease conditions. This report analyzes the mutual relationships among telomere length, telomere length distribution, and aging-related change of subtelomeric methylation of the normal Japanese population with a wide age range. The TRF length measurement in easily accessible specimens such as peripheral blood has been suggested to be useful as a surrogate parameter for the relative telomere length in other tissues (33), and peripheral blood leukocytes are an excellent source for investigating how telomeres shorten (34). Therefore, the present study investigated the telomere changes associated with aging in blood cells in normal adults.
MATERIALS AND METHODS
Subjects
All participants completed an in-person interview that ascertained information about factors that may be related to blood cell/peripheral blood mononuclear cell telomere length. From 147 participants (89 men and 58 women) ranging in age from 20 to 68 years (Table 1), blood samples (taken using 10 mL Vacutainer tubes containing ethylenediaminetetraacetic acid/heparinized syringes) were drawn and stratified into 10-year age groups. The groups were very similar with respect to smoking status, family income, level of physical activity, gender makeup, and socioeconomic status. This research was performed, following approval by the Conjoint Health Research Ethics Board of Kyushu University.
Table 1.
Age and Gender Profiles of the Participants
| Age Range | Men | Women | p value (age) |
| 20s | 8 (25.75 ± 1.56) | 9 (25.50 ± 2.75) | .862 |
| 30s | 21 (35.90 ± 1.56) | 10 (35.90 ± 2.50) | .996 |
| 40s | 38 (45.42 ± 2.42) | 21 (45.14 ± 2.15) | .706 |
| 50s | 16 (54.73 ± 2.19) | 12 (54.33 ± 1.89) | .692 |
| 60s | 6 (63.24 ± 2.40) | 6 (60.83 ± 0.83) | .223 |
| 20s–60s | 89 (43.93 ± 7.64) | 58 (44.35 ± 8.66) | .815 |
Note: The numbers of the participants and their mean ages are shown. Data are given as mean age, with SD in parenthesis.
Telomere Detection
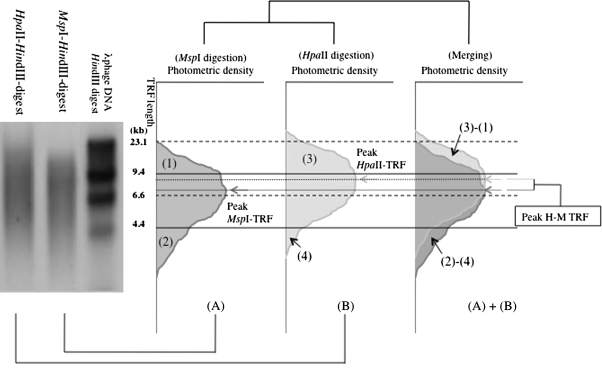
Telomere detection was performed as previously described with a modification (10,20,21,35). Methylation-sensitive and -insensitive isoschizomers HpaII and MspI were used. Both enzymes recognize and cut tetranucleotide CCGG, but HpaII does not cut CCGG with methylated cytosine of the dinucleotide CG in the center of CCGG. Briefly, genomic DNA was extracted from peripheral leukocyte specimens using PureGene DNA Extraction Kits (Gentra Systems, Minneapolis, MN), and the quality was assessed by agarose gel electrophoresis. The DNA (0.1 μg) was digested at 37°C with 1 U MspI or HpaII for 2 hours. The digests (10 μL) were resolved by agarose gel electrophoresis and transferred by Southern blotting to a positively charged nylon membrane (Roche Diagnostics, Mannheim, Germany). The blotted DNA fragments were hybridized to a long (TTAGGG)n digoxigenin-labeled probe specific for telomeric repeats. The telomeric repeat probe used here is 500 bp long. This length is much longer than that of conventional oligonucleotide telomere probes commonly used. This long probe yields dense signals and enables one to clearly detect telomeres shorter than 4.4 kb according to a Southern blot analysis. The blotted membranes were incubated with anti-digoxigenin-alkaline phosphatase-specific antibody. The telomere probe was visualized by CSPD (C18H20ClO7PNa2; Boehringer Mannheim GmbH, Mannheim, Germany). The membrane was then exposed to Fuji XR film with an intensifying screen (FUJIFILM Corporation, Tokyo, Japan). The smears of the autoradiogram were captured on an Image Master (Trioptics Japan, Shizuoka, Japan), and then, the telomere length was quantitatively assessed (Figure 1). Each Southern blot experiment was repeated twice, and the mean value was used.
Figure 1.
A schematic drawing of a densitometric analysis of MspI- and HpaII-digested genomic Southern blot probed with a telomeric sequence repeat. DNA digested with MspI and HpaII appeared as smear on the left photo. The densitometry of the smears of MspI digest (A) and HpaII digest (B) is described in the right. The densitometry of the smears is fractioned according to the molecular weight standard, HindIII-digested l phage DNA, showing bands of 23.1, 9.4, 6.6, and 4.4 kb. The standard sizes for the fractionation are shown as horizontal broken (23.1 and 6.6 kb) and solid (9.4 and 4.4 kb) lines. The peak density (dotted lines and arrows) of the smears described as MspI-TRF (dark gray arrow) and HpaII-TRF (pale gray arrow) are used in the following study as representative telomere lengths of the individuals. The subtracted length of peak-MspI-TRF from peak-HpaII-TRF (peak-H-M-TRF) of an individual is used as a representative length of unmethylated subtelomeric region of the individual. (1), (2), (3), and (4) depict restricted densitometric areas of the smears as follows: (1) longer than 9.4 kb in MspI digestion, (2) shorter than 4.4 kb in MspI digestion, (3) longer than 9.4 kb in HpaII digestion, and (4) shorter than 4.4 kb in HpaII digestion. Each of the areas is described as a percentage to the whole smear as 100%, and the subtracted figures (3) − (1) and (2) − (4) are used in the following analysis. {(3) − (1)}/(3) and {(2) − (4)}/(2), which are described as H-M/H (>9.4 kb) and M-H/M (4.4 kb>) in text and the following figures, are used as indices to evaluate how much subtelomeric regions of telomeres shorter than 9.4 kb and shorter than 4.4 kb are methylated, respectively. TRF = terminal restriction fragment.
TRF Analysis
Telomere length distribution was analyzed by comparing the telomere length using a telomere percentage analysis with three intervals of length as defined by a molecular weight standard. The intensity (photostimulated luminescence [PSL]) was quantified as follows: Each telomeric sample was divided into grid squares as follows according to the molecular size ranges: >9.4, 9.4 ³ ³ 4.4, and 4.4 kb>. The percentage of PSL in each molecular weight range was measured (%PSL = intensity of a defined region − background × 100/total lane intensity − background). Peak telomere fragment lengths (peak-TRF) were used as representatives of TRF in this study. Subtelomeric methylation was assessed by comparing peak-MspI-TRF and peak-HpaII-TRF (peak-H-M-TRF) and by comparing MspI telomere length distribution and HpaII telomere length distribution as described in Figure 1.
Combined Bisulfite Restriction Analysis
The bisulfite treatment of the extracted genomic DNA was performed using the Epitect bisulfite kit (Qiagen, Tokyo, Japan) according to the manufacturer’s protocol. The bisulfite-treated DNA was subjected to a polymerase chain reaction (PCR) to amplify the subtelomere region with the primers specific for chromosome arm 17p and Xp/Yp. The amplified products were digested with MspI, and the digestion patterns were compared with those of the corresponding DNA amplified without bisulfite treatment according to the principle introduced previously (36). The primers used are as follows: 17p-5′: GAATCCACGGATTGCT TTGTGTACTT, 17p-3′: CCTCAGCCTCTCAACCTGCTTGG, X/Yp-5′: RCRRTACCARRRACCRRRACAA ATARA, X/Yp-3′: CCCTCTRAAARTRRACCTAT (R = A or G).
Statistical Analysis
The normality of the data was examined using the Kolmogorov–Sminov test and homogeneity of variance using the Levene Median test. The difference in the mean TRF length and a telomere percentage analysis with age and gender were analyzed using a two-way analysis of variance (ANOVA) test, followed by all pairwise multiple comparison procedures using Tukey’s post hoc test. If the normality and variance of the data were not acceptable in the first test, then logarithmic or square root transformations were used to normalize the data for fitting to the two-way ANOVA. We found no significant difference in interaction between age and gender, and other factors, such as age on mean TRF or telomere loss, were assessed using line regression models. The data are shown using the mean ± standard error bars. The criterion for significance is p < .05. All analyses were carried out using a Sigma Statistical Analysis Software (Sigma 2.03, 2001; Sigma, St Louis, MO).
RESULTS
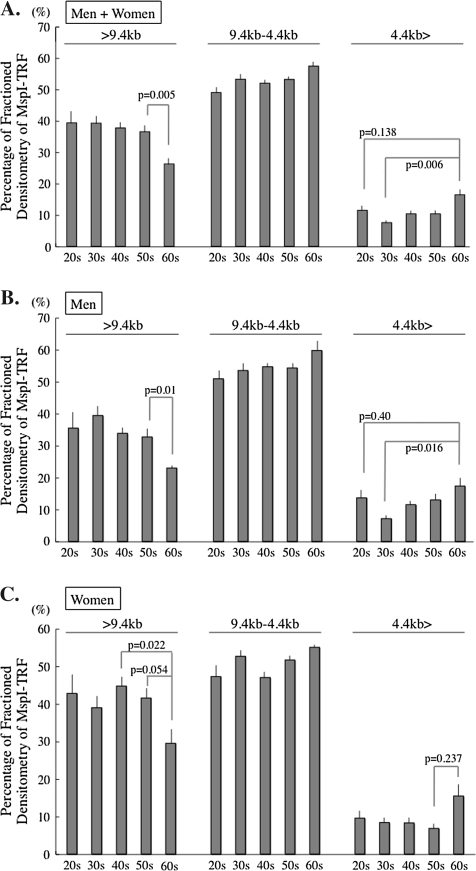
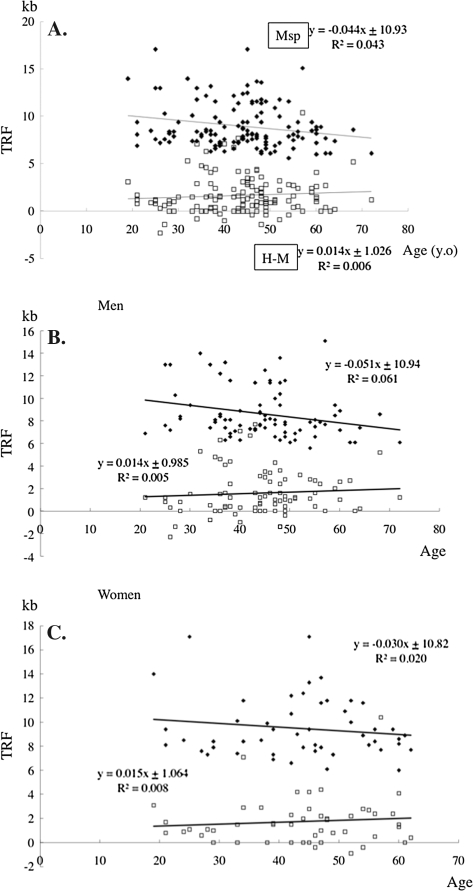
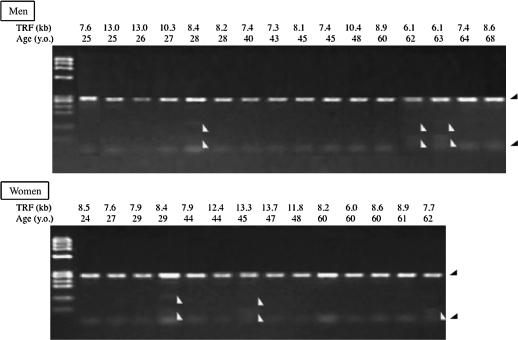
A difference between an MspI-TRF length and an HpaII-TRF length near a same chromosomal end indicates the distance from the methylated CCGG tetranucleotide to the unmethylated CCGG both closest to the terminal end of the chromosome. The mean difference between peak-MspI-TRF length and peak-HpaII-TRF length (peak-H-M-TRF length) of peripheral blood cells of an individual seemed to be an indicator of a mean length of the most distal methylated subtelomeric region of the somatic cells of the individual. Peak-MspI-TRF was reduced by −44 bp/y with aging in the normal Japanese population (Figure 2). Peak-H-M-TRF increased slowly by +14 bp/y with aging in the peripheral leukocytes in both genders (men, +14 bp/y; women, +15 bp/y). The telomere attrition rate was faster in men (−51 bp/y) than in women (−30 bp/y), but the changing rate of subtelomeric methylation with aging was similar in both genders. Unexpectedly, some minus peak-H-M values were observed. This may occur when the telomeres shorter than the peak-MspI-TRF bear longer methylated subtelomeric region and the telomeres longer than the peak-MspI-TRF bear shorter subtelomeric methylated region, thus resulting in the peak-HpaII-TRF being shorter than the peak-MspI-TRF. The existence of the minus peak-H-M values suggested that the length of the subtelomeric methylated region is not correlated with telomere length. Next, the combined bisulfite restriction analysis (COBRA) revealed chromosome arm–specific subtelomeric regional methylation status. The bisulfite treatment interfered MspI digestion of PCR-amplified 17p subtelomeric DNA fragment in some of the participants (Figure 3). No association, however, was observed between the age or the TRF and hypomethylation of the subtelomeric region as indicated by MspI-undigested band appearing of the participants. The COBRA data from Xp/Yp chromosome subtelomere also did not support the association (data not shown). The analysis of chromosomes 17p and Xp/Yp did not seem to support the methylation status evaluated by H-M-TRF in the present study. The H-M distribution analysis therefore seems to be better in this analysis to evaluate the subtelomeric methylation status closely adjacent to the telomeres of all chromosomes in a cell population. Finally, changes in the three-divided MspI- and HpaII-TRF length distribution with aging were determined to analyze subtelomeric methylation with different TRFs. The difference in the densitometric telomere length distribution between MspI and HpaII digestion can thus be an indicator for the subtelomeric methylation, especially, in the longest and the shortest range of size-fractioned densitometry (Figure 4). Analyzing the MspI-TRF length distribution revealed that a reduction of telomeres longer than 9.4 kb (>9.4 kb) and an increase of telomeres shorter than 4.4 kb (4.4 kb>) beyond the age of 60 and higher percentages of 4.4 kb> telomere in those in their 20s than in those in their 30s in both genders (Figure 4). The HpaII–MspI subtracted telomere length distribution in Figure 5 revealed increased values above the age of 60 both in >9.4 and 4.4 kb> ranges of apparent methylated status of the subtelomeric region. The methylation level of the subtelomere region is reflected more accurately in the ratio between the HpaII–MspI subtracted telomere length distribution and MspI area than in the HpaII–MspI subtracted area (compare Figures 5 and 6).
Figure 2.
Peak-MspI-TRF and HpaII–MspI subtracted (H-M) TRF distribution with aging. Closed diamonds, peak-MspI-TRF; open squares, H-M-TRF. (A, Whole population; B, Men; C, Women) H-M values are regarded as approximately the mean values of the methylated lengths of subtelomeric region. The MspI-TRF gradually decreased with aging, but the H-M values did not. Note that some H-M values are negative. TRF = terminal restriction fragment.
Figure 3.
COBRA of the subtelomeres specific for chromosome 17p in healthy Japanese. Bisulfite-treated genomic DNA of the participants was polymerase chain reaction–amplified with chromosome arm–specific primers and digested with MspI. Ladder of HaeI-digested φX174 DNA is shown in the left of the each gel. A complete digestion of the amplified bisulfite-untreated product with MspI yields the bands, which are indicated by black arrowheads. The age, TRF, and relative subtelomeric methylation rate are shown on the top of each panel. Note that 6 of the 30 participants (2 in their 20s, 1 in the 40s, and 3 in the 60s) represent extra bands indicated by white arrowheads, which contain thymine residue changed from unmethylated cytosine residue by the bisulfite treatment. Almost all participants, except for those with extra bands, seem to show the subtelomeric region on chromosome 17p to be well methylated. COBRA = combined bisulfite restriction analysis; TRF = terminal restriction fragment.
Figure 4.
Changes of the subdivided MspI-TRF distribution with aging. Southern blotting smear of MspI-TRF was divided into three portions (>9.4, 9.4 ≥ ≥ 4.4, and 4.4 >). The percentages of densitometry of each portion are shown as columns. (A, Whole population; B, Men; C, Women) The longest telomeric portion decreases with aging and the shortest increases beyond the age of 60. The middle subdivided part (9.4 > > 4.4 kb) did not reveal a clear aging-related change. Horizontal bars depict the standard errors. TRF = terminal restriction fragment.
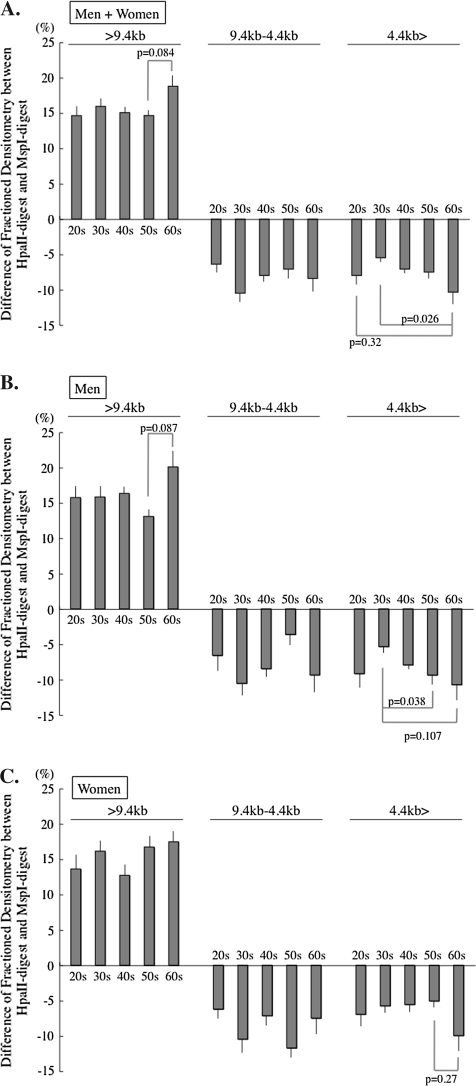
Figure 5.
H-M-TRF length distribution with aging. Subtracted value of the MspI-TRF from the HpaII-TRF densitometry in the three subdivided parts are shown as columns. (A, Whole population; B, Men; C, Women) The individuals in their 60s apparently have methylated telomeres both in long and in short telomeres. Horizontal bars depict the standard errors. TRF = terminal restriction fragment.
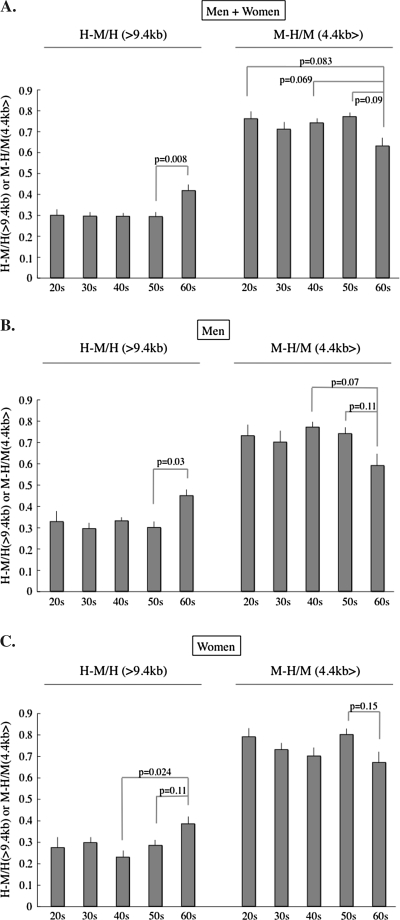
Figure 6.
Change of the relative methylation status of subtelomere with aging. The H-M area/HpaII area of >9.4 kb portion and the M-H area/MspI area of the 4.4 kb>, portion are used as indices indicating the subtelomeric methylation of longer and shorter (than 4.4 kb) telomeres, respectively. (A, Whole population; B, Men; C, Women). In participants in their 60s, the longer (>9.4 kb) telomeres contain more methylated subtelomeres and the shorter telomere (<4.4 kb) contains less methylated subtelomeres. This pattern is opposite to the pattern of the MspI columns >9.4 and <4.4 kb in the participants in their 60s in Figure 1. Horizontal bars depict the standard errors.
An HpaII–MspI/HpaII ratio of >9.4 kb was constant in all the age ranges, except the 60s. This implies that >9.4 kb telomeres contained a consistent length of the methylated subtelomeric region until the age of 60. A significant increase in the ratio was observed in the telomere length range of >9.4 kb and a decreasing tendency of the ratio in the range of 4.4 kb> in participants in their 60s in both genders, indicating that the subtelomeric region revealed more methylated longer telomeres and less methylated of shorter telomeres beyond the age of 60.
The increased 4.4 kb> telomeres contained fewer telomeres with a hypermethylated subtelomeric region in participants in their 60s. On the other hand, the MspI telomere area decreased, but the HpaII–MspI area and HpaII–MspI/HpaII ratio increased in the >9.4 kb area, suggesting that only telomeres with a hypomethylated subtelomeric region decreased and telomeres with hypermethylated subtelomeric region were retained in the >9.4 kb area beyond the age of 60. In summary, the telomere length distribution and the subtelomeric methylated state were different between before and after the age of 60. Telomere length shortening seemed to be accelerated after the age of 60, in association with an increase of short telomeres with a hypomethylated subtelomeric region in both genders.
DISCUSSION
Contrary to our expectation that long telomeres decrease in number, whereas short telomeres increase in somatic cells with aging, it seemed exceptional in the current analysis that the individuals in their 20s bore more of the shortest range of telomeres than some of the older age ranges. One of the hypotheses explaining this observation is that some of the short telomeres (4.4 kb>) in individuals in their 20s might result from short telomeres accumulated during growth until age 20 and after the halt of growth before the age of 20 the accumulated cells containing the short telomeres fell into a cell senescence state. As a result, those in their 30s bore a relatively lower percentage of 4.4 kb> telomeres. This hypothesis is supported by the observation of a rapid drop of telomere length of peripheral blood cells early in life due to cell expansion from stem cells in primates (37). Peripheral blood cells of age 30 or older can be better materials to elucidate the genomic alterations associated with aging-related telomere attrition. We pursue a simple and easy method to detect an extent of subtelomeric methylation of human somatic cells, which help evaluate an individual’s somatic aging. For this purpose, we chose at first the difference of densitometric peak between MspI digest and HpaII digest of telomeric Southern blot using peripheral blood cells. Peak-H-M-TRF revealed few tendencies regarding aging. The difference between the peak-MspI-TRF and peak-HpaII-TRF, however, did not seem suitable to sensitively detect aging-associated hypomethylation of subtelomeric regions because unexpectedly a peak-MspI-TRF was longer than the peak-HpaII-TRF in some participants. This indicated that the subtelomeric methylation is heterogeneously distributed. The extent of telomeric methylation may vary with telomere length. The minus value of H-M-TRF indicated a nonlinear correlation between telomere length distribution and the regional extent of subtelomeric methylation. The chromosome arm–specific analysis using the COBRA method did not reveal any clear association regarding the subtelomeric methylation of some chromosomes with the aging-related telomere length change. An analysis of a wider subtelomeric range and more chromosome arms may therefore be necessary to detect any such association. The detectable regions in the H-M analysis seemed to be different from those in the current COBRA method. Finally, the aging-associated changes of subtelomeric methylation were detectable based on the difference between MspI digest and HpaII digest of the densitometry of Southern blot smears. The difference between the MspI and the HpaII densitometry longer than 9.4 kb and shorter than 4.4 kb thus appears to be obvious and applicable as an indicator of subtelomeric methylation. The differences of >9.4 and 4.4 kb> ranges are derived from the subtelomeric methylation of telomeres shorter than 9.4 kb and those shorter than 4.4 kb, respectively.
It is not clear in the present study whether the increased subtelomeric methylation of long telomeres in the elderly is associated with a decrease in the number of long telomeres. The mechanism regarding the increased subtelomeric methylation of long telomeres may be independent of the mechanism regarding the aging-associated decrease in long telomeres. However, the decrease of MspI telomeres of >9.4 kb and the increase of the HpaII–MspI/HpaII ratio of >9.4 kb in participants in their 60s is possibly explained by that shortening of >9.4 kb telomeres preferentially occurred in those with a hypomethylated state of subtelomeric region. The increase of the MspI telomeres shorter than 4.4 kb and the decrease of the MspI–HpaII/MspI ratio of 4.4 kb> telomeres suggested an increasing tendency of telomeres with hypomethylated subtelomeric region in the telomeres shorter than 4.4 kb in participants in their 60s. This can also be explained by that shorter telomeres tend to have a shorter range of subtelomeric methylation. Those telomeric and subtelomeric changes in their 60s seemed to have longer telomeres with a more methylated subtelomeric region and shorter telomeres with a less methylated subtelomeric region. This tendency was observed in both genders. With aging, the TRF of telomeres with hypomethylated subtelomeric region seemed to be shifted from longer to shorter and hypermethylated telomeres seemed to be left behind in the longer area, and these events seemed to be accelerated after the age of 60. These observations can be derived from telomeres with less methylated subtelomeric regions that tend to become shortened faster during the aging process independent of gender.
The observed aging-related change of telomere length and subtelomeric methylation are consistent with the idea of telomere-shortening acceleration accompanying widened subtelomeric hypomethylation. This also indicates the possibility that subtelomeric hypomethylation is an indicator of the shortening state of the telomeres. A ratio of methylated and nonmethylated TRF of >9.4 and 4.4 kb> can therefore be a candidate marker for aging acceleration. DNA methylation is an epigenetic marker for an important chromatin modification in mammals, thus leading to transcriptional regulation and regulation of the accessibility of DNA-binding factors of the region (38). The hypomethylation of the subtelomeres is associated with increased accessibility of DNA-binding proteins for suppression of the subtelomeric and telomeric position effects (39,40). With a hypomethylated state of the telomeric and subtelomeric region, stress-induced harmful factors may have access to these regions and alter the expression of telomere-binding proteins (41,42). In mammals, highly repetitive genomic DNA regions are highly methylated, mainly to prevent homologous recombination, which can possibly result in harmful genomic alterations (43–45). Longer telomeres give rise to silencing of genes located closely to the telomere, accompanying subtelomeric hypermethylation, whereas shorter telomeres are inversely correlated with a less methylated state of their subtelomeric regions (39,40). In embryonic stem cells and cancer cells, DNA hypomethylation in telomeric regions is reported to be associated with suppression of telomere elongation mediated by telomerase and with aberrant telomeric elongation mediated by a sister chromatid exchange mechanism, called alternative lengthening of telomeres (32,46). In contrast to cancer cells and ES cells, somatic cells, such as human peripheral blood cells, express little telomerase activity. The present study suggests that subtelomeric hypomethylation takes place in association with telomere attrition or acceleration of the attrition in somatic cells. A report of hypomethylated subtelomeres linked to an enhanced telomeric shortening in ICF syndrome also supports this idea (29). The current data support the possibility that subtelomeric hypomethylation may be a biomarker for the aging-associated acceleration of telomere shortening. Contrary to the TRF attrition that proceeds quite slowly (50–200 bp/y), the epigenetic modification of the subtelomeric region is thought to change more rapidly, and alterations of subtelomeric methylation are more easily detectable than telomere attrition by a Southern blotting analysis using a methylation-sensitive tetranucleotide-recognizing restriction enzyme, for the average length of tetranucleotide-recognizing restriction fragments is theoretically 44 bp (256 bp).
Subtelomeric hypomethylation also reflects telomeric hypomethylation without telomere elongating in the absence of telomerase activity, thus resulting in telomere shortening. Telomeres with a high accessibility of DNA-binding molecules may allow hazardous factors to easily bind to telomeric regions, resulting in accelerating telomere shortening in somatic cells. The present study suggests an association between subtelomeric hypomethylation and aging-related telomere shortening. Therefore, subtelomeric methylation may be a predisposing condition for the acceleration of telomere shortening with aging in human peripheral blood cells.
Further investigation is necessary to confirm the hypothetical relationship between the acceleration of telomere shortening and the subtelomeric methylation in various conditions including aging and other diseases, that induce telomere attrition.
FUNDING
This work was supported, in part, by a Grant-in-Aid from the Ministry of Education, Science, and Culture of Japan (#20590703).
Acknowledgments
We thank Ms Ueda and Ms Taguchi for their valuable technical assistance. We are grateful to Mr Brian Quinn for his linguistic advice.
References
- 1.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblast. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 4.Ju Z, Rudolph KL. Telomeres and telomerase in cancer stem cells. Eur J Cancer. 2006;42:1197–1203. doi: 10.1016/j.ejca.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Hallmarks of telomeres in ageing research. J Pathol. 2007;211:114–123. doi: 10.1002/path.2090. [DOI] [PubMed] [Google Scholar]
- 6.Satyanarayana A, Wiemann SU, Buer J, et al. Telomere shortening impairs organ regeneration by inhibiting cell cycle re-entry of a subpopulation of cells. EMBO J. 2003;22:4003–4013. doi: 10.1093/emboj/cdg367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HW, Blasco MA, Gottlieb GJ, Horner JW, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 8.Vaziri H, Schächter F, Uchida I, et al. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda K, Khan MY, Skurnick J, Kimura M, Aviv H, Aviv A. Telomere attrition of the human abdominal aorta: relationships with age and atherosclerosis. Atherosclerosis. 2000;152:391–398. doi: 10.1016/s0021-9150(99)00482-7. [DOI] [PubMed] [Google Scholar]
- 10.Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci U S A. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kappei D, Londoño-Vallejo JA. Telomere length inheritance and aging. Mech Ageing Dev. 2008;129:17–26. doi: 10.1016/j.mad.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 13.Bischoff C, Graakjaer J, Petersen HC, et al. The heritability of telomere length among the elderly and oldest-old. Twin Res Hum Genet. 2005;8:433–439. doi: 10.1375/183242705774310141. [DOI] [PubMed] [Google Scholar]
- 14.Epel ES, Blackburn EH, Lin F, et al. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci U S A. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdes AM, Andrew T, Gardner JP, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 16.Uziel O, Singer JA, Danicek V, et al. Telomere dynamics in arteries and mononuclear cells of diabetic patients: effect of diabetes and of glycemic control. Exp Gerontol. 2007;42:971–978. doi: 10.1016/j.exger.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Brouilette S, Singh RK, Thompson JR, Goodall AH, Samani NJ. White cell telomere length and risk of premature myocardial infarction. Arterioscler Thromb Vasc Biol. 2003;23:842–846. doi: 10.1161/01.ATV.0000067426.96344.32. [DOI] [PubMed] [Google Scholar]
- 18.Okuda K, Khan MY, Skurnick J, et al. Telomere attrition of the human abdominal aorta: relationships with age and atherosclerosis. Atherosclerosis. 2000;152:391–398. doi: 10.1016/s0021-9150(99)00482-7. [DOI] [PubMed] [Google Scholar]
- 19.Panossian LA, Porter VR, Valenzuela HF, et al. Telomere shortening in T cells correlates with Alzheimer’s disease status. Neurobiol Aging. 2003;24:77–84. doi: 10.1016/s0197-4580(02)00043-x. [DOI] [PubMed] [Google Scholar]
- 20.Guan JZ, Maeda T, Sugano M, et al. A percentage analysis of the telomere length in Parkinson’s disease patients. J Gerontol A Biol Sci Med Sci. 2008;63:467–473. doi: 10.1093/gerona/63.5.467. [DOI] [PubMed] [Google Scholar]
- 21.Guan JZ, Maeda T, Sugano M, et al. An analysis of telomere length in sarcoidosis. J Gerontol A Biol Sci Med Sci. 2007;62:1199–1203. doi: 10.1093/gerona/62.11.1199. [DOI] [PubMed] [Google Scholar]
- 22.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 23.Koering CE, Pollice A, Zibella MP, et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 25.Yoder JA, Waish CP, Bestor TH. Cytosine methylation & the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 26.Steinert S, Shay JW, Wright WE. Modification of subtelomeric DNA. Mol Cell Biol. 2004;24:4571–4580. doi: 10.1128/MCB.24.10.4571-4580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benetti R, García-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 28.Brock GJ, Charlton J, Bird A. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- 29.Cross S, Lindsey J, Fantes J, McKay S, McGill N, Cooke H. The structure of a subterminal repeated sequence present on many human chromosomes. Nucleic Acids Res. 1990;18:6649–6657. doi: 10.1093/nar/18.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lange T, Shiue L, Myers RM, et al. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 33.Friedrich U, Gries E, Schwab M, Fritz P, Thon K, Klotz U. Telomere length in different tissues of elderly patients. Mech Ageing Dev. 2000;119:89–99. doi: 10.1016/s0047-6374(00)00173-1. [DOI] [PubMed] [Google Scholar]
- 34.Frenck RW, Jr, Blackburn EH, Shannon KM. The rate of telomere sequence loss in human leukocytes varies with age. Proc Natl Acad Sci U S A. 1998;95:5607–5610. doi: 10.1073/pnas.95.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherif H, Tarry JL, Ozanne SE, Hales CN. Ageing and telomeres: a study into organ- and gender-specific telomere shortening. Nucleic Acids Res. 2003;31:1576–1583. doi: 10.1093/nar/gkg208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baerlocher GM, Rice K, Vulto I, Lansdorp PM. Longitudinal data on telomere length in leukocytes from newborn baboons support a marked drop in stem cell turnover around 1 year of age. Aging Cell. 2007;6:121–123. doi: 10.1111/j.1474-9726.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- 38.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 39.van Overveld PG, Lemmers RJ, Sandkuijl LA, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 40.Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, Murnane JP. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco R, Muñoz P, Flores JM, Klatt P, Blasco MA. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 2007;21:206–220. doi: 10.1101/gad.406207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Multani AS, He H, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 43.Domínguez-Bendala J, McWhir J. Enhanced gene targeting frequency in ES cells with low genomic methylation levels. Transgenic Res. 2004;13:69–74. doi: 10.1023/b:trag.0000017176.77847.80. [DOI] [PubMed] [Google Scholar]
- 44.Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 1998;12:1381–1389. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bender J. Cytosine methylation of repeated sequences in eukaryotes: the role of DNA pairing. Trends Biochem Sci. 1998;23:252–256. doi: 10.1016/s0968-0004(98)01225-0. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]